Abstract
Genomic stability in eukaryotic cells is maintained by the coordination of multiple cellular events including cell cycle checkpoint, DNA repair, transcription, and apoptosis after DNA damage. Pax2 transactivation domain interaction protein (PTIP), a protein that contains six BRCT domains, has been implicated in DNA damage response. In this study we showed that recruitment of PTIP to damaged chromatin depends on DNA damage signaling proteins γH2AX·MDC1·RNF8, which in turn facilitates sustained localization of PA1 (PTIP-associated protein 1) to sites of DNA break. Similar to PTIP, depletion of PA1 increases cellular sensitivity to ionizing radiation. Furthermore, we demonstrated that the N-terminal PA1 binding domain and the C-terminal focus-localization domain of PTIP are critical for PTIP function in DNA damage repair. Interestingly, although PTIP and PA1 associate with MLL (mixed lineage leukemia) complexes and participate in transcriptional regulation, this function of PTIP·PA1 in DNA damage response is likely to be independent of the MLL complexes. Taken together, we propose that a subset of PTIP·PA1 complex is recruited to DNA damage sites via the RNF8-dependent pathway and is required for cell survival in response to DNA damage.
The genome of all living cells constantly suffers a variety of genomic insults, which if not fixed would lead to genomic instability. Therefore, in response to DNA damage, cells elicit an elaborated signaling network, which is collectively known as the DNA damage response pathway (1). Through a cascade of sensors, transducers, and effectors, the DNA damage response pathway coordinates a process that include cell cycle checkpoints, DNA repair, cellular senescence, and apoptosis (2, 3).
The regulation of this pathway is best studied after ionizing radiation (IR).2 In response to IR, the histone variant H2AX is phosphorylated by ATM or ATR (4) and serves as a platform for the recruitment of MDC1, which further facilitates the loading of many checkpoint and repair proteins to sites of DNA damage to form IR-induced foci (IRIF) (5, 6). More recent studies revealed that phosphorylation of MDC1 binds directly to and accumulates the E3 ubiquitin ligase complex, RNF8·Ubc13 at DNA damage sites, which ubiquitinates H2AX, H2A, and possibly additional proteins and allows the recruitment of 53BP1 and the ubiquitin-interacting motif domain-containing protein RAP80 at sites of breaks after DNA damage (7-13). Although RAP80 specifically recruits BRCA1 to sites of DNA break, it remains to be determined how 53BP1 localizes to DNA damage sites via this ubiquitination-dependent cascade and whether additional DNA damage checkpoint and repair proteins would localize to sites of DNA break through the same or a similar mechanism.
PTIP was originally identified as Pax2 transactivation domain interaction protein in a yeast two-hybrid screen (14). Subsequently, several studies suggest that PTIP is an essential component of histone H3K4 methyltransferase complexes, which may be involved in the regulation of gene expression through modulation of H3K4 methylation (15, 16). The importance of PTIP functions in vivo is revealed by the observations that PTIP null embryos die at E9.5 due to widespread cell death (17). In addition, PTIP null cells show a very high number of DNA breaks during S-phase and an inability to progress through mitosis, suggesting a defect in DNA repair (17). A possible function of PTIP in DNA damage repair is also suggested by the presence of six BRCT domains in PTIP, as BRCT domains are phosphoprotein binding domains that preferentially bind to Ser(P)/Thr(P) motifs, especially those generated by ATM and ATR (18). Indeed, PTIP has been shown to interact with 53BP1 in response to DNA damage, and this interaction requires ATM-dependent phosphorylation of 53BP1 (18, 19). The four C-terminal BRCT domains of PTIP are required for its foci formation after IR and its interaction with 53BP1 via 53BP1 Ser-25 phosphorylation (19). However, the exact function of PTIP in the DNA damage-responsive pathway and where it fits in the well defined DNA damage-signaling cascade remain elusive.
Here we report that PTIP acts downstream of γH2AX·MDC1·RNF8 in the DNA damage signal transduction cascade. In addition, we showed that PTIP forms a stable complex with PTIP-associated protein 1 (PA1), and this complex is required for cell survival after IR.
MATERIALS AND METHODS
Cell Culture and Plasmids—HeLa, 293T, and U2OS cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified incubator with 5% CO2 (v/v). Mouse embryonic fibroblast (MEF) cells were cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in 5% CO2 (v/v). SV40T-immortalized PTIPflox/flox MEFs (15) were infected with adenovirus expressing Cre to permanently delete PTIP gene. After serial dilution, single colonies were isolated, and deletion of the PTIP gene was confirmed by real time PCR. H2AX-/- MEFs, H2AX-/- MEFs reconstituted with wild type H2AX, MDC1-/-, 53BP1-/-, RNF8-/-, UBC13-/-, PTIP-/-, and their respective wild-type MEFs, NBS1-deficient ILB1 cells and its derivative cells reconstituted with wild-type NBS1, ATM-deficient (FT169A), and reconstituted (YZ5) cells, and HCC1937 and HCC1937-BRCA1, FANCD2-reconstituted and -deficient cells were previously reported (6, 7, 20-22). RAD18+/+ and RAD18-/- MEFs were a generous gift of Masaru Yamaizumi at Kumamoto University. FANCN-deficient VU1341F cells were provided by Dr. de Winter.
Mouse PTIP cDNA was subcloned into pDONR201 entry vector and then transferred to destination vectors containing N-terminal triple-epitope tag SFB (S protein, FLAG, and streptavidin binding peptide tag) or HA- or Myc-epitope tag using the gateway system (Invitrogen). All deletion mutants were generated by site-directed mutagenesis (Stratagene) and verified by sequencing. Transfections were performed using FuGENE 6 or Lipofectamine according to the manufacturer's instructions.
Antibodies—Rabbit polyclonal anti-PA1 antibodies were raised by immunizing rabbits with glutathione S-transferasefused PA1 recombinant protein. The antibody was affinity-purified using AminoLink Plus Immobilization and a purification kit (Pierce). Phospho-H2AX, MDC1, RNF8, MLL3 antibodies were described previously (6, 7, 15, 23). Both monoclonal anti-FLAG M2 and anti-β-actin antibodies were purchased from Sigma.
Retrovirus Production and Infection—Full-length PTIP was cloned into pEF1A-HA-FLAG retroviral vector using the gateway system. Virus-containing supernatant was collected 48 and 72 h after co-transfection of pEF1A-HA-FLAG PTIP and pcl-ampho into BOSC23 packaging cells and were used to infect MEF cells in the presence of Polybrene. Two days later MEF cells were either irradiated as indicated or cultured in medium containing puromycin for the selection of stable clones. The clones stably expressing HA-FLAG-tagged PTIP were identified and verified by Western blotting and immunostaining using anti-FLAG antibodies.
Immunofluorescence Staining—Cells grown on coverslips were mock-treated or irradiated with a JL Shepherd Cs137 source (10 Gy) and allowed to recover for 6 h. Cells were fixed in 3% paraformaldehyde solution for 10 min and then permeabilized in 0.5% Triton X-100-containing solution for 5 min at room temperature. Cells were incubated with primary antibodies diluted in 5% goat serum at 37 °C for 30 min. After washing with phosphate-buffered saline twice, cells were incubated either with fluorescein isothiocyanate-conjugated or rhodamine-conjugated secondary antibodies for 20 min at 37 °C. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and then mounted onto glass slides with anti-Fade solution. Images were taken with a Nikon Eclipse E800 fluorescence microscope.
Co-immunoprecipitation and Western Blotting—Cells were lysed with NTEN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) containing 20 mm NaF and 1 μg/ml of pepstatin A and aprotinin on ice for 20 min. After removal of cell debris by centrifugation, the soluble fractions were collected and incubated with either protein A-agarose beads coupled with anti-PTIP, PA1 antibodies, or streptavidin-Sepharose beads (Amersham Biosciences) for 3 h at 4 °C. The precipitates were then washed 4 times with NTEN buffer and boiled in 2× SDS loading buffer. Samples were resolved on SDS-PAGE and transferred to polyvinylidene difluoride membranes, and immunoblotting was carried out with antibodies as indicated.
Fluorescence-activated Cell Sorting—For cell cycle analysis, cells were washed twice with phosphate-buffered saline, resuspended in 300 μl of phosphate-buffered saline, and then fixed with the addition of 700 μl of 100% ethanol. After being stored at -20 °C overnight, fixed cells were washed and incubated in sodium citrate buffer containing RNase A for 30 min and then stained with propidium iodide for 30 min. Cells were then run on a FACScan, and cell cycle analysis was performed.
Tandem Affinity Purification—293T cells stably expressing SFB-PTIP were used for tandem affinity purification. The SFB-PTIP stable cells were lysed with NETN buffer (see above) on ice for 20 min. After removal of cell debris by centrifugation, crude lysates were incubated with streptavidin-Sepharose beads for 1 h at 4 °C. The bead-bound proteins were washed 3 times with NETN buffer and eluted with 2 mg/ml biotin (Sigma) for 1 h twice at 4 °C. The eluates were further incubated with S-protein-agarose (Novagen) for 1 h at 4 °C and washed 3 times with NETN buffer. The proteins bound to S-protein-agarose beads were subjected to SDS-PAGE and visualized by Coomassie Blue staining. The identities of eluted proteins were revealed by mass spectrometry analysis, performed by the Taplin Biological Mass Spectrometry Facility (Harvard University).
RNA Interference—Small interfering RNAs (siRNAs) against human PTIP or PA1 were purchased from Dharmacon, Inc. A non-targeting siRNA was used as control. The sequences of PA1 siRNA#1 and siRNA#2 are CUGAUUGACCGGAGACGCAUU and AUGAGGAGCCGGAGGCCAAUU, respectively. Sequences of siRNA#1 and siRNA#2 against PTIP are ACACUGAGGAAUAUUACUAdTdT and UGUUUGCAAUUGCGGAUUAUU, respectively. The sequences of MLL3 siRNA#1 and siRNA#2 are GCAAUGGUCUUUCUGGAUAUU and CCAGGUCAAUCAACAGUUAUU, respectively. The sequence of H2AX siRNA is CAACAAGAAGACGCGAAUCdTdT. The sequences for MDC1 and RNF8 siRNA were previously described (7). The sequence of control siRNA is UUCAAUAAAUUCUUGAGGUUU. HeLa cells were transfected twice at 24-h time intervals with the indicated siRNAs using Oligofectamine (Invitrogen) to achieve efficient siRNA-mediated down-regulation of their target genes.
Clonogenic Survival Assays—IR sensitivity assays were carried out as described previously (24). Briefly, cells were seeded onto 60-mm dish in triplicate and treated with ionizing radiation with indicated doses. Cells were left for 14 days to allow colonies to form. Colonies were fixed and stained with Coomassie Blue and then counted using a GelDoc with Quantity One software (Bio-Rad). Results were the averages of data obtained from three independent experiments.
G2/M Checkpoint Assay—PTIP MEFs, siRNA knockdown PTIP, or siRNA knockdown PA1 U2OS cells were seeded onto 10-cm dishes, irradiated with 3 Gy of ionizing radiation, and incubated for 1 h before collection. G2/M checkpoint assay were performed as described previously (24).
RESULTS
PA1 Is a Major PTIP-associated Protein—In an attempt to understand PTIP functions in DNA damage response, we established a 293T derivative cell line stably expressing a triple-tagged (S-protein, FLAG, and streptavidin binding peptide) PTIP for the identification of potential PTIP binding partners. After tandem affinity purification, the identities of PTIP-associated proteins were revealed by mass spectrometry analysis (supplemental Table 1). In agreement with our recent report (15), we reproducibly identified PA1 as a major PTIP-associated protein (Fig. 1A). We performed co-immunoprecipitation experiments and were able to confirm an interaction not only between overexpressed PTIP and PA1 (Fig. 1B) but also between endogenous proteins (Fig. 1C), suggesting that these two proteins indeed associate with each other in vivo. Moreover, the binding of PA1 to PTIP was not changed after cells were exposed to IR (Fig. 1C).
FIGURE 1.
PTIP interacts with PA1. A, Coomassie Blue staining of affinity-purified PTIP-containing protein complexes. Cell extracts prepared from 293T cells stably expressing SFB-PTIP were subjected to tandem affinity purification. The final eluent was subjected to SDS-PAGE and visualized by Coomassie Blue staining. Proteins were identified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry analysis and summarized in supplemental Table 1. Lines indicate protein bands corresponding to PTIP and PA1. M, molecular mass standards. B, 293T cells were transfected with plasmids encoding SFB-tagged PTIP together with plasmids encoding Myc-tagged PA1. Immunoprecipitation (IP) reactions were performed using S-protein beads and then subjected to Western blot analyses using indicated antibodies. C, endogenous interaction between PTIP and PA1 in the absence or presence of IR. For a control, anti-PTIP or anti-PA1 immunoprecipitates were immunoblotted with indicated antibodies. D, cell cycle-independent expression of PTIP and PA1. HeLa cells were arrested overnight with 0.5 μg/ml nocodazole (Noc). Mitotic cells were “shaken off” and then released into normal media (upper). T24 cells were allowed to grow to confluency for 96 h and then trypsinized and released into fresh media (lower). Samples were taken at the indicated time points and analyzed by fluorescence-activated cell sorting and Western blotting. E and F, mapping of the corresponding regions required for PTIP·PA1 interaction. Immunoprecipitation reactions were performed using S-protein beads and then subjected to Western blot analyses using antibodies as indicated. Schematic diagrams of wild-type (WT) and deletion mutants of PTIP (E) and PA1 (F) used in this study are also included. NT, N terminus; CT, C terminus.
To understand how these two proteins may function together, we examined whether PTIP and PA1 expression would be coordinately regulated in cells. We carried out mitotic “shake off” experiments using HeLa cells. In addition, we also arrested T24 cells in G0 phase by contact inhibition. In both cases we allowed these synchronized cells to enter the cell cycle by changing them into fresh media at appropriate density. As shown in Fig. 1D, the levels of PTIP and PA1 only varied slightly, indicating that both PTIP and PA1 are ubiquitously expressed throughout cell cycle.
To identify the region(s) on PTIP required for its interaction with PA1, we co-expressed a series of internal deletion mutants of PTIP (Fig. 1E) with Myc-tagged PA1 in 293T cells. Results revealed that the interaction between PTIP and PA1 was dramatically decreased by the deletion of the second BRCT domain of PTIP, suggesting that this N-terminal BRCT2 domain of PTIP is involved in its interaction with PA1 (Fig. 1E). Conversely, we found that the N terminus of PA1, but not its C terminus, is critical for PA1-PTIP interaction (Fig. 1F). Using bacterially expressed and purified proteins, we showed that PA1 binds directly to PTIP BRCT1/2, but not to PTIP BRCT3/4 or PTIP BRCT5/6 (supplemental Fig. S1). This result indicates that although PTIP·PA1 interaction requires the BRCT2 domain of PTIP, this interaction occurs in a phosphorylation-independent manner.
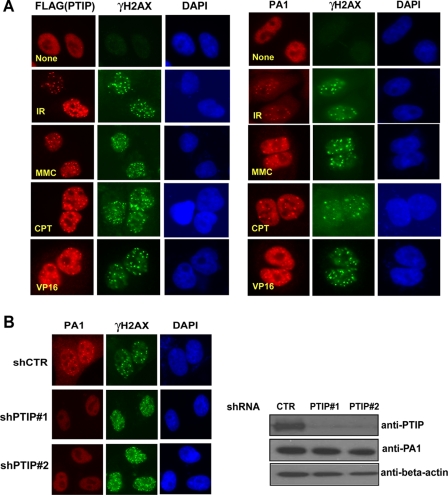
PTIP Targets PA1 to Sites of DNA Damage—Previous studies suggested that PTIP forms nuclear foci in response to ionizing radiation (19). Because of the limited sensitivity of our anti-PTIP antibodies, we repeated these experiments using 293T cells stably expressing SFB-tagged PTIP. Indeed, damage-induced PTIP foci formation was readily detected after ionizing radiation (Fig. 2A). We also observed PTIP foci formation in cells treated with various agents leading to DNA damage, including camptothecin (CPT), mitomycin C (MMC), and etoposide (VP16) (Fig. 2A), suggesting that PTIP responds to a variety of DNA-damaging agents. Similarly, we detected PA1 foci formation in cells treated with the same agents that induce DNA damage (Fig. 2A). The fact that DNA damage-induced PTIP and PA1 foci co-localized with those containing γH2AX (Fig. 2A) indicates that both PTIP and PA1 relocate to sites of DNA breaks.
FIGURE 2.
PTIP accumulates PA1 at DNA damage sites. A, PTIP and PA1 form foci after DNA damage. SFB-PTIP 293T stable cell lines or U2OS cells grown on coverslips were treated with 10 Gy IR, 100 μg/ml mitomycin (MMC), 25 nm camptothecin (CPT), or 2.5 μm VP16. 8 h later cells were fixed, and immunostaining was carried out using indicated antibodies. B, U2OS cells with PTIP knockdown (shPTIP#1, shPTIP#2) or transfected with control vector (shCTR) were irradiated (10 Gy), allowed to recover for 6 h, fixed, and immunostained using antibodies as indicated. Western blot analysis was performed to verify the stable knockdown of PTIP (right panel).
IR-induced PA1 foci formation was reduced in U2OS cells with PTIP knockdown (Fig. 2B), whereas PTIP IRIF remained the same in cells with or without PA1 knockdown (Fig. 3). These data suggest that PTIP targets PA1 to sites of DNA damage. We further confirmed these results using overexpressed PA1 and PTIP. Ectopically expressed PA1 (FLAG-PA1-NLS) localized to nuclei but could not form visible DNA damage-induced foci (supplemental Fig. S2). However, when co-expressed with PTIP, PA1 foci were readily detected after IR (supplemental Fig. S2). This PTIP-dependent localization of exogenously expressed PA1 also requires the intact PA1/PTIP interaction, as PTIP deleted of its N-terminal BRCT2 domain, which has reduced PA1 binding activity, failed to target PA1 to damage-induced foci (supplemental Fig. S2), again supporting that a direct interaction between PTIP and PA1 is required for PTIP ability to target PA1 to sites of DNA breaks.
FIGURE 3.
PTIP·PA1 localization depends on a signaling cascade involves H2AX, MDC1, and RNF8. HeLa cells or HeLa cells stably expressing HA-FLAG-PTIP were transfected with control siRNA or H2AX-, MDC1-, RNF8-, PTIP-, PA1-specific siRNAs. Cells were irradiated (10 Gy), fixed, and immunostained with anti-FLAG, anti-PA1, and pH2AX antibodies. Western blot analysis was carried out to verify the knockdown of H2AX, MDC1, and RNF8 (lower panel).
PTIP Acts Downstream of the H2AX·MDC1·RNF8-signaling Pathway in Response to DNA Damage—Having established that PTIP is required for PA1 localization after DNA damage, we next explored what would be the upstream signaling molecules that facilitate PTIP localization to DNA damage sites. We performed experiments using siRNA knockdown in HeLa cells. PTIP and PA1 IRIF were abolished in cells with H2AX, MDC1, or RNF8 knockdown (Fig. 3). We also used a panel of cell lines defective in various components known to be involved in DNA damage checkpoint response. As shown in Fig. 4A, in contrast to their respective wild type counterparts, PTIP foci formation was abolished in H2AX-/-, MDC1-/-, and RNF8-/- MEFs, indicating that the damage-induced focus localization of PTIP depends on a signaling cascade involves H2AX, MDC1, and RNF8. On the other hand, we observed normal PTIP localization in ATM-deficient FT169A cells (Fig. 4A), suggesting that ATM is not essential for PTIP localization after DNA damage. Similarly, there is no evident difference of PTIP foci formation in BRCA1-deficient HCC1937 cells or NBS1-deficient ILB1 fibroblast cells when compared with the same cells reconstituted with wild-type BRCA1 or NBS1 (Fig. 4A). Collectively, these data establish that PTIP acts downstream of H2AX, MDC1, and RNF8 but is independent of BRCA1 or NBS1 in the DNA damage-signaling cascade.
FIGURE 4.
PTIP foci formation depends on γH2AX·MDC1·RNF8 but not other DNA damage checkpoint or repair proteins. A, PTIP acts downstream of H2AX·MDC1·RNF8 signaling pathway in response to DNA damage. Cells deficient for various DNA damage checkpoint or repair proteins and their respective wild-type counter-parts or reconstituted cells were infected with retrovirus expressing FLAG-tagged PTIP. Cells were then irradiated (10 Gy), and immunostaining experiments were performed using anti-FLAG and anti-γH2AX antibodies 6 h later. B, both the FHA domain and RING domain of RNF8 are required for PTIP IRIF formation. RNF8-/- MEFs stably expressing HA-tagged PTIP were infected with retroviruses expressing SFB-RNF8, SFB-RNF8-ΔFHA, or SFB-RNF8-ΔRING mutants. Immunostaining experiments were performed using anti-γH2AX and anti-HA antibodies. C, UBC13 is required for PTIP localization at or near the sites of DNA breaks. Wild-type or UBC13-/- MEFs were infected with retroviruses expressing SFB-PTIP. Cells were irradiated (10 Gy), fixed, and immunostained with anti-FLAG and pH2AX antibodies.
PTIP foci formation would depend on several DNA repair proteins. We employed RAD18-/- MEFs, FANCD2-deficient PD20 cells, BRCA2-deficient CHOVC8 cells, PALB2/FANCN-deficient VU1341F cells, and their wild-type counterpart or reconstituted cells. As shown in Fig. 4A, PTIP IRIF formed normally in all these cells, implying that PTIP foci formation is independent of these repair proteins after IR.
Given that PTIP IRIF requires RNF8, we next determined whether RNF8-dependent ubiquitination events are instrumental for PTIP accumulation at sites of DNA breaks. Because we do not have any anti-mouse PTIP antibody that works for immunostaining, we established RNF8-/- MEF cells stably expressly HA-epitope-tagged PTIP. Interestingly, relocalization of PTIP to DNA damage sites was observed only in RNF8-/- cells when these cells were reconstituted with wild-type RNF8 but not when they were reconstituted with RNF8 FHA or RING domain deletion mutants (Fig. 4B), indicating that appropriate loading of RNF8 and its subsequent ability to promote protein ubiquitination at the vicinity of double-strand breaks are pre-requisite events for sustained accumulation of PTIP.
UBC13 was recently proposed to function with RNF8 in a DNA damage-signaling pathway to mediate focal accumulation of a number of checkpoint proteins at sites of DNA breaks (7, 12). Consistent with this notion, we noticed that PTIP foci formation was abrogated in UBC13-/- MEFs (Fig. 4C). Thus, RNF8/UBC13 act together and are required for the concentration of PTIP at or near the sites of DNA breaks.
MLL Complexes May Not Be Involved in DNA Damage Response Together with PTIP·PA1—Recent reports have shown that PTIP associates physically with the mixed lineage leukemia 3 (MLL3)-, and MLL4-containing histone methyltransferases complexes (15, 25) and raised the possibility that similar to PTIP, these MLL complexes might also participate in DNA damage response. To explore a role for MLL complexes in DNA damage response, we first examined whether MLL3 protein or other MLL components would be able to form IRIF. As shown in Fig. 5A, neither MLL3 nor UTX, one of component of MLL3 complex, could localize to IRIF. In addition, knockdown of endogenous MLL3 has no effect on PTIP or PA1 IRIF formation (Fig. 5B). Moreover, using a gel filtration assay and a common MLL subunit WDR5 as a marker, we detected a distinct subcomplex of PTIP and PA1 that is not associated with MLL complexes (Fig. 5C), indicating that separate PTIP pools with different functions are present in the cell. Taken together, we conclude that PTIP participates in DNA damage response in a manner that is largely independent of the MLL protein complexes.
FIGURE 5.
MLL3 complex may not be involved in DNA damage response. A, MLL3 and UTX do not form IRIF. U2OS cells were transfected with plasmids encoding SFB-tagged UTX. Immunostaining was carried out with anti-MLL3, anti-FLAG, and anti-γH2AX antibodies 6 h after cells exposed to IR (10 Gy). CTR, control. B, PTIP or PA1 IRIF formation occurs independently of MLL3. HeLa cells or HeLa cells stably expressing HA-FLAG-PTIP were transfected with control siRNA or MLL3 specific siRNAs (siMLL3#1 and siMLL3#2). Cells were irradiated (10 Gy), fixed, and immunostained with anti-FLAG, anti-PA1, and pH2AX antibodies. Western blot analysis was carried out to verify the knockdown of MLL3 (left panel). C, FLAG-PTIP-associated proteins isolated from HeLa S cell nuclear extracts were fractionated on Superose 6 gel filtration column (Amersham Biosciences) as described in Ref. 15. Fractions were analyzed by Western blot with indicated antibodies. Positions of protein molecular weight standards are indicated at the bottom.
Targeting PTIP to DNA Damage Sites Requires Its BRCT3 and BRCT4 Domains—Previous report have documented that the C-terminal BRCT domains of PTIP are important for its localization to double-strand breaks (19). To fine map the focus localization region of PTIP, we used a series of internal deletion mutants of PTIP. Deletion of the N-terminal BRCT domains (BRCT1 and BRCT2) and the very C-terminal BRCT domains (BRCT5 and BRCT6) as well as the poly glutamine region of PTIP had no effect on PTIP foci formation, whereas deletion of BRCT3 domain or BRCT4 domain abolished PTIP focus localization after IR (Fig. 6A). These observations suggest that this pair of BRCT domains, BRCT 3 and BRCT4, is essential for PTIP localization to DNA breaks.
FIGURE 6.
Distinct C-terminal BRCT domains are involved in PTIP foci formation and PTIP/53BP1 interaction. A, PTIP foci formation requires its BRCT3 and BRCT4 domains. U2OS cells transfected with plasmids encoding SFB-tagged wild-type (WT) or deletion mutants of PTIP were exposed to 10 Gy of ionizing radiation. Cells were fixed and immunostained with anti-FLAG and anti-γH2AX antibodies. B, U2OS cells were transfected with plasmids encoding SFB-tagged wild-type (WT) or mutant PTIP. Immunostaining was carried out with anti-FLAG and anti-γH2AX antibodies 6 h after cells were exposed to IR (10 Gy). C, PTIP does not bind to ubiquitin. Ubiquitin-agarose beads were incubated lysates containing either SFB-PTIP or SFB-USP3 (positive control) for 2 h, and proteins associated with ubiquitin were subjected to Western blotting with anti-FLAG antibody. D, PTIP and 53BP1 form foci independent of each other. Wild-type, 53BP1-/-, or PTIP-/- MEFs were irradiated, fixed, and immunostained with antibodies as indicated. E, binding to 53BP1 requires all four C-terminal BRCT domains of PTIP. 293T cells were transfected with plasmids encoding Myc-tagged 53BP1 together with plasmids encoding wild-type or deletion mutants of SFB-tagged PTIP. Cells were irradiated and immunoprecipitation (IP) reactions were conducted using S protein beads and then subjected to Western blotting using antibodies as indicated.
The dependence of PTIP localization on RNF8/UBC13 implies the involvement of an ubiquitin-binding protein in the localization of PTIP. Using ubiquitin-agarose pulldown assay, we showed that PTIP could not bind to ubiquitin (Fig. 6B). A previous study has already shown that the tandem BRCT domain (BRCT3/4) of PTIP has phosphopeptide binding activity (18), suggesting that an unknown phosphorylated protein may be required for PTIP localization after DNA damage. To further confirm that the integrity of this tandem BRCT domain is required for PTIP IRIF formation, we mutated the highly conserved Trp residue on the α3 helix of BRCT domain. We observed normal focus localization of W75R (mutation in BRCT1), W165R (mutation in BRCT2), and W916R (mutation in BRCT6) mutants of PTIP but failed to detect foci formation of PTIP W663R mutant (mutation in BRCT3) after DNA damage (Fig. 6C). This is consistent with our deletion analysis and supports that the intact BRCT3/4 pair of PTIP is required for PTIP focus localization in response to DNA damage. Furthermore, in agreement with our hypothesis that PTIP is required for PA1 foci formation, PTIP mutant with deletion of either its BRCT3 or BRCT4 domain failed to target exogenously expressed PA1 to IRIF (supplemental Fig. S2), suggesting that PTIP localization to sites of DNA break is a prerequisite for the damage-induced focus localization of PA1.
Accumulation of PTIP and 53BP1 at DNA Damage Sites Occurs Independently of Each Other—It is well established that PTIP interacts with 53BP1 after IR (18, 19, 26). Because the C-terminal BRCT domains of PTIP have been shown to mediate its interaction with 53BP1 as well as its localization after DNA damage (18, 19, 26), we examined whether PTIP localization would depend on 53BP1. As shown in Fig. 6D, PTIP foci formation was similar in 53BP1-/- and wild-type MEFs. Likewise, we also found typical 53BP1 localization after IR in PTIP-/- cells, suggesting that although both act downstream of RNF8/UBC13, these two proteins are recruited to DNA damage sites through independent mechanisms.
Using a series of internal deletion mutants of PTIP, we showed that the interaction between PTIP and 53BP1 was abolished by deletion of any of the four C-terminal BRCT domains of PTIP (Fig. 6E), suggesting that all four C-terminal BRCT domains of PTIP are indispensable for its interaction with 53BP1. Thus, the requirement for 53BP1 binding is distinctly different from its focus formation, which only requires BRCT3 and BRCT4.
PTIP and PA1 Are Required for Cell Survival after Double-strand Breaks—Although PTIP is clearly involved in DNA damage response, its function in this process remains to be determined. We first examined the role of PTIP in IR-induced G2/M checkpoint control. As a control, ATM-/- MEFs displayed a clear defect in IR-induced G2/M checkpoint (Fig. 7A). However, PTIP-/- cells showed modest G2/M checkpoint deficiency. PTIP knockdown in human cells also led to a partial defect in G2/M checkpoint control, which is very similar to those observed in cells with PA1 knockdown (Fig. 7A), indicating that PTIP and PA1 may play a minor role in this G2/M checkpoint control after DNA damage.
FIGURE 7.
PTIP is important for proper DNA damage response after ionizing radiation. A, PTIP and PA1 play a minor role in G2/M checkpoint control after DNA damage. PTIP+/+, or PTIP-/-, and ATM-/- MEFs were mock-treated or irradiated (3 Gy). Similarly, U2OS cells transfected with control siRNA (siCTR), PTIP specific siRNAs (siPTIP#1 and siPTIP#2), or PA1-specific siRNAs (siPA1#1 and siPA1#2) were also mock-treated or irradiated. Cells were harvested 1 h later, and a G2/M checkpoint assay was carried out (see “Materials and Methods”). The percentages of cells stained with phospho-H3 antibody before and after IR treatment were obtained from three individual experiments. Error bars indicate S.D. B, PTIP and PA1 are required for cell survival after ionizing radiation. PTIP-/- cells were reconstituted with wild-type or various PTIP deletion mutants. Cell survival after irradiation was measured by clonogenic assay according to the “Materials and Methods.” The expression of PTIP or its deletion mutants in these cells were confirmed by Western blotting using indicated antibodies (lower panel). Similarly, U2OS cells were treated with control or PA1 specific siRNAs. Cell survival after IR was measured by clonogenic assay (upper panel), and Western blot analysis was performed to verify the depletion of PA1 after siRNA transfection (right panel). WT, wild type. C, a proposed model of the DNA damage responsive pathway involving PTIP. See “Discussion” for details.
Next we examined whether PTIP would be required for cell survival after IR. We established PTIP-/- MEF cells that stably express wild-type or various deletion mutants of PTIP as indicated (Fig. 7B). Although PTIP-/- cells reconstituted with wild-type PTIP was able to restore cell survival after IR similar to wild-type cells or PTIP-/- cells reconstituted with the BRCT1, BRCT5, BRCT6, or glutamine region deletion mutants of PTIP, PTIP-/- cells reconstituted with the BRCT2, BRCT3, or BRCT4 domain deletion mutants failed to do so (Fig. 7B). These results suggest that PTIP is required for cell survival after IR and that the N-terminal BRCT2 domain and C-terminal tandem BRCT3 or BRCT4 domains are critical for this function of PTIP. Similar to PTIP, depletion of PA1 increased IR sensitivity, suggesting that the PTIP·PA1 complex is required for cell survival in response to DNA damage.
DISCUSSION
PTIP is known to be involved in DNA damage response (19, 26). In this study we demonstrated that PTIP acts downstream of the well defined γH2AX·MDC1·RNF8 DNA damage signaling pathway (Fig. 7C). The loading of PTIP at DNA damage foci relies on the RNF8/UBC13-dependent protein ubiquitination events. Once at the sites of DNA breaks, PTIP can further load its binding partner PA1 via a direct interaction between PTIP and PA1. Moreover, we showed that the PTIP·PA1 complex is required for cell survival and optimal G2/M checkpoint control after IR. These studies firmly establish a role of PTIP in the known DNA damage-signaling cascade.
Our study also demonstrated that PTIP is required for cell survival and G2/M checkpoint control after ionizing radiation, albeit its role in G2/M checkpoint is limited. Moreover, we show that the focus-localization of PTIP is critical for its function in promoting cell survival following IR and re-enforces a notion that sustained accumulation of DNA damage check-point and repair proteins at sites of DNA breaks are important for optimal DNA repair after DNA damage. Currently, we do not know exactly how PTIP accumulates at DNA damage sites. Early studies suggest that RNF8-dependent ubiquitin chains formed at DNA damage sites can serve as docking sites for the recruitment of downstream proteins or protein complexes (7, 11, 12). For instance, ubiquitin-interacting motif-containing protein RAP80 can be recruited to DNA damage sites via its ability to bind to ubiquitin chains and itself serves as an “adaptor” for the further recruitment of BRCA1 (8-10, 13). The focus localization region of PTIP is mapped to a tandem BRCT domain, which is best known for its ability to bind to phosphorylated proteins (18). It is unlikely that this tandem BRCT domain of PTIP would interact directly with ubiquitin chains formed at DNA damage sites. Indeed, we failed to obtain any ubiquitin binding activity of PTIP (Fig. 6B). We suspect that PTIP recruitment may be very similar to the recruitment of BRCA1 as discussed above. There is another yet-to-be identified mediator protein that has ubiquitin binding capability similar to RAP80, which allows its own localization to DNA damage sites. This unidentified protein may be phosphorylated and, therefore, recruit PTIP through a direct protein-protein interaction via the PTIP BRCT3/4 domains. The goal in the near future is to identify this and other specific ubiquitin-binding proteins and elucidate how these proteins facilitate DNA damage signal transduction and regulate proteins including PTIP in the DNA damage response.
Besides binding to PA1, PTIP also interacts with 53BP1. In this study we showed that although PTIP and 53BP1 associate with each other (18, 19, 26) and both position downstream of RNF8 in the DNA damage-signaling cascade (7, 11, 12), they localize to sites of DNA damage independently. We speculate that their interaction at DNA damage sites may be important for certain DNA repair process. However, we did not observe an obvious defect in cell survival after ionizing radiation in cells expressing PTIP mutants specifically defective in 53BP1-binding (e.g. PTIP BRCT5 or BRCT6 deletion mutants). This could suggest that an interaction between PTIP and 53BP1 may only be essential for a particular repair process (for example, class-switch recombination), and thus, its functional significance cannot be revealed by IR sensitivity assay used here, which measures combined capacity of multiple DNA repair pathways. Indeed, we examined whether PTIP or PA1 would be involved in a particular DNA repair pathway, for example homologous recombination repair. As shown in supplemental Fig. S3, we observed normal damage-induced replication protein A foci formation, Rad51 foci formation, and gene conversion in PTIP- or PA1-depleted cells, suggesting that this complex may not play a major role in homologous recombination repair. We speculate that similar to 53BP1, PTIP·PA1 complex may participate in certain aspect of NHEJ. Further studies using PTIP conditional knock-out mice will reveal whether similar to 53BP1, PTIP (and the PTIP/53BP1 interaction) would be involved in class-switch recombination and/or other specific DNA repair processes.
Supplementary Material
Acknowledgments
We thank all colleagues in the Chen laboratory, especially Dr. Jun Huang for insightful discussion and technical assistance. We also thank Dr. Michael S. Y. Huen for proofreading the manuscript and the Harvard Taplin Biological Mass Spectrometry Facility for mass spectrometry analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants CA089239, CA092312, and CA100109 (to J. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1-S3.
Footnotes
The abbreviations used are: IR, ionizing radiation; IRIF, IR-induced foci; PTIP, Pax2 transactivation domain interaction protein; PA1, PTIP-associated protein 1; siRNA, small interfering RNAs; MLL, mixed lineage leukemia; DAPI, 4′,6-diamidino-2-phenylindole; MEF, mouse embryonic fibroblast; HA, hemagglutinin; Gy, gray; UTX, ubiquitously transcribed tetratricopeptide repeat, X chromosome; ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3related; BRCT, BRCA1 carboxyl termini; FANCN, Fanconi anemia, complementation group N; DSB, double strand break.
References
- 1.Harper, J. W., and Elledge, S. J. (2007) Mol. Cell 28 739-745 [DOI] [PubMed] [Google Scholar]
- 2.Sancar, A., Lindsey-Boltz, L. A., Unsal-Kacmaz, K., and Linn, S. (2004) Annu. Rev. Biochem. 73 39-85 [DOI] [PubMed] [Google Scholar]
- 3.Zhou, B. B., and Elledge, S. J. (2000) Nature 408 433-439 [DOI] [PubMed] [Google Scholar]
- 4.Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S., and Bonner, W. M. (1998) J. Biol. Chem. 273 5858-5868 [DOI] [PubMed] [Google Scholar]
- 5.Stucki, M., and Jackson, S. P. (2006) DNA Repair (Amst) 5 534-543 [DOI] [PubMed] [Google Scholar]
- 6.Lou, Z., Minter-Dykhouse, K., Franco, S., Gostissa, M., Rivera, M. A., Celeste, A., Manis, J. P., van Deursen, J., Nussenzweig, A., Paull, T. T., Alt, F. W., and Chen, J. (2006) Mol. Cell 21 187-200 [DOI] [PubMed] [Google Scholar]
- 7.Huen, M. S., Grant, R., Manke, I., Minn, K., Yu, X., Yaffe, M. B., and Chen, J. (2007) Cell 131 901-914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim, H., Chen, J., and Yu, X. (2007) Science 316 1202-1205 [DOI] [PubMed] [Google Scholar]
- 9.Wang, B., and Elledge, S. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 20759-20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, B., Matsuoka, S., Ballif, B. A., Zhang, D., Smogorzewska, A., Gygi, S. P., and Elledge, S. J. (2007) Science 316 1194-1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mailand, N., Bekker-Jensen, S., Faustrup, H., Melander, F., Bartek, J., Lukas, C., and Lukas, J. (2007) Cell 131 887-900 [DOI] [PubMed] [Google Scholar]
- 12.Kolas, N. K., Chapman, J. R., Nakada, S., Ylanko, J., Chahwan, R., Sweeney, F. D., Panier, S., Mendez, M., Wildenhain, J., Thomson, T. M., Pelletier, L., Jackson, S. P., and Durocher, D. (2007) Science 318 1637-1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobhian, B., Shao, G., Lilli, D. R., Culhane, A. C., Moreau, L. A., Xia, B., Livingston, D. M., and Greenberg, R. A. (2007) Science 316 1198-1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lechner, M. S., Levitan, I., and Dressler, G. R. (2000) Nucleic Acids Res. 28 2741-2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho, Y. W., Hong, T., Hong, S., Guo, H., Yu, H., Kim, D., Guszczynski, T., Dressler, G. R., Copeland, T. D., Kalkum, M., and Ge, K. (2007) J. Biol. Chem. 282 20395-20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel, S. R., Kim, D., Levitan, I., and Dressler, G. R. (2007) Dev. Cell 13 580-592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho, E. A., Prindle, M. J., and Dressler, G. R. (2003) Mol. Cell Biol. 23 1666-1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manke, I. A., Lowery, D. M., Nguyen, A., and Yaffe, M. B. (2003) Science 302 636-639 [DOI] [PubMed] [Google Scholar]
- 19.Jowsey, P. A., Doherty, A. J., and Rouse, J. (2004) J. Biol. Chem. 279 55562-55569 [DOI] [PubMed] [Google Scholar]
- 20.Maser, R. S., Zinkel, R., and Petrini, J. H. (2001) Nat. Genet. 27 417-421 [DOI] [PubMed] [Google Scholar]
- 21.Yu, X., Fu, S., Lai, M., Baer, R., and Chen, J. (2006) Genes Dev. 20 1721-1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rappold, I., Iwabuchi, K., Date, T., and Chen, J. (2001) J. Cell Biol. 153 613-620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chini, C. C., Wood, J., and Chen, J. (2006) Oncogene 25 4165-4171 [DOI] [PubMed] [Google Scholar]
- 24.Lou, Z., Minter-Dykhouse, K., Wu, X., and Chen, J. (2003) Nature 421 957-961 [DOI] [PubMed] [Google Scholar]
- 25.Issaeva, I., Zonis, Y., Rozovskaia, T., Orlovsky, K., Croce, C. M., Nakamura, T., Mazo, A., Eisenbach, L., and Canaani, E. (2007) Mol. Cell. Biol. 27 1889-1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoz, I. M., Jowsey, P. A., Toth, R., and Rouse, J. (2007) Nucleic Acids Res. 35 5312-5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.