Abstract
An efficient route for delivering specific proteins and peptides into neurons could greatly accelerate the development of therapies for various diseases, especially those involving intracellular defects such as Parkinson disease. Here we report the novel use of polybutylcyanoacrylate nanoparticles for delivery of intact, functional proteins into neurons and neuronal cell lines. Uptake of these particles is primarily dependent on endocytosis via the low density lipoprotein receptor. The nanoparticles are rapidly turned over and display minimal toxicity to cultured neurons. Delivery of three different functional cargo proteins is demonstrated. When primary neuronal cultures are treated with recombinant Escherichia coli β-galactosidase as nanoparticle cargo, persistent enzyme activity is measured beyond the period of nanoparticle degradation. Delivery of the small GTPase rhoG induces neurite outgrowth and differentiation in PC12 cells. Finally, a monoclonal antibody directed against synuclein is capable of interacting with endogenous α-synuclein in cultured neurons following delivery via nanoparticles. Polybutylcyanoacrylate nanoparticles are thus useful for intracellular protein delivery in vitro and have potential as carriers of therapeutic proteins for treatment of neuronal disorders in vivo.
Many neurodegenerative diseases involve accumulation of intracellular aggregates of misfolded protein, implying that defects in protein processing may be linked to their development and pathogenesis (1, 2). Molecular genetic studies have identified a number of intracellular proteins involved in these processes, and these may ultimately prove useful as targets for therapeutic drug development (3). Engineered proteins themselves could be useful as treatments or experimental reagents (4, 5). A major barrier, however, is delivery of such proteins across the membrane of neuronal cells. Traditional means of transfection, protein transduction, and macromolecular delivery have proven difficult in neurons due to their terminally differentiated state (6). A number of commercially available methods rely on use of liposomes or other charged lipid formulations, which have limited complex stability in serum and often high toxicity over time (7). Electroporation-based techniques often yield higher rates of transfection but are only effective when performed during a specific window of development in young, healthy, and undifferentiated cells, with eventual loss of expression or bioactivity over time (8). Viral-based vectors and fusions have demonstrated only limited efficacy in humans and animals while raising a number of concerns regarding safety, and typically require invasive procedures such as direct injection into the brain to achieve targeted delivery (9).
Here we describe an approach for protein delivery into neurons using polybutylcyanoacrylate (PBCA)2 nanoparticles (NPs) as carriers. These particles have been shown to mediate significant transport of chemotherapeutics across the blood-brain barrier (BBB) in live animals (10, 11). Based on the observation that they absorb multiple proteins, particularly apolipoprotein E, onto their surfaces following injection into the bloodstream and that artificial coating of the particles with apolipoprotein E improves their uptake across the BBB, it was suggested that these NPs may be taken up via lipoprotein receptor-mediated endocytosis through mimicry of endogenous low density lipoproteins (LDLs) (12). Lipoprotein transport across the BBB is crucial for the delivery of essential lipids to brain cells, and direct evidence has been provided for the existence of a LDL receptor in vivo on the BBB (13). LDLs have further been shown to undergo transcytosis across the brain capillary endothelium without apparent degradation by endothelial cells, and they may therefore gain entry into other tissues following passage across the BBB (14). However, whether NPs actually gain entry into neuronal cells has not been demonstrated.
We hypothesized that neurons, which are known to express a number of lipoprotein-binding receptors on their surface, would be capable of NP uptake. Here, we report that PBCA NPs are taken up by primary hippocampal cultures, and that this uptake is dependent on the LDL receptor. The particles are capable of delivering intact, functional proteins into neurons and other mammalian cells. These particles may represent a new and significantly improved approach over existing, often invasive methods of drug transport.
EXPERIMENTAL PROCEDURES
Preparation of PBCA NPs—Nanoparticles were synthesized as previously described by Kreuter et al. (15). Briefly, 1% (v/v) PBCA (GluStitch, Point Roberts, WA) was slowly added to an acidic polymerization medium consisting of 1% dextran 70,000 (Sigma-Aldrich) in 0.01 n HCl and stirred for 4 h with a magnetic stirrer. For production of 3H-labeled and FITC-nanoparticles, tritiated dextran 70,000 (250 mCi/mg, American Radiolabeled Chemicals, St. Louis, MO) or anionic, lysine fixable fluorescein (FITC) dextran 70,000 (excitation = 494 nm, emission = 521 nm, Invitrogen) were utilized as stabilizers instead. Polymerization was terminated by neutralization with 0.1 n NaOH, and the suspension was filtered through a glass fiber filter of pore size 0.45 μm (Millipore, Billerica, MA). 3% (w/v) mannitol was added as a cryoprotectant, and the solution was lyophilized using a Savant Novalyphe NL500 freeze-dryer (Savant, Farmingdale, NY). For preparation of protein-loaded NPs, 5 mg of lyophilized NPs was resuspended in phosphate-buffered saline and incubated with 5 mg of protein and 0.08% Polysorbate 80 (Sigma-Aldrich) for 60 min with stirring. Proteins utilized were Escherichia coli β-galactosidase (β-gal, Sigma-Aldrich), purified recombinant rhoG, and the mouse anti-α-synuclein monoclonal antibody H3C (16). The solution was then stirred for 1 h and filtered once more through a glass fiber filter of pore size 0.45 μm. The total amount of protein absorbed onto the particles was calculated by filtering the suspension through a hydrophilic, 0.1-μm pore polyvinylidene fluoride membrane filter (Millipore) and measuring the amount of free protein in the filtrate via UV spectroscopy (DU 640 spectrophotometer, Beckman Coulter Instruments, Fullerton, CA). Protein loading was determined to be ∼0.7–0.8 μg of protein per μg of NPs. Mean particle size was determined to be 200–250 nm by dynamic light scattering using a Brookhaven Instruments BI-200 SM goniometer (Holtsville, NY), and by scanning electron microscopy using an Hitachi S-4700 SE microscope (Schaumburg, IL).
Culture and Treatment of Cells—E18 rat hippocampi (Brain-Bits Inc., Springfield, IL) were dissociated and seeded at a density of 1.6 × 104 cells per cm2 onto poly-d-lysine-coated labware. When grown in Neurobasal supplemented with B27, cultures contained 95 ± 2%. Cultures were maintained in Neurobasal/B-27 (Invitrogen) supplemented with 0.5 mm glutamine and 25 μm glutamate at 37 °C, 5% CO2. Half the volume of medium (plus glutamine, but no glutamate) was replaced every 3–4 days. Experiments were performed on neurons at age 14 DIV. PC12 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% horse serum, 5% fetal bovine serum, 50 units/ml penicillin, 50 μm streptomycin, and 200 mm l-glutamine at 37 °C, 5% CO2. For neurite outgrowth experiments, some cells were treated with 2.5 S mouse natural nerve growth factor (NGF, Fisher Scientific) at a concentration of 50 ng/ml. Neurite outgrowth was quantified as the percentage of cells in each field exhibiting one or more projections at least twice the length of the cell body. A minimum of 50 fields per sample was examined and counted double-blind.
Fluorescence and Light Microscopy Studies—Nanoparticles were added directly to the cell medium at a final concentration of 250 μg/ml. For cells pre-treated with IgG-8H6 (generous gift of Dr. Hiroaki Hattori and Dr. Tadao Iawaki, BML Inc., Saitama, Japan), a monoclonal blocking antibody against LDL receptor (17, 18), antibody was added directly to the cell medium at a concentration of 100 μg/ml, and the samples were incubated for 2 h then washed three times with PBS prior to addition of the nanoparticles plus fresh medium. Anti-EGF receptor antibody 151-8AE4 (Developmental Studies Hybridoma Bank, University of Iowa) was utilized at a concentration of 250 μg/ml (19) as above. For fluorescence microscopy, cells were fixed with 4% paraformaldehyde for 15 min and permeabilized for 10 min at room temperature using 0.25% Triton X-100 (Sigma). Cells were then blocked with 10% bovine serum albumin for 1 h at 37 °C, incubated with Anti-NeuN (Millipore) in 2% bovine serum albumin for 1 h at 37 °C, and then incubated with Alexa Fluor 594 anti-mouse (Invitrogen). Cells were mounted with 2% PVA-DABCO (Sigma-Aldrich) prior to visualization. Some cultures were incubated with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen) following fixation, to allow fluorescence visualization of cell nuclei.
Primary neurons treated with β-gal-loaded NPs were washed three times with PBS to remove surface-bound particles and enzyme, then incubated with 33 μm ImaGene Green C12FDG substrate (Invitrogen) in culture medium for 30 min at 37 °C and 5% CO2. Cells were then washed with PBS and incubated for 30 min at room temperature with Live/Dead™ Red fluorescent dye (Invitrogen), washed twice with PBS, and visualized as above. For samples stained with X-gal (5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside), cells were washed and fixed as before, then incubated with 2 ml of β-Galactosidase Staining Kit Complete Staining Solution (Active Motif, Carlsbad, CA) per well for 2 h at 37 °C.
For measurements of fluorescence resonance energy transfer (FRET), H3C or a non-synuclein antibody control (mouse anti-Myc, Invitrogen) were delivered via nanoparticles (250 μg/ml) and indirectly labeled with a secondary antibody (rabbit anti-mouse, Invitrogen) conjugated to Alexa Fluor 488 (excitation = 488–500 nm, emission = 519–550 nm). The N terminus of synuclein was labeled with the SC-7012 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) followed by a secondary antibody (donkey anti-goat, Chemicon) conjugated to Cy3 (excitation = 532–555 nm, emission = 570–600 nm). Endogenous galactosidase (GLB1, EC 3.2.1.23) and heat shock protein 70 (hsp70) were detected using mouse anti-GLB1 (Abcam Inc., Cambridge, MA) or mouse anti-hsp70 (Affinity BioReagents, Inc., Golden, CO), respectively, and Alexa Fluor 488-labeled anti-mouse (Invitrogen). Goat IgG purified from normal, non-immunized animal serum (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) was detected with Alexa Fluor 488 anti-goat as an additional negative control. Fixed and stained cells were visualized using a Leica DM IRE2 fluorescence microscope. The fluorescence intensity of Alexa Fluor 488 (the “donor”) was measured using OpenLab™ software (Improvision Inc., Lexington, MA) before and after photobleaching of Cy3 (the “acceptor”). Energy transfer was calculated as the ratio of D (donor fluorescence after photobleach) to DA (donor fluorescence in the presence of the acceptor, before photobleach); a value greater than 1.0 represents a positive FRET signal. A minimum of 20 measurements was made per sample.
Measurements of FITC- and 3H-Nanoparticle Uptake and Turnover—For measurements of temperature dependence, cells were grown in black 96-well plates (Nunc, Rochester, NY) and treated with FITC-labeled NPs (final concentration of 250 μg/ml) for the indicated times. Before and after washing with PBS, fluorescence intensity (excitation = 494 nm, emission = 521 nm) was measured on a SpectraMax M2 microplate reader (Molecular Devices, Sunnyvale, CA). Background fluorescence was subtracted from each sample, and fluorescence incorporation was calculated as a percentage of the total dose of nanoparticles. For measurements of 3H-nanoparticle uptake, cells were grown on 12- to 13-mm round coverslips with NPs (final concentration of 250 μg/ml) for the indicated time, the coverslips were washed three times with PBS and transferred to scintillation vials filled with 10 ml of ScintiVerse BD mixture (Fisher Chemicals, Fairlawn, NJ), and counted on a Beckman LS-1701 counter. Background counts were subtracted from each sample, and 3H incorporation was calculated as a percentage of the total dose of nanoparticles.
Immunoprecipitation and Western Blot Analysis of Cell Lysates—Cells grown in 25-cm2 flasks were incubated with β-gal-loaded or empty nanoparticles or β-gal plous 0.08% Polysorbate 80 alone (final concentration of 250 μg/ml) for 2 h then washed once with PBS prior to detachment with trypsin-EDTA (Cambrex Bioproducts, East Rutherford, NJ). The cells were pelleted and lysed with 1 ml of ice-cold non-denaturing lysis buffer (1% Triton X-100, 50 mm Tris-Cl, pH 7.4, 300 mm NaCl, 5 mm EDTA, and 0.02% sodium azide) plus 1 μl of Sigma Protease Inhibitor Mixture P-8340, incubated on ice for 30 and at 4 °C for 15 min at 16,000 × g.20 μl of supernatant from each sample was then subjected to SDS-PAGE and blotted with rabbit anti-β-gal-horseradish peroxidase (Abcam, Cambridge, MA), mouse anti-β-actin (Abcam), and horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies (Amersham Biosciences).
For immunoprecipitation of cell lysates treated with H3C-loaded or empty NPs, samples were treated as above except that, prior to SDS-PAGE and Western blot analysis, 500 μl of supernatant from each sample was incubated with 50 μl of Protein G beads (Sigma) that had been pre-washed two times with non-denaturing lysis buffer. Samples were then incubated on an end-over-end rotator at 4 °C for 2 h, and washed four times with 1 ml of non-denaturing lysis buffer and one time with ice-cold PBS. 20 μl of each immunoprecipitate and 20 μl of each total cell lysate were then analyzed via SDS-PAGE and Western blot analysis using goat anti-synuclein (SC7012) and horseradish peroxidase-conjugated rabbit and sheep antibodies against goat and mouse Ig, respectively (as above).
Affinity purification of active (GTP-bound) rho from PC12 cells was performed using the E-Z Detect™ Rho Activation Kit (Pierce) according to the manufacturer's instructions. Briefly, cells were lysed and incubated with GTPγS to activate rho. GST fused to the binding domain of rhotekin (GST-rhotekin-RBD), which specifically binds GTP-bound rho, was immobilized on a glutathione resin and incubated with cell lysates for 1 h at 4 °C. The samples were then washed, eluted in sample buffer, and analyzed by SDS-PAGE. GTP-bound rho was subsequently detected via Western blot using an anti-rho mouse monoclonal antibody (Pierce). Whole lysates were also probed with mouse anti-Myc antibody (Invitrogen) and mouse anti-β-actin (Abcam) as a loading control.
Measurement of β-gal Activity—Cells were incubated with 250 μg/ml β-gal nanoparticles for 2 h then washed with PBS and switched to non-particle-containing medium. The neurons were then harvested at the time points given by scraping into 1 ml of PBS and analyzed using the Invitrogen β-Galactosidase Assay Kit (catalogue no. K1455-01). Hydrolysis of ortho-nitrophenyl-d-galactopyranoside was then measured as per the manufacturer's protocol. The β-gal activity was quantitated by measuring absorbance at 420 nm using a SpectraMax M2 microplate reader (Molecular Devices). The enzyme activity (after background subtraction) observed immediately after the 2-h treatment was set as 100%, and all subsequent activity observed after this period at specific assayed time points was divided by this amount to determine the “percent activity” remaining over time.
Purification of Myc-Tagged rhoG—mammalian expression vector pEF-BOS containing the coding sequence for Myc-tagged human rhoG was kindly provided by M. Negishi (Kyoto University, Japan). HEK-293 cells were cultured in 150-cm2 flasks containing Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum, 50 units/ml penicillin, and 50 μm streptomycin at 37 °C, 5% CO2. Each flask was transfected with 50 μg of plasmid using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions. Cells were harvested 48 h after transfection and lysed using M-PER Mammalian Protein Extraction Reagent (Pierce). Myc-rhoG was then purified from the lysates using the μMACS Anti-c-Myc Isolation Kit (Miltenyi Biotec, Auburn, CA) and quantified using a BCA Protein Assay Kit (Pierce).
RESULTS
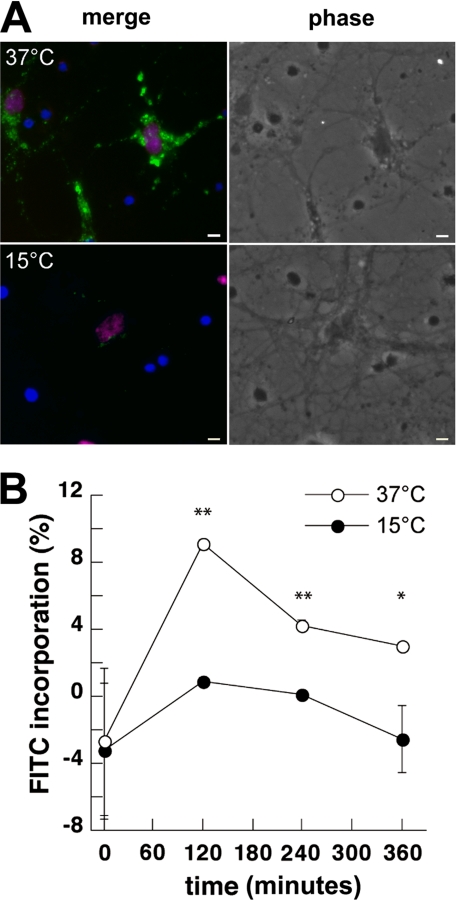
Neuronal Uptake of PBCA Nanoparticles by Endocytosis—To assess the uptake of PBCA NPs into cultured neurons, fluorescent PBCA NPs were synthesized utilizing dextran coupled to FITC. Primary E18 rat hippocampal neurons were incubated with 250 μg/ml NP for 2 h at either 15° or 37 °C, then fixed and stained with a mouse antibody against NeuN (20), a protein specifically expressed in neuronal cell nuclei (Fig. 1A). At 37 °C, extensive green fluorescence was observed in 74 ± 6% of NeuN-positive cells, indicating uptake of FITC-NPs. The percentage of NeuN-positive cells in our cultures at 14 DIV was 95 ± 2%. Images depicting a larger fraction of non-neuronal cells (i.e. NeuN-negative, DAPI-positive) are shown to demonstrate that FITC-NPs localize mainly to neurons and not other cell types (Fig. 1A). When cells were incubated with FITC-NPs for 2 h at 15 °C, little to no uptake was detected, indicating that NP uptake was likely via endocytosis rather than diffusion or non-specific adhesion. Staining was restored when cells incubated at 15 °C were brought back to physiological temperature (data not shown).
FIGURE 1.
Uptake of FITC-labeled PBCA NPs by primary neurons is temperature-dependent. A, fluorescence and phase-contrast micrographs of primary rat hippocampal neurons incubated with 250 μg/ml FITC-NPs (green) at 37°C versus 15 °C. Cells were fixed and counterstained with DAPI (blue) and the neuron-specific marker NeuN (red). Only neurons (purple nuclei), are co-labeled with FITC, while glia (blue nuclei) are unlabeled. Scale bar = 10 μm. B, fluorescence intensity was measured for each group of cells over time. FITC incorporation was calculated as a percentage of the total nanoparticle dose in medium alone, and plotted as mean ± S.D. **, p < 0.0001; *, p < 0.01, by one-way analysis of variance with post-hoc Tukey test.
Fluorescence intensity was also measured over a period of 6 h in live primary neurons incubated with 250 μg/ml nanoparticles at 37 °C versus 15 °C (Fig. 1B). Maximum incorporation was noted in both samples at 2 h post-treatment but was substantially reduced in cells incubated at 15 °C (p < 0.0001).
Nanoparticle Uptake Is Mediated by LDL Receptor—It was previously hypothesized that transport of PBCA NPs across vascular endothelial cells might be mediated by the LDL receptor (12). We tested whether uptake into cultured neurons might require LDL receptor by blocking the receptor prior to treatment with nanoparticles and examining the effects on FITC-NP uptake. Pre-treatment of primary neuron cultures with a monoclonal blocking antibody against LDL receptor (17, 18) inhibited incorporation of fluorescence, whereas treatment with a blocking antibody against the EGF receptor, which is also known to be specifically endocytosed (21, 22), resulted in no change (Fig. 2A).
FIGURE 2.
LDL receptor blockade inhibits uptake of PBCA NPs. A, fluorescence and phase-contrast microscopy images of primary hippocampal neurons treated with FITC-NPs, following pretreatment with anti-LDL receptor antibody (middle), anti-EGF receptor antibody (bottom), or without antibody pretreatment (top). Cells were counterstained with anti-NeuN (red). Scale bar = 10 μm. B, time course of incorporation of 3H-nanoparticles in the presence or absence of blocking antibodies against LDL receptor and EGF receptor is plotted as mean ± S.D. At the 45-min time point and beyond, LDL receptor blockade resulted in significantly reduced uptake relative to EGF receptor blockade or no antibody treatment (p < 0.05, by one-way analysis of variance with post-hoc Tukey test).
This effect of anti-LDL receptor antibodies on NP uptake was quantitated in cultures treated with 3H-labeled NP (Fig. 2B). As with FITC-NPs, maximum incorporation of label was achieved at ∼2 h for 3H-labeled NPs, and pre-treatment of the cultures with anti-EGF receptor antibody did not affect the time course of incorporation. However, pretreatment with anti-LDL receptor resulted in a nearly complete blockade of nanoparticle uptake. Similar data were obtained when measuring fluorescence intensity in live FITC-NP-treated neurons over time (not shown).
As further confirmation of the role of LDL receptors in NP uptake, we analyzed CHO LDL-A7 cells, one of a limited number of viable mammalian cell lines lacking functional LDL receptors (23). Normal CHO cells demonstrated efficient nanoparticle uptake that was blocked by anti-LDL receptor antibody, but CHO LDL-A7 cells exhibited impaired uptake of both 3H- and FITC-NPs as compared with normal controls (supplemental Fig. S1). Our data are thus consistent with the hypothesis that LDL receptor-mediated endocytosis is the primary mechanism of neuronal uptake for PBCA NPs.
Kinetics of Nanoparticle Uptake and Turnover—The time course of nanoparticle delivery was analyzed over a range of doses from 50 to 500 μg/ml. FITC incorporation (which reflects both particle uptake and turnover) was strongly dose-dependent at the lower particle concentrations, but began to saturate beyond ∼300 μg/ml (supplemental Fig. S2). Turnover of 3H-nanoparticles was also assessed, and the half-life of the particles was estimated to be ∼27 min (supplemental Fig. S3). Neuronal viability was measured for a range of nanoparticle doses; NPs were well tolerated at concentrations up to 500 μg/ml (supplemental Fig. S4). We selected 250 μg/ml as the standard dose for subsequent experiments.
PBCA Nanoparticles Deliver Proteins into Primary Neuronal Cells—We next utilized the particles for delivery of an exogenous and otherwise membrane-impermeant protein, E. coli β-gal, into primary neuronal cultures (Fig. 3). β-Galactosidase has well known activity that is readily detectable in mammalian cells and is used extensively as a marker of gene and protein expression. NP-mediated delivery of β-gal into neurons produces readily visible enzymatic activity throughout the cell body and neurites, detectable using the fluorescent substrate C12FDG in live cells, or the colorimetric substrate X-gal in fixed samples (Fig. 3A). Positive staining was observed in 69 ± 7% of neurons. Cells incubated with only β-gal plus Polysorbate 80 (which is used as a surfactant coating for nanoparticles) demonstrated no green fluorescence after treatment with C12FDG and were negative for staining with X-gal (Fig. 3A). The live neurons were also counterstained with Live/Dead™ Red to demonstrate viability of β-gal-positive cells. The percentage of dead cells (i.e. positively stained with Live/Dead™ Red) was determined to be 19 ± 8%, which is similar to the toxicity we measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide test (supplemental Fig. S4). Of the C12FDG-positive cells, however, only 4 ± 3% were also positive for Live/Dead™ Red.
FIGURE 3.
Intraneuronal delivery of functional protein. A, fluorescence and phase-contrast microscopy images of live primary neurons treated with β-gal loaded nanoparticles (top row) or β-gal only with Polysorbate 80 (bottom row) and stained for enzymatic activity using the substrate C12FDG; viability of assessed with Live/Dead™ Red, which is excluded from live cells. Fixed cells were stained with X-gal and visualized via light microscopy. B, decay of β-gal activity in neurons after 2 h of treatment with β-gal-loaded NPs. Data are plotted as mean ± S.D. B (inset), Western blot analysis of lysates from cells treated with β-gal-loaded NPs (lane 1), β-gal alone with surfactant (lane 2), and β-gal-loaded nanoparticles after pre-treatment with a blocking antibody against the LDL receptor (lane 3). Only cells treated with β-gal-loaded NPs in the absence of anti-LDL receptor contained detectable levels of β-gal enzyme. β-Actin was also probed for as a loading control. Scale bar = 10 μm.
Following NP-mediated delivery into neuronal cells, β-gal activity persists for several hours after treatment. Enzyme activity was measured by ortho-nitrophenyl-d-galactopyranoside assay, and the half-life was estimated to be 122 min by exponential curve fit (R2 = 0.957), as compared with 27 min for the NPs themselves, indicating that the β-gal cargo was degraded independently of the carrier NPs (Fig. 3B). Extracts of β-gal-treated cells were also assessed at 2 h by immunoblot, with β-actin as a loading control. The β-gal immunoreactivity was detected in cells treated with β-gal nanoparticles, but not in cells pretreated with anti-LDL receptor blocking antibodies or with β-gal alone (Fig. 3B, inset).
Nanoparticle-mediated Delivery of rhoG Produces Neurite Outgrowth and Differentiation in PC12 Cells—We next investigated whether we could deliver a protein with overt functional and morphological activity into neurons. For this purpose we selected rhoG, a small GTPase which, when genetically expressed in PC12 cells, causes terminal differentiation into catecholaminergic cells with neuronal morphology (24).
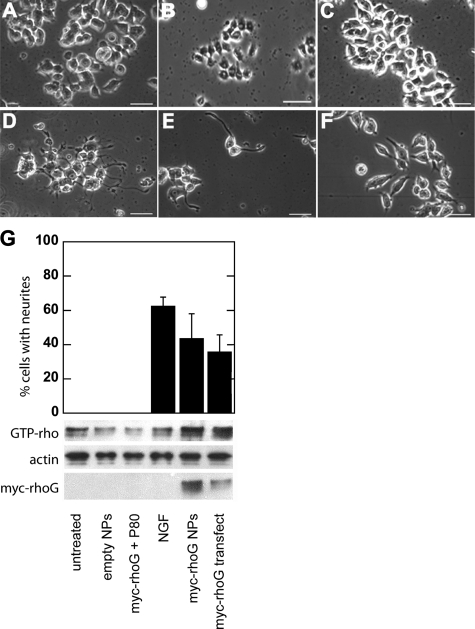
Purified Myc-tagged rhoG was loaded onto the nanoparticles, and cells were treated with the NPs daily for up to 10 days. Beginning at 2–5 days post-treatment, cells began sprouting neurite-like projections (Fig. 4E), similar to cells treated with nerve growth factor (Fig. 4D). After 10 days treatment, 43 ± 14% of PC12 cells treated with rhoG-loaded nanoparticles exhibited outgrowth of neurites as compared with 36 ± 10% of rhoG-transfected cells and 62 ± 5% of cells treated with NGF alone (Fig. 4G). Significant differences were observed among cells treated with rhoG-NPs versus empty particles (p < 0.0001) and rhoG protein alone (p < 0.0001). Transfected cells began extending processes ∼4 days after treatment as opposed to only 2 days for cells administered NGF or rhoG protein-loaded NPs.
FIGURE 4.
Delivery of rhoG into PC12 cells produces neurite outgrowth and differentiation. A, phase-contrast microscopy images of untreated PC12 cells; B, PC12 cells treated with empty nanoparticles; C, with Myc-rhoG protein plus Polysorbate 80, with NGF (D), treated with nanoparticles loaded with Myc-rhoG protein (E), and transfected with rhoG DNA (F). G (top), quantification of neurite outgrowth in cultures treated as above. Values are reported as mean ± S.D. (*, p < 0.01, by one-way analysis of variance with post-hoc Tukey test). G (bottom), Western blot analyses of PC12 cell lysates treated as above; lysates were incubated with GTPγS to activate rho, subjected to pull-down with GST-rhotekin-RBD, and probed with anti-rho (first row). Whole lysates were also probed with an anti-Myc antibody to detect exogenous Myc-rhoG (third row) with β-actin as a loading control (second row). All samples were assayed at 5 days. Scale bar = 50 μm.
To verify that the exogenously delivered rhoG protein was capable of interacting with a downstream effector, GTP-bound rho was affinity-purified from lysates using GST-tagged rhotekin, a specific target of members of the rho small GTPase family. The results indicate increased levels of active rho only in the cultures that were treated with rhoG NPs or directly transfected with rhoG (Fig. 4G, top row). Purified (not shown) and whole lysates were also probed with anti-Myc to specifically detect NP-delivered or transfected Myc-rhoG (Fig. 4G, center row), with β-actin as a loading control (Fig. 4G, bottom row).
Antibodies Delivered via Nanoparticles Can Interact with Their Intracellular Targets—We examined whether a mouse monoclonal (H3C), raised against the α-synuclein C terminus (16), maintains its ability to bind its target antigen in neurons following delivery via PBCA NPs. Because α-synuclein is normally found in the cytosolic compartment, successful delivery of H3C antibody to the neuronal cytosol should result in a specific interaction between the two proteins.
To detect intracellular delivery of H3C, cells were treated with H3C-loaded nanoparticles and subsequently stained using a Alexa Fluor 594-labeled secondary antibody to H3C. Diffuse staining was detected throughout the cytosol and neurites (Fig. 5A) in 71 ± 9% of H3C-NP-treated cells but not in those exposed to H3C plus Polysorbate 80 in the absence of NPs (Fig. 5B). A co-immunoprecipitation assay also supported the NP-mediated, LDL receptor-dependent delivery of functional H3C into these cells: the endogenous synuclein and the NP-delivered H3C could be co-immunoprecipitated from cell lysates using Protein G (Fig. 5C). Protein G failed to immunoprecipitate endogenous synuclein from cells that had not been exposed to H3C-loaded nanoparticles, or from cells in which NP uptake had been blocked using anti-LDL receptor antibody.
FIGURE 5.
Delivery of anti-synuclein antibody H3C into primary neurons. A, fluorescence microscopy images of primary neurons treated with nanoparticles loaded with a monoclonal antibody H3C against α-synuclein or (B) with H3C and Polysorbate 80 alone and stained with a Alexa Fluor 594-conjugated secondary and co-stained with DAPI. C, Western blot analysis of whole-cell lysates (W) and protein-G pulldown assays (G) from neurons treated with H3C-NPs (lanes 3 and 4), empty nanoparticles (lane 5 and 6), or H3C-NPs after preincubation with anti-LDL receptor antibody (lanes 7 and 8). Lane 1 = H3C antibody (50 ng), lane 2 = recombinant human α-synuclein (50 ng). Blot was probed with a goat antibody (SC7012) against N-terminal α-synuclein. D, immunocytochemistry of neurons treated with H3C-NPs then fixed and stained with SC7012, a goat antibody against the N terminus of α-synuclein. The primary antibodies were detected using Alexa Fluor 488-conjugated anti-mouse and Cy3-conjugated anti-goat, respectively. NP-delivered H3C colocalizes with SC7012. Scale bar = 50 μm.
To confirm that the NP-delivered H3C is closely associated with endogenous synuclein in intact cells, we performed immunocytochemistry (Fig. 5D) and measured fluorescence resonance energy transfer (FRET, Table 1) using a previously established acceptor photobleach technique (25, 26). H3C was labeled with Alexa Fluor 488, and the N terminus of α-synuclein was labeled indirectly using the SC7012 antibody (27, 28) and a secondary antibody conjugated to Cy3. Because the emission wavelengths of Alexa Fluor 488 and the excitation wavelengths of Cy3 overlap, these two fluorophores can serve as a “FRET pair,” with Alexa Fluor 488 serving as the fluorescent “donor” and Cy3 acting as the “acceptor.” The fluorescence intensity of the donor is measured before and after photobleaching of the acceptor. A positive FRET signal is measured as an increase in donor fluorescence after receptor photobleach, which only occurs when the two fluorophores are <10 nm apart. With the indirect immunolabeling approach used here, the antibody epitopes themselves must be within 30 nm distance of each other (27).
TABLE 1.
Quantitation of FRET between NP-delivered H3C and α-synuclein in neurons Primary antibodies were either delivered as nanoparticle cargo or utilized as labeling antibodies after fixation, as indicated. Anti-mouse Alexa Fluor 488 and anti-goat Cy3 were used as secondary antibodies. D/DA is the ratio of donor fluorescence intensity after (D) and before (DA) acceptor photobleach, reported as mean ± S.D. p values were determined by comparison to the null hypothesis D/DA = 1.00 by one-way analysis of variance, with post-hoc analysis by Tukey test.
| Primary antibodies | D/DA | p |
|---|---|---|
| H3C (cargo) + SC7012 | 2.67 ± 0.54 | <0.001 |
| Anti-myc (cargo) + SC7012 | 0.70 ± 0.22 | NSa |
| H3C (cargo) + goat IgG | 0.63 ± 0.18 | NS |
| H3C (cargo) | 0.80 ± 0.19 | NS |
| Anti-hsp70 + SC7012 | 2.04 ± 0.06 | <0.0005 |
| Anti-GLB1 + SC7012 | 0.77 ± 0.14 | NS |
NS, not significant
Efficient FRET was measured between NP-delivered H3C and labeling antibody SC7012 (D/DA = 2.67 ± 0.54, p < 0.001, Table 1). These antibodies interact with the C terminus and N terminus of α-synuclein, respectively. This result indicates that NP-delivered H3C associates with α-synuclein in intact cells, because NP treatment occurred prior to fixation and processing for immunocytochemistry. SC7012 also exhibits FRET with heat shock protein 70 (D/DA = 2.04 ± 0.06, p < 0.0005, Table 1), a chaperone shown to bind α-synuclein in vitro and closely associated with α-synuclein aggregates in animal models of Parkinson disease (29, 30). Neither immunocytochemical colocalization (supplemental Fig. S5) nor FRET were detected between SC7012 and an anti-Myc antibody that was delivered via NP treatment (Table 1). Likewise, no FRET was observed between NP-delivered H3C and endogenous galactosidase (GLB1), a nonspecific protein in the neuronal cytosol. H3C does not demonstrate FRET with Cy3 anti-goat alone, nor with nonspecific goat IgG plus Cy3 anti-goat, indicating that the apparent association of labeled H3C and α-synuclein was not a consequence of nonspecific interactions among the labeling antibodies.
DISCUSSION
Neurons are relatively difficult targets for genetic manipulation, presenting obstacles for both basic research and therapeutic development (31). In culture, neurons are difficult to transfect at high efficiency (32). Methods for direct protein delivery exist, but they generally require either manipulation of the protein sequence or direct injection of the protein (33). In vivo protein delivery is even more challenging, because the BBB prevents access of most large molecules to the brain parenchyma (6).
Direct physical methods of protein expression such as microinjection are typically labor-intensive and require precise manual control and can therefore only be performed on small numbers of mammalian neurons at a time (32). Electroporation and nucleofection require membrane perturbation and can only be performed on young, freshly isolated, undifferentiated cells (34–36). Viral vectors can yield infection rates of up to 95% in cultured neurons (12) but are limited by constraints such as: 1) the time and effort necessary for constructing recombinant viruses, 2) size restrictions on the DNA expression cassette and, finally, 3) the need for additional safety precautions during handling and processing (6, 37–40).
PBCA NPs have potential to circumvent these barriers. Prior studies suggest that these NPs can be transcytosed across the brain capillary endothelium via an LDL receptor-dependent pathway, perhaps by forming a complex with LDL in the bloodstream (12, 14, 41). These studies did not evaluate uptake of PBCA NPs into neurons and glia, however. Because neurons are also known to express LDL receptor (42), we asked whether PBCA NPs could mediate uptake of proteins into cultured neurons. Such uptake could have immediate impact in studies of protein function in vitro and also represent an important first step in the development of vectors for protein delivery to neurons in the brain following peripheral administration. We achieved successful intracytoplasmic delivery of three functionally diverse proteins: E. coli β-gal (EC 3.2.1.23), recombinant human Myc-tagged rhoG (EC 3.6.5.2), and the anti-α-synuclein mouse monoclonal antibody H3C.
E. coli β-gal does not normally cross the plasma membrane. Its activity is readily detected with colorimetric or fluorescent substrates, and it is commonly used as a marker of exogenous gene delivery in vertebrate cells. When primary hippocampal cultures were treated with β-gal-loaded NPs, positive staining for β-gal activity was observed, indicating that the delivered protein was sufficiently intact to retain its enzymatic activity. Control cultures were treated with recombinant β-gal and Polysorbate 80 (with no NPs), to show that the surfactant itself was not directly permeabilizing the cell membrane. Only those cultures treated with β-gal-loaded NPs demonstrated significant β-gal activity.
We indirectly compared the NP-mediated delivery of protein to other means of protein transduction by relying on published reports in the research literature. The duration of detectable enzymatic activity that we measured (8 h) was less than what has been reported for cells genetically transfected with β-gal (at least 24 h (43)) but comparable to other means of direct protein delivery. A study by Barka et al. (44) found that transduction of a TAT-HA-β-gal fusion protein into salivary gland acinar and ductal cells following extracellular administration resulted in enzyme activity over the course of 10 min to 6 h, which is similar to what we observed for primary neurons incubated with β-gal NPs. Smith et al. (45) reported that the half-life for histochemical staining of β-gal after direct pressure injection of the protein into sensory neurons is between 24 and 48 h. They used a 10× higher concentration of protein (2.5 mg/ml) in their experiments, however, which may have altered the kinetics of turnover. PBCA NPs have the advantage of requiring lower dosages of protein without the need for sequence modifications or special equipment.
NGF-induced differentiation of PC12 cells (46) is mediated by the activity of rhoG, and overexpression of rhoG in PC12 cells can mimic the effects of NGF administration (24). We sought to test whether direct delivery of recombinant Myc-tagged rhoG protein via PBCA NPs could likewise induce differentiation. Our results demonstrate that similar morphological changes can be induced by treatment with rhoG-loaded NPs, with roughly equivalent numbers of cells sprouting neurite-like projections over time as those genetically transfected with rhoG through conventional lipofection-based means. A major advantage of direct protein delivery, however, was more rapid onset of neuritogenesis. Cells administered rhoG-loaded nanoparticles began extending processes after only 2 days, half the time required for those genetically transfected with rhoG. The latter also exhibited visibly shorter neurites and a less differentiated phenotype by 10 days post-treatment.
Antibodies are also typically impermeant to cell membranes, yet they hold great potential for manipulation of protein folding and turnover in vitro and in vivo. In some cases single domain and single chain Fv antibodies have been engineered for intracellular expression (47). Such “intrabodies” directed against the protein α-synuclein have been shown to alter α-synuclein aggregation in cultured cells (48). This approach is limited in vivo by the availability of a robust gene delivery system for neuronal cells. We found that the mouse monoclonal antibody H3C is capable of binding α-synuclein following delivery to cultured neurons. Given prior studies demonstrating migration of PBCA NPs across the BBB, these particles have potential utility for targeting of therapeutic proteins to brain neurons in vivo. We are currently investigating ways that these nanoparticles may be modified for enhanced uptake into specific neuronal populations.
We show that uptake of PBCA NPs into neurons is dependent on endocytosis via the LDL receptor. The particles are internalized via energy-dependent, receptor-mediated endocytosis, a process that is selectively inhibited by exposure to low temperatures (49, 50). In the current study, incubation of primary neurons at 15 °C dramatically decreased uptake of FITC-labeled NPs. If nanoparticles were simply bound to the plasma membrane rather than actively endocytosed, fluorescence staining would be similar regardless of temperature. Incorporation of the NPs is also saturable and dose-dependent, findings consistent with a receptor-mediated endocytic pathway (51). In all cells tested here (primary neurons, PC12 cells, and CHO cells) uptake was specifically dependent on LDL receptor, because pre-treatment of cells with an LDL receptor-specific blocking antibody (17, 18) results in significantly reduced NP uptake. Incubation with a blocking antibody against EGF receptor, another endocytosed receptor (21, 22), does not affect NP uptake. Moreover, mutant CHO cells lacking functional LDL receptor (23) incorporate significantly fewer NPs than wild-type, LDL receptor-expressing CHO cells.
Several observations suggest that the NP cargo proteins are somehow delivered to the cytosolic compartment. The pattern of β-gal staining is diffuse, unlike the punctate pattern typical of the endolysosomal system. Both the H3C antibody and rhoG cargo proteins interact with cytosolic targets following NP-mediated delivery. There is a precedent for such behavior with anionic nanoparticles formulated from copolymers of poly(dl-lactide-co-glycolide) (52). Poly(dl-lactide-co-glycolide) nanoparticles are proposed to act as a “proton-sponge” within endolysosomal organelles, leading to osmotic disruption as these compartments become increasingly acidified, a process termed “endolysosomal escape” (53). We speculate that the cargo of PBCA NPs reaches the cytosol via a similar mechanism, and once present in this compartment the degradation of the released protein is separate from that of the polymer. This could also account for our finding that the half-life of β-gal activity was nearly five times that of the nanoparticle carriers (i.e. 122 versus 27 min). Cytosolic proteins are normally degraded via complex, multistep conduits such as the ubiquitin-proteasome pathway and chaperone-mediated autophagy (54–56), whereas alkylcyanoacrylates are thought to be rapidly hydrolyzed by nonspecific esterases (57).
Our data indicate that in primary cultured neurons, PBCA NPs coated with Polysorbate 80 are well tolerated at doses at which uptake saturation occurs; in some cases, even lower toxicity is observed for protein-loaded NPs than for empty NPs. At the concentration used in our studies (250 μg/ml), cells treated with protein-loaded nanoparticles had ∼80% viability, as compared with 65% for empty NPs (supplemental Fig. S4). This is consistent with the observation that absorbed proteins can considerably influence the toxicity and distribution of nanomaterials in vivo and in vitro (58–60). Although cytotoxicity of alkylcyanacrylate-based drug delivery systems has occasionally been reported (60, 61), these effects are highly dependent on polymer alkyl side-chain length, and polymers with longer alkyl chains (like PBCA) demonstrate little toxicity toward multiple cell types (62, 63). There is a report that PBCA NPs coated with Polysorbate 80 gain entry to the brain due to nonspecific opening of the BBB (64), although later findings suggest that no disruption of endothelial cell tight junctions nor nonspecific toxic effect occurs at therapeutic doses of the NPs (65). Nevertheless, several surface modifications of polyalkylcyanoacrylate carriers continue to be explored in ongoing efforts to improve safety and biocompatibility (61, 66).
Comparable examples of molecular delivery to neurons are scarce in the research literature, particularly for protein delivery. In many cases, uptake efficiency is reported with only a qualitative assessment of toxicity (67–69). Viral-derived peptides such as the TAT protein transduction domain have only been described as having “highly inefficient” uptake by neurons and “demonstrated toxicity” (70).
Nucleofection and calcium phosphate co-precipitation have resulted in 75 and 10% transfection in primary rat hippocampal neurons, respectively, but the percent cell death observed after treatment was not stated (68). In differentiated neurons derived from mesencephalic stem cells, however, viability following nucleofection was observed to be 40% (71). Cationic lipids such as Lipofectamine 2000™ demonstrated only 27% efficiency in E18 hippocampal neurons at 4 DIV but nearly 100% viability; transfection in cells older than 4 DIV, however, was significantly reduced (72). Using a modified electroporation technique, Bucsher et al. (73) similarly achieved 17% transfection in primary hippocampal neurons but observed only 37% viability after 2 days.
Engineered viral vectors based on HIV-1 were previously shown to infect up to 30% of rat cerebellar granule neurons in culture with recombinant β-gal, but no assays of toxicity were performed (74). Only 50% transduction was observed by Bender et al. (75) for recombinant lentiviruses in cultured embryonic mouse motoneurons using the least toxic multiplicity of infection (80% viability). It should be noted, however, that these neurons were spinal in origin rather than from the brain. Up to 60% transduction has been reported for specific adeno-associated virus serotypes in primary hippocampal neurons treated at 7 DIV, but cytotoxicity was not quantified (67).
Nearly 80% transduction of enhanced green fluorescent protein DNA was observed for carbon nanotube “spearing” in primary cortical neurons, but retraction of neurites also resulted from this invasive technique, which involves direct and physical penetration of neuronal cell membranes (69). Although recovery was said to have occurred by 48 h post-treatment, no toxicity data were given (69). PBCA NPs, however, successfully delivered protein in ∼70% of neuronal cells at the main dose (250 μg/ml) used for our studies with 80–90% viability.
In summary, PBCA NPs are useful vectors for protein delivery in vitro. They can efficiently deliver purified protein at physiologically significant concentrations to the cytosol of neurons and other cells in culture, without the need for modifications to the protein sequence itself, and without the use of special equipment. They are relatively non-toxic and compatible with a range of protein cargoes. Ultimately they may also prove valuable as tools for in vivo protein delivery.
Supplementary Material
Acknowledgments
We thank Vania Petrova, Subramanian Ramakrishnan, Charles Zukoski, Tzumin Lee, David Nelson, and Feng Sheng Hu at the University of Illinois Urbana-Champaign for technical assistance and advice; Manabu Negishi at Kyoto University, Japan, for providing myc-rhoG construct; Monty Krieger at the Massachusetts Institute of Technology for providing CHO LDL-A7 cells; and Andrew Belmont at the University of Illinois Urbana-Champaign for providing wild-type CHO cells.
This work was supported, in whole or in part, by National Institutes of Health Cell Molecular Biology Training Grant PHS5T32 GM07283. This work was also supported by the Branfman Family Foundation, the Parkinson's Disease Foundation, and National Institute on Aging Grant NIA R01 AG13762. Scanning electron microscopy was carried out in the Center for Microanalysis of Materials, University of Illinois, which is partially supported by the U.S. Dept. of Energy under Grant DEFG02-91-ER45439. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: PBCA, polybutylcyanoacrylate; BBB, blood-brain barrier; β-gal, β-galactosidase; DAPI, 4′-6-diamidino-2-phenylindole; EGF, epidermal growth factor; GLB1, acid β-galactosidase 1; LDL, low density lipoprotein; NGF, nerve growth factor; NP, nanoparticle; PVA-DABCO, polyvinyl alcohol-1;4-diazabicyclo(2.2.2)octane; X-gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; DIV, days in vitro; FRET, fluorescence resonance energy transfer; CHO, Chinese hamster ovary; PBS, phosphate-buffered saline; GTPγS, guanosine 5′-3-O-(thio)triphosphate; FITC, fluorescein isothiocyanate; C12FDG, 5-dodecanoylaminofluoroscein di-β-d-galactopyranoside.
References
- 1.Soto, C., and Estrada, L. D. (2008) Arch. Neurol. 65 184-189 [DOI] [PubMed] [Google Scholar]
- 2.Windisch, M., Wolf, H., Hutter-Paier, B., and Wronski, R. (2008) Neurodegener. Dis. 5 218-221 [DOI] [PubMed] [Google Scholar]
- 3.Winklhofer, K. F., Tatzelt, J., and Haass, C. (2008) EMBO J. 27 336-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Windisch, M., Hutter-Paier, B., Schreiner, E., and Wronski, R. (2004) J. Mol. Neurosci. 24 155-165 [DOI] [PubMed] [Google Scholar]
- 5.Zhou, C., Emadi, S., Sierks, M. R., and Messer, A. (2004) Mol. Ther. 10 1023-1031 [DOI] [PubMed] [Google Scholar]
- 6.Berges, B. K., Wolfe, J. H., and Fraser, N. W. (2007) Mol. Ther. 15 20-29 [DOI] [PubMed] [Google Scholar]
- 7.Whittlesey, K. J., and Shea, L. D. (2006) Biomaterials 27 2477-2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gartner, A., Collin, L., and Lalli, G. (2006) Methods Enzymol. 406 374-388 [DOI] [PubMed] [Google Scholar]
- 9.Luo, D., and Saltzman, W. M. (2000) Nat. Biotechnol. 18 33-37 [DOI] [PubMed] [Google Scholar]
- 10.Alyautdin, R. N, Gothier, D., Petrov, V., Kharkevich, D. A., and Kreuter, J. (1995) Eur. J. Pharm. Biopharm. 41 44-48 [Google Scholar]
- 11.Gulyaev, A. E., Gelperina, S. E., Skidan, I. N., Antropov, A. S., Kivman, G. Y., and Kreuter, J. (1999) Pharm. Res. 16 1564-1569 [DOI] [PubMed] [Google Scholar]
- 12.Kreuter, J. (2001) Adv. Drug Delivery Rev. 47 65-81 [DOI] [PubMed] [Google Scholar]
- 13.Meresse, S., Delbart, C., Fruchart, J. C., and Cecchelli, R. (1989) J. Neurochem. 53 340-345 [DOI] [PubMed] [Google Scholar]
- 14.Dehouck, B., Fenart, L., Dehouck, M. P., Pierce, A., Torpier, G., and Cecchelli, R. (1997) J. Cell Biol. 138 877-889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreuter, J., Alyautdin, R. N., Kharkevich, D. A., and Ivanov, A. A. (1995) Brain Res. 674 171-174 [DOI] [PubMed] [Google Scholar]
- 16.George, J. M., Jin, H., Woods, W. S., and Clayton, D. F. (1995) Neuron 15 361-372 [DOI] [PubMed] [Google Scholar]
- 17.Kosaka, S., Takahashi, S., Masamura, K., Kanehara, H., Sakai, J., Tohda, G., Okada, E., Oida, K., Iwasaki, T., Hattori, H., Kodama, T., Yamamoto, T., and Miyamori, I. (2001) Circulation 103 1142-1147 [DOI] [PubMed] [Google Scholar]
- 18.Takahashi, M., Ikeda, U., Takahashi, S., Hattori, H., Iwasaki, T., Ishihara, M., Egashira, T., Honma, S., Asano, Y., and Shimada, K. (2001) Clin. Genet. 59 290-292 [DOI] [PubMed] [Google Scholar]
- 19.Chandler, L. P., Chandler, C. E., Hosang, M., and Shooter, E. M. (1985) J. Biol. Chem. 260 3360-3367 [PubMed] [Google Scholar]
- 20.Mullen, R. J., Buck, C. R., and Smith, A. M. (1992) Development 116 201-211 [DOI] [PubMed] [Google Scholar]
- 21.Fallon, L., Belanger, C. M., Corera, A. T., Kontogiannea, M., Regan-Klapisz, E., Moreau, F., Voortman, J., Haber, M., Rouleau, G., Thorarinsdottir, T., Brice, A., van Bergen En Henegouwen, P. M., and Fon, E. A. (2006) Nat. Cell Biol. 8 834-842 [DOI] [PubMed] [Google Scholar]
- 22.Almeida, C. G., Takahashi, R. H., and Gouras, G. K. (2006) J. Neurosci. 26 4277-4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingsley, D. M., and Krieger, M. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 5454-5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh, H., Yasui, H., Yamaguchi, Y., Aoki, J., Fujita, H., Mori, K., and Negishi, M. (2000) Mol. Cell. Biol. 20 7378-7387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenworthy, A. K. (2001) Methods 24 289-296 [DOI] [PubMed] [Google Scholar]
- 26.Day, R. N., Voss, T. C., Enwright, J. F., 3rd, Booker, C. F., Periasamy, A., and Schaufele, F. (2003) Mol. Endocrinol. 17 333-345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma, N., Hewett, J., Ozelius, L. J., Ramesh, V., McLean, P. J., Breakefield, X. O., and Hyman, B. T. (2001) Am. J. Pathol. 159 339-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Agnaf, O. M., Salem, S. A., Paleologou, K. E., Cooper, L. J., Fullwood, N. J., Gibson, M. J., Curran, M. D., Court, J. A., Mann, D. M., Ikeda, S., Cookson, M. R., Hardy, J., and Allsop, D. (2003) FASEB J. 17 1945-1947 [DOI] [PubMed] [Google Scholar]
- 29.McLean, P. J., Kawamata, H., Shariff, S., Hewett, J., Sharma, N., Ueda, K., Breakefield, X. O., and Hyman, B. T. (2002) J. Neurochem. 83 846-854 [DOI] [PubMed] [Google Scholar]
- 30.Huang, C., Cheng, H., Hao, S., Zhou, H., Zhang, X., Gao, J., Sun, Q. H., Hu, H., and Wang, C. C. (2006) J. Mol. Biol. 364 323-336 [DOI] [PubMed] [Google Scholar]
- 31.Bergen, J. M., Park, I. K., Horner, P. J., and Pun, S. H. (2008) Pharm. Res. 25 983-998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Washbourne, P., and McAllister, A. K. (2002) Curr. Opin. Neurobiol. 12 566-573 [DOI] [PubMed] [Google Scholar]
- 33.Nagel, F., Falkenburger, B. H., Tonges, L., Kowsky, S., Poppelmeyer, C., Schulz, J. B., Bahr, M., and Dietz, G. P. (2008) J. Neurochem. 105 853-864 [DOI] [PubMed] [Google Scholar]
- 34.Teruel, M. N., Blanpied, T. A., Shen, K., Augustine, G. J., and Meyer, T. (1999) J. Neurosci. Methods 93 37-48 [DOI] [PubMed] [Google Scholar]
- 35.Gresch, O., Engel, F. B., Nesic, D., Tran, T. T., England, H. M., Hickman, E. S., Korner, I., Gan, L., Chen, S., Castro-Obregon, S., Hammermann, R., Wolf, J., Muller-Hartmann, H., Nix, M., Siebenkotten, G., Kraus, G., and Lun, K. (2004) Methods 33 151-163 [DOI] [PubMed] [Google Scholar]
- 36.Leclere, P. G., Panjwani, A., Docherty, R., Berry, M., Pizzey, J., and Tonge, D. A. (2005) J. Neurosci. Methods 142 137-143 [DOI] [PubMed] [Google Scholar]
- 37.Amalfitano, A., and Parks, R. J. (2002) Curr. Gene Ther. 2 111-133 [DOI] [PubMed] [Google Scholar]
- 38.Ehrengruber, M. U., Hennou, S., Bueler, H., Naim, H. Y., Deglon, N., and Lundstrom, K. (2001) Mol. Cell. Neurosci. 17 855-871 [DOI] [PubMed] [Google Scholar]
- 39.Janson, C. G., McPhee, S. W., Leone, P., Freese, A., and During, M. J. (2001) Trends Neurosci. 24 706-712 [DOI] [PubMed] [Google Scholar]
- 40.Slack, R. S., and Miller, F. D. (1996) Curr. Opin. Neurobiol. 6 576-583 [DOI] [PubMed] [Google Scholar]
- 41.Kreuter, J., Shamenkov, D., Petrov, V., Ramge, P., Cychutek, K., Koch-Brandt, C., and Alyautdin, R. (2002) J. Drug Target 10 317-325 [DOI] [PubMed] [Google Scholar]
- 42.Beffert, U., Stolt, P. C., and Herz, J. (2004) J. Lipid Res. 45 403-409 [DOI] [PubMed] [Google Scholar]
- 43.Altman-Hamamdzic, S., Groseclose, C., Ma, J. X., Hamamdzic, D., Vrindavanam, N. S., Middaugh, L. D., Parratto, N. P., and Sallee, F. R. (1997) Gene Ther. 4 815-822 [DOI] [PubMed] [Google Scholar]
- 44.Barka, T., Gresik, E. W., and van Der Noen, H. (2000) J. Histochem. Cytochem. 48 1453-1460 [DOI] [PubMed] [Google Scholar]
- 45.Smith, R. L., Geller, A. I., Escudero, K. W., and Wilcox, C. L. (1995) J. Virol. 69 4593-4599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mills, J. C., Kim, L. H., and Pittman, R. N. (1997) Exp. Cell Res. 231 337-345 [DOI] [PubMed] [Google Scholar]
- 47.Emadi, S., Liu, R., Yuan, B., Schulz, P., McAllister, C., Lyubchenko, Y., Messer, A., and Sierks, M. R. (2004) Biochemistry 43 2871-2878 [DOI] [PubMed] [Google Scholar]
- 48.Messer, A., and McLear, J. (2006) BioDrugs 20 327-333 [DOI] [PubMed] [Google Scholar]
- 49.Vasile, E., Simionescu, M., and Simionescu, N. (1983) J. Cell Biol. 96 1677-1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haylett, T., and Thilo, L. (1991) J. Biol. Chem. 266 8322-8327 [PubMed] [Google Scholar]
- 51.Yu, L., Nielsen, M., Han, S. O., and Wan Kim, S. (2001) J. Control Release 72 179-189 [DOI] [PubMed] [Google Scholar]
- 52.Panyam, J., Zhou, W. Z., Prabha, S., Sahoo, S. K., and Labhasetwar, V. (2002) FASEB J. 16 1217-1226 [DOI] [PubMed] [Google Scholar]
- 53.Pack, D. W., Hoffman, A. S., Pun, S., and Stayton, P. S. (2005) Nat. Rev. Drug Discov. 4 581-593 [DOI] [PubMed] [Google Scholar]
- 54.Elkabetz, Y., Kerem, A., Tencer, L., Winitz, D., Kopito, R. R., and Bar-Nun, S. (2003) J. Biol. Chem. 278 18922-18929 [DOI] [PubMed] [Google Scholar]
- 55.Kaushik, S., and Cuervo, A. M. (2008) Methods Mol. Biol. 445 227-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vogiatzi, T., Xilouri, M., Vekrellis, K., and Stefanis, L. (2008) J. Biol. Chem. 283 23542-23556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vauthier, C., Dubernet, C., Fattal, E., Pinto-Alphandary, H., and Couvreur, P. (2003) Adv. Drug Deliv. Rev. 55 519-548 [DOI] [PubMed] [Google Scholar]
- 58.Douglas, S. J., Davis, S. S., and Illum, L. (1987) Crit. Rev. Ther. Drug Carrier Syst. 3 233-261 [PubMed] [Google Scholar]
- 59.Moghimi, S. M., Muir, I. S., Illum, L., Davis, S. S., and Kolb-Bachofen, V. (1993) Biochim. Biophys. Acta 1179 157-165 [DOI] [PubMed] [Google Scholar]
- 60.Goppert, T. M., and Muller, R. H. (2005) J. Drug Target. 13 179-187 [DOI] [PubMed] [Google Scholar]
- 61.Blasi, P., Giovagnoli, S., Schoubben, A., Ricci, M., and Rossi, C. (2007) Adv. Drug Deliv. Rev. 59 454-477 [DOI] [PubMed] [Google Scholar]
- 62.Kubiak, C., Couvreur, P., Manil, L., and Clausse, B. (1989) Biomaterials 10 553-556 [DOI] [PubMed] [Google Scholar]
- 63.Müller, R. H., Lherm, C., Herbort, J., Blunk, T., and Couvreur, P. (1992) Int. J. Pharm. 84 1-11 [Google Scholar]
- 64.Olivier, J. C., Fenart, L., Chauvet, R., Pariat, C., Cecchelli, R., and Couet, W. (1999) Pharm. Res. 16 1836-1842 [DOI] [PubMed] [Google Scholar]
- 65.Kreuter, J., Ramge, P., Petrov, V., Hamm, S., Gelperina, S. E., Engelhardt, B., Alyautdin, R., von Briesen, H., and Begley, D. J. (2003) Pharm. Res. 20 409-416 [DOI] [PubMed] [Google Scholar]
- 66.Chauvierre, C., Leclerc, L., Labarre, D., Appel, M., Marden, M. C., Couvreur, P., and Vauthier, C. (2007) Int. J. Pharm. 338 327-332 [DOI] [PubMed] [Google Scholar]
- 67.Royo, N. C., Vandenberghe, L. H., Ma, J. Y., Hauspurg, A., Yu, L., Maronski, M., Johnston, J., Dichter, M. A., Wilson, J. M., and Watson, D. J. (2008) Brain Res. 1190 15-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeitelhofer, M., Vessey, J. P., Xie, Y., Tubing, F., Thomas, S., Kiebler, M., and Dahm, R. (2007) Nat. Protoc. 2 1692-1704 [DOI] [PubMed] [Google Scholar]
- 69.Cai, D., Mataraza, J. M., Qin, Z. H., Huang, Z., Huang, J., Chiles, T. C., Carnahan, D., Kempa, K., and Ren, Z. (2005) Nat. Methods 2 449-454 [DOI] [PubMed] [Google Scholar]
- 70.Reis, S. A., Willemsen, R., van Unen, L., Hoogeveen, A. T., and Oostra, B. A. (2004) J. Mol. Histol. 35 389-395 [DOI] [PubMed] [Google Scholar]
- 71.Cesnulevicius, K., Timmer, M., Wesemann, M., Thomas, T., Barkhausen, T., and Grothe, C. (2006) Stem Cells 24 2776-2791 [DOI] [PubMed] [Google Scholar]
- 72.Ohki, E. C., Tilkins, M. L., Ciccarone, V. C., and Price, P. J. (2001) J. Neurosci. Methods 112 95-99 [DOI] [PubMed] [Google Scholar]
- 73.Buchser, W. J., Pardinas, J. R., Shi, Y., Bixby, J. L., and Lemmon, V. P. (2006) BioTechniques 41 619-624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mochizuki, H., Schwartz, J. P., Tanaka, K., Brady, R. O., and Reiser, J. (1998) J. Virol. 72 8873-8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bender, F. L., Fischer, M., Funk, N., Orel, N., Rethwilm, A., and Sendtner, M. (2007) Histochem. Cell Biol. 127 439-448 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.