Abstract
KSR1 (kinase suppressor of Ras 1) is a molecular scaffold and positive regulator of the Raf/MEK/ERK phosphorylation cascade. KSR1 is required for maximal ERK activation induced by growth factors and by some cytotoxic agents. We show here that KSR1 is also required for maximal ERK activation induced by UV light, ionizing radiation, or the DNA interstrand cross-linking agent mitomycin C (MMC). We further demonstrate a role for KSR1 in the reinitiation of the cell cycle and proliferation following cell cycle arrest induced by MMC. Cells lacking KSR1 underwent but did not recover from MMC-induced G2/M arrest. Expression of KSR1 allowed KSR1–/– cells to re-enter the cell cycle following MMC treatment. However, cells expressing a mutated form of KSR1 unable to bind ERK did not recover from MMC-induced cell cycle arrest, demonstrating the requirement for the KSR1-ERK interaction. In addition, constitutive activation of ERK was not sufficient to promote cell cycle reinitiation in MMC-treated KSR1–/– cells. Only cells expressing KSR1 recovered from MMC-induced cell cycle arrest. Importantly, MMC-induced DNA damage was repaired in KSR1–/– cells, as determined by resolution of γ-H2AX-containing foci. These data indicate that cell cycle reinitiation is not actively signaled in the absence of KSR1, even when DNA damage has been resolved. These data reveal a specific role for the molecular scaffold KSR1 and KSR1-mediated ERK signaling in the cellular response to DNA interstrand cross-links.
Maintenance of genomic integrity is critical to cell survival. To prevent potentially damaging DNA mutations, which may lead to either cell death or carcinogenesis, cells employ specific damage-sensing pathways that sense and respond to different types of DNA damage (1). Cells must halt proliferation until the damage is repaired to prevent passing damaged or mutated DNA to daughter cells. These cellular mechanisms respond to both mutations incurred by endogenous causes, such as DNA replication, and damage induced by ectopic agents.
DNA damage sensors, such as ATM (ataxia telangiectasia mutated) and ATR (ATM and Rad3 related), detect damage caused by genotoxic agents and trigger signal transduction pathways in which MAPK6 pathways play a prominent role (2, 3). The evolutionarily conserved Raf/MEK/ERK MAPK cascade mediates signaling downstream of the proto-oncogene Ras and promotes cell survival and proliferation (4–6). The MAPKs ERK, p38, and JNK can be activated by mitogen stimulation (7–9). However, p38 and JNK are primarily activated in response to cellular stress (10, 11). In addition to mitogenic stimulation, ERK is also activated in response to multiple types of DNA damage including UV photoproducts induced by UV irradiation (12), DNA interstrand cross-links (ICLs) generated by cisplatin and MMC (13–15), and double strand breaks (DSBs) introduced by IR, hydroxyurea, and etoposide (16–18). Depending on the cell type, the stimulus used, and the duration of activation, ERK activation is able to promote a variety of biological responses, such as proliferation, apoptosis, cell cycle arrest, or differentiation (19–23).
Damage caused by ectopic agents can differentially stimulate ERK signaling and may result in a variety of cellular outcomes. For example, whereas JNK and p38 MAPK are transiently activated at early time points by DNA-damaging agents, biphasic or sustained ERK activation is observed (3, 9). MMC has been shown to activate JNK, p38, and ERK in corneal fibroblasts (14). Similar to the response to IR, JNK, and p38 are activated within minutes, whereas ERK is activated several hours following MMC treatment. In response to different stimuli, ERK can mediate both pro-survival and pro-apoptotic responses. ERK activation is necessary for IR-induced G2/M arrest in MCF-7 cells (24). Also, inhibition of ERK1/2 increases the sensitivity of cells to DNA damage (18, 25). ERK activity enhances apoptosis caused by cytotoxic doses of cisplatin (13). ERK activation is required for mitochondrial membrane depolarization, cytochrome c release, and caspase 3 activation after cisplatin exposure (13). These data demonstrate the wide array of biological responses produced by MAPK signaling in response to different types of DNA damage.
KSR1 (kinase suppressor of Ras 1) is a molecular scaffold for the Raf/MEK/ERK phosphorylation cascade (19, 26, 27). KSR1 binds Raf, MEK, and ERK and positively regulates ERK activation (19, 26–30). Deletion of KSR1 results in impaired ERK activation in both magnitude and duration in response to growth factor stimulation (19). Deletion of KSR1 impairs proliferation, differentiation into adipocytes, H-RasV12-induced and replicative senescence, and H-RasV12-induced transformation (19, 20, 31, 32). Loss of KSR1 in mouse intestinal epithelial cells and adult mouse colon cells increases tumor necrosis factor-α-mediated apoptosis (33, 34), and KSR1 is also critical for the suppression of DNA damage-induced apoptosis mediated by ERK in cortical neurons (35). In contrast, KSR1 enhances cisplatin-induced ERK activation and cisplatin sensitivity (13). Recently, McKay and Morrison (36) showed that KSR1 undergoes caspase-dependent cleavage in apoptotic cells. As a result, the cleaved C-terminal KSR1 product inhibits ERK activation by preventing the KSR1-ERK scaffolding interaction (36). These data demonstrate that KSR1 is a potent regulator of DNA damage-induced signaling and survival.
ICLs are highly toxic to mammalian cells (37, 38). ICLs, such as those induced by cisplatin, MMC, or psoralen, covalently link the two strands of DNA and inhibit the strand separation required for replication and transcription (37, 38). As such, DNA cross-linking agents are potent chemotherapeutic agents for certain types of cancers. ICLs are sensed primarily in the S phase during DNA replication (1, 39, 40). The replication machinery stalls upon encountering an ICL, resulting in recruitment of the sensor protein ATR to the site of damage. ATR phosphorylates and activates the effector kinase Chk1 (38, 40). Chk1 then phosphorylates and inactivates the phosphatase Cdc25 to cause cell cycle arrest (41, 42). Cdc25 proteins normally dephosphorylate and activate cyclin·Cdk complexes to promote cell cycle progression (38, 43). The arrest in cell cycle progression provides time for repair of the ICLs by a mechanism that requires XPF-ERCC1 (1, 38, 44–47). However, the mechanism by which cell cycle arrest is lifted and the cells reenter the cell cycle following the repair of damaged DNA has not been well characterized.
Here we present data that KSR1-mediated ERK activation is required for the efficient reinitiation of the cell cycle following induction of ICLs. Fibroblasts deficient in expression of KSR1 are impaired in cell cycle reinitiation and proliferation following treatment with the DNA cross-linking agent MMC. These data couple a mitogenic signaling cascade to the release from cell cycle arrest following DNA damage.
EXPERIMENTAL PROCEDURES
Cell Culture and Generation of Stable Cell Lines—Mouse embryo fibroblasts (MEFs) were generated from day 13.5 embryos from KSR1–/– and KSR1+/+ mice on a DBA1/LacJ background and were immortalized by a 3T9 protocol (26). MEFs were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Tissue Culture Biologicals), 1% penicillin/streptomycin (Invitrogen), 100 nm minimum Eagle's medium nonessential amino acids (Invitrogen), and 2 mm l-glutamine (Mediatech, Inc.) and were incubated at 37 °C in a 6% CO2 atmosphere.
To re-express KSR1 in KSR1–/– MEFs (19), KSR1–/– MEFs were transduced with the MSCV bicistronic retrovirus encoding green fluorescent protein and KSR1 separated by an internal ribosomal entry site (a generous gift of Dr. Eric Gosink, St. Jude Children's Research Hospital). GFP-expressing cells were isolated by fluorescence-activated cell sorting. The cells were excited at 488 nm and separated at 530/30 nm using a FACStar or FACSAria (BD Biosciences), with base-line fluorescence of uninfected cells having a mean intensity of 6 ± 0.5 (range 0–15). Post-sorted cells were assessed by fluorescence-activated cell sorter. Expression of ectopic KSR1 was detected by Western blot (19). The average expression of ectopic KSR1 was within 3-fold of endogenous KSR1 expression in KSR1+/+ MEFs. KSR1.AAAP cells were generated similarly and as described previously (31).
The constitutively activated MEK-ERK fusion protein (ERK2-MEK1-LA, a generous gift of Dr. Melanie Cobb, University of Texas Southwestern Medical Center) (48) was digested from the pCMV5 vector and ligated into the pGEMT TA shuttle vector. XhoI restriction sites were added using polymerase chain reaction, and MEK-ERK was ligated into MSCV-IRES-YFP at XhoI restriction digest sites. MSCV-[MEK-ERK]-IRES-YFP was transfected alone or with MSCV-KSR1-IRES-GFP and an ecotropic packaging vector into 293T cells. Retroviral supernatant was collected and filtered 48 and 72 h post-transfection and was used to infect KSR1–/– MEFs. YFP positive and GFP/YFP double positive cells were isolated by flow cytometry using a FACSAria. The cells were excited at 488 nm, and GFP and YFP signals were discriminated with 505- and 535-nm long pass dichroic filters, respectively. Cells demonstrating the appropriate levels of fluorescence through both 513/30 nm (GFP) and 560/40 nm (YFP) were collected.
Western Blotting—The cells were lysed in buffer containing 20 mm Tris-HCl (pH 8.0), 137 mm NaCl, 10% glycerol, 1% Igepal (Sigma), 1 mm phenylmethylsulfonyl fluoride, 5 mm sodium vanadate, 5 μg/ml aprotinin, and 10 μg/ml leupeptin (49). Alternatively, some cells were lysed in 1× Laemmli sample buffer (48.75 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.1 m dithiothreitol) and were sonicated three times for 5 s each. For Western blot analysis, the lysates were resolved on SDS-PAGE gels. The proteins were transferred to nitrocellulose membranes, and the membranes were blocked with Odyssey blocking buffer (LI-COR, 1:1 with PBS). The membranes were probed with primary antibodies diluted in Odyssey blocking buffer (1:1 with PBS). Chk1 (Santa Cruz Biotechnology), ERK1/2, phospho-ERK Thr202/Tyr204, phospho-Chk1 Ser345 or Ser317, p38, phospho-p38 Thr180/Try182, SAPK/JNK, phospho-SAPK/JNK Thr183/Try185 (Cell Signaling Technology), and KSR1 (BD Biosciences) were used as primary antibodies. The membranes were probed with secondary antibodies diluted in Odyssey blocking buffer (1:1 with PBS) containing 0.1% Tween 20. Secondary antibodies used were anti-rabbit or anti-mouse IgG AlexaFluor 680 (Molecular Probes) and anti-mouse or anti-rabbit IgG IRDye 800 (Rockland Immunochemicals). Blots were scanned using the Odyssey system (LI-COR). Immunoreactive proteins were quantified using Odyssey software and analyzed using Microsoft Excel.
Mitogenic Stimulation—MEFs were serum-starved for 4 h and treated with 100 ng/ml epidermal growth factor in Dulbecco's modified Eagle's medium containing 0.5% bovine serum albumin for the indicated times. The cells were lysed, and Western blot analysis was performed.
DNA-damaging Agents—MEFs were treated with 0.5 μg/ml MMC (Sigma) for 2 h. MMC was washed out four times with PBS, and fresh medium was replaced for the indicated times. The cells were treated with UV-C irradiation (254 nm) at a dose of 5 J/m2 using a Spectroline ENF-280C irradiator. The cells were treated with IR at a dose of 8 Gy using a Mark I 68A irradiator and an X-2 attenuator.
To determine cell viability and survival, MTT assays, Trypan blue staining, and colony forming assays were performed. For the MTT assay, the cells were plated in 96-well plates at 1.5 × 103 cells/well and were allowed to incubate for 24 h prior to treatment. The cells were treated with the indicated concentrations of MMC and incubated for 5 days. On day 5, MTT was added to each well and incubated for 4 h at 37 °C. MMC and MTT were removed, and Me2SO was added. Absorbance at 570 nm was determined by spectrometry. Cytotoxicity was expressed as the percentage of A570 of treated cells relative to untreated cells. For Trypan blue staining, MEFs were seeded at low density in 12-well plates. After 24 h, the cells were treated with MMC (0.5 μg/ml) for 2 h. MMC was removed by washing four times with PBS, and normal culture medium was replaced. After 3 days the cells and medium supernatant were collected, and the cells were counted in the presence of Trypan blue. The number of Trypan blue-positive (dead) cells was divided by the total number of cells to determine the percentage of cell death.
For the colony forming assay, MEFs were seeded at 4.0 × 103 cells/10-cm dish 24 h prior to treatment. The cells were then treated with 0.5 μg/ml MMC for 2 h. MMC was removed by washing cells with PBS four times, and normal culture medium was replaced. The cells were stained using Giemsa stain after 1 week. Three fields per plate were analyzed by phase contrast microscopy, and individual colonies were evaluated for the number of cells present.
To assess Chk1 phosphorylation, MEFs were treated with varying concentrations of MMC (0.5–50 μg/ml) for 8 h. The increased dose and time of treatment were necessary to detect Chk1 activation in our pool of asynchronous cells.
Cell Cycle Analysis—Cell cycle analysis was performed by propidium iodide staining and flow cytometry. The cells were treated with MMC (0.5 μg/ml) for 6 h or IR (8 Gy). Treatment for 6 h with MMC was necessary to detect cell cycle arrest by flow cytometry. MMC was removed by washing four times with PBS, and fresh medium was replaced. The cells were harvested using trypsin 0–48 h post-treatment, fixed in cold 70% ethanol, and stained for DNA content using Telford reagent. DNA was analyzed by flow cytometry (FACSCalibur).
The cytokinesis block proliferation index (CBPI) checkpoint assay was used to assess cell cycle arrest in S or G2 phase by scoring cells as mono- or binucleated (50, 51). The cells were plated on coverslips at very low density. After 24 h, the cells were treated with either MMC (0.5 μg/ml for 2 h), UV (5 J/m2), IR (8 Gy), or left untreated. MMC was removed by washing four times with PBS. The cells were then fed with fresh medium containing the actin inhibitor cytochalasin B (5 μg/ml) to inhibit cytokinesis. After 0, 24, or 48 h of cytochalasin B treatment, the cells were swollen with 75 mm KCl for 10 min and were fixed with cold methanol:acetone (1:1) at –20 °C for 30 min. The cells were treated with 0.5% Triton X-100 for 2 min at room temperature, and the nuclei were stained using Hoechst dye 33258. The cells were analyzed using phase contrast and fluorescent microscopy (filter set UV-2E/C; excitation 330–380 nm, dichromatic mirror 400, barrier filter emission 435–485 nm) and were scored as having one or two nuclei. Binucleated cells were those that had entered mitosis and were arrested prior to cytokinesis by cytochalasin B. Mononucleated cells were those that had arrested in the cell cycle because of treatment with exogenous agents and had not yet entered mitosis. The percentage of binucleated cells was determined for each condition.
H2AX Detection—The formation of nuclear foci containing phosphorylated histone H2AX was assessed by immunofluorescence. The cells were seeded at low density on coverslips. After 24 h the cells were treated with MMC (0.5 μg/ml) for 2 h. MMC was removed by washing four times with PBS, and fresh medium was replaced. The coverslips were processed 0, 2, 24, or 48 h after MMC treatment. The cells were fixed in 2% paraformaldehyde for 15 min and then washed twice with PBS. The cells were permeabilized with 0.1% Triton X-100, and nonspecific binding was blocked with 50% Odyssey blocking buffer in PBS for 1 h at room temperature. The cells were probed with an anti-γ-H2AX primary antibody (Upstate) and an anti-goat-Cy3 secondary antibody. The nuclei were counterstained using Hoechst dye 33258. The nuclei were visualized by fluorescence microscopy using the appropriate filters, and photomicrographs were obtained using IPLab software. The number of foci per nucleus was determined in at least 100 cells/condition.
Proliferation Studies—The cells were seeded at 4.0 × 104 cells/35-mm-diameter dish. Triplicate dishes were counted every 48 h for total cell number on a Beckman Coulter counter.
Transformation Assay—To assess lack of contact-inhibited growth, MEFs were seeded at 105 cells/10-cm-diameter dish. The medium was changed every 2–3 days. The photomicrographs were taken after 1 week.
Statistical Analysis—The data are presented as the means ± standard deviation. Statistical analyses were performed using a Student's t test where p < 0.05 was considered significant.
RESULTS
KSR1 Is Required for ERK Activation Induced by Genotoxic Stress—ERK activation is a critical step in the cellular response to treatment with genotoxic agents, regulating apoptosis, survival, and proliferation. The activation of ERK has been demonstrated following UV, IR, MMC, and cisplatin treatment (12–14, 16). KSR1 is required for maximal ERK activation induced by growth factor stimulation and cisplatin treatment (7, 13). We assessed whether KSR1 was required for ERK activation induced by different types of DNA-damaging agents. To assess KSR1 function, we compared MEFs from KSR1–/– mice to KSR1–/– MEFs in which ectopic KSR1 had been re-expressed.
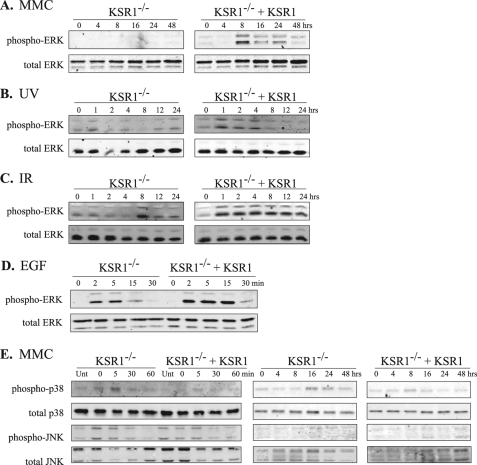
KSR1–/– or KSR1–/– MEFs expressing KSR1 at levels that recapitulate the responsiveness of wild type MEFs (19) were treated with UV irradiation (5 J/m2), IR (8 Gy), or the DNA cross-linking agent MMC (for 2 h, 0.5 μg/ml). These doses were chosen because they were sufficient to induce DNA damage and cell cycle arrest without causing significant cell death. The cells were lysed 0–48 h after treatment, and ERK activation was assessed by Western blot. ERK phosphorylation was detected following treatment with all of the genotoxic agents tested in KSR1-expressing cells. MEFs expressing ectopic KSR1 demonstrated ERK phosphorylation within 8 h following MMC treatment, within 1 h following UV irradiation, and within 1 h following IR (Fig. 1, A–C). However, KSR1–/– MEFs exhibited markedly decreased ERK activation following treatment with MMC, UV irradiation, or IR. Similar results were obtained with higher doses of UV (10 J/m2) or IR (12 Gy) treatment (data not shown). These data indicate that KSR1 mediates ERK activation in response to genotoxic stress induced by UV irradiation, IR, or MMC.
FIGURE 1.
KSR1 is required for DNA damage-induced ERK phosphorylation. KSR1–/– or KSR1–/– MEFs expressing ectopic KSR1 were treated with MMC (0.5 μg/ml) for 2 h followed by incubation for the indicated times (A), UV radiation (5 J/m2) (B), or IR (8 Gy) (C) and lysed at the indicated times following treatment. D, for comparison, KSR1–/– and KSR1–/– MEFs expressing ectopic KSR1 were serum-starved for 4 h and treated with epidermal growth factor (100 ng/ml) for the indicated times. Total ERK and phospho-ERK were detected by Western blotting. E, KSR1 is not required for p38 or JNK MAPK activation in response to MMC. KSR1–/– or KSR1–/– MEFs expressing ectopic KSR1 were treated with MMC (0.5 μg/ml for 2 h), washed with PBS, and incubated for the indicated times. The cells were lysed, and total and phopsho-p38 and JNK were detected by Western blotting.
ERK phosphorylation is impaired in both intensity and duration in KSR1-deficient cells following epidermal growth factor stimulation (Ref. 19 and Fig. 1D). KSR1 has not been shown to regulate MAPK signaling through JNK (19). However, because both JNK and p38 are involved in cellular responses to DNA-damaging agents, we tested whether KSR1 was required for maximal p38 or JNK phosphorylation following treatment with MMC. JNK and p38 were phosphorylated within minutes following MMC treatment in both KSR1–/– MEFs and KSR1–/– MEFs expressing ectopic KSR1 (Fig. 1E). At later time points, we detected some phosphorylation of p38, but not JNK, 8–16 h after MMC treatment in both KSR1–/– MEFs and KSR1–/– MEFs expressing ectopic KSR1. These data indicate that KSR1 is not required for MMC-induced JNK or p38 phosphorylation and suggest that ERK is the primary MAPK regulated by KSR1 following MMC treatment.
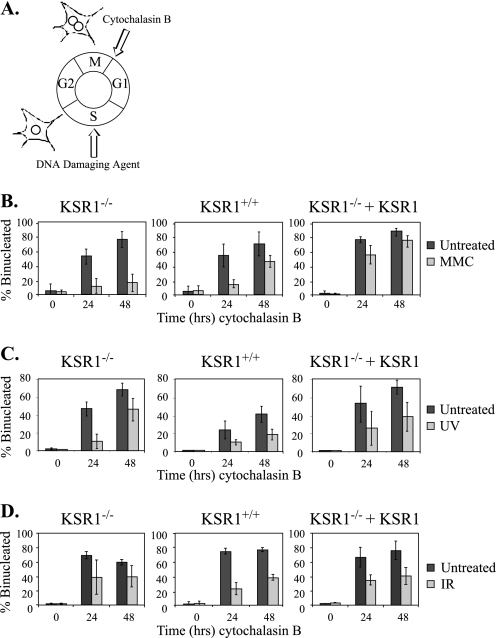
KSR1 Is Required for the Reinitiation of the Cell Cycle in Response to ICLs—Because KSR1 is required for ERK activation following genotoxic stress, we tested whether KSR1 is required for the cellular response to treatment with DNA-damaging agents. To investigate whether KSR1 is required for cell cycle arrest following DNA damage, we performed a CBPI assay (51). We used this assay to assess the ability of cells to undergo cell cycle arrest following DNA damage, as well as to recover from DNA damage and resume cell cycle progression. The CBPI assay utilizes the actin inhibitor cytochalasin B to monitor the accumulation of cells at cytokinesis. Normally, cells treated with cytochalasin B to block actin polymerization will undergo cell cycle arrest at cytokinesis and will become binucleated. However, cytochalasin B-treated cells that have undergone cell cycle arrest at an earlier stage of the cell cycle (e.g. because of DNA damage) and have not yet replicated their nuclei will have one nucleus (Fig. 2A). We tested KSR1–/– MEFs and KSR1–/– MEFs expressing ectopic KSR1, as well as MEFs generated from KSR1+/+ mice, to assess the requirement for KSR1 on cell cycle arrest and re-entry. KSR1–/–, KSR1+/+, or KSR1–/– MEFs expressing KSR1 were treated with UV irradiation (5 J/m2), IR (8 Gy), or MMC (0.5 μg/ml for 2 h). Following treatment, the cells were fed with fresh medium containing cytochalasin B to inhibit cytokinesis. After 0, 24, or 48 h, the number of nuclei was scored in each cell. Untreated cells contained predominantly two nuclei, indicating that these cells had undergone mitosis but had then been arrested by cytochalasin B before completion of cytokinesis. In contrast, cells treated with UV, IR, or MMC were predominantly mononucleated after 24 h, suggesting that cell cycle progression had been halted prior to mitosis because of DNA damage (Fig. 2, B–D). The cell cycle checkpoint was intact in KSR1–/–, KSR1+/+, and KSR1–/– MEFs expressing KSR1, because all three displayed a decreased number of binucleated cells following treatment with the DNA-damaging agents.
FIGURE 2.
KSR1 is required for cell cycle re-entry following MMC treatment. A, diagram representing cell cycle arrest and the CBPI assay. The cells that complete mitosis and undergo cell cycle arrest at cytokinesis because of cytochalasin B treatment are binucleated. The cells that have undergone arrest earlier in the cell cycle and have not yet completed mitosis are mononucleated. KSR1–/–, KSR1+/+, or KSR1–/– MEFs expressing ectopic KSR1 were treated with DNA-damaging agents (light gray bars) or left untreated (dark gray bars). The cells were treated with 0.5 μg/ml MMC for 2 h (B), 5 J/m2 UV radiation (C), or 8 Gy IR (D). Following treatment, the cells were incubated with cytochalasin B for the indicated times and analyzed by CBPI assay. The percentage of binucleated cells is indicated. The values are the averages ± standard deviations of at least three independent experiments.
We also used the CBPI assay to assess resumption of the cell cycle following DNA damage-induced cell cycle arrest. Forty-eight hours after MMC treatment, the number of binucleated cells increased, indicating that the cells were recovering from the damage-induced arrest and had reinitiated the cell cycle to enter mitosis. Compared with KSR1–/– MEFs, the percentage of binucleated cells was ∼5-fold higher in KSR1+/+ MEFs and KSR1–/– MEFs expressing KSR1 48 h after MMC treatment (Fig. 2B). These data indicate that KSR1–/– MEFs had impaired or delayed cell cycle re-entry following MMC treatment.
KSR1 deletion did not impair cell cycle re-entry in response to treatment with UV or IR at levels that activate ERK (Fig. 2, C and D, left panels). UV irradiation causes UV photoproducts (DNA intrastrand cross-links), whereas IR causes DNA double strand breaks (1, 52, 53). In contrast, MMC causes ICLs (1, 37, 38, 54). These data indicate that KSR1 is not required for resumption of the cell cycle following arrest because of UV photoproducts (DNA intrastrand cross-links) or DSBs but that KSR1 specifically regulates the response to ICLs.
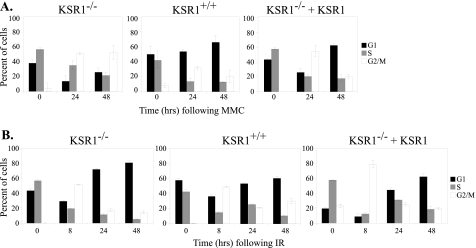
To further test the role of KSR1 in cell cycle re-entry, we performed cell cycle analysis of MMC-treated cells by flow cytometry. KSR1–/–, KSR1+/+, and KSR1–/– MEFs expressing ectopic KSR1 were treated with 0.5 μg/ml MMC for 6 h. MMC was removed, the cells were washed with PBS and fed with fresh medium and were incubated for 24–48 h. The cells were collected, and DNA was stained with propidium iodide for flow cytometric analysis. Within 24 h, all three cell lines showed an increase in the G2/M population, indicating arrest in the cell cycle (Fig. 3A). Within 48 h after MMC treatment, there was a decrease in the G2/M population and an increase in the G1 population in KSR1-expressing cells, indicating that these cells were reinitiating the cell cycle. However, the G2/M arrest persisted in cells deficient for KSR1 (Fig. 3A, left panel). These data are consistent with the results of the CBPI assay (Fig. 2), suggesting that KSR1 is required for cell cycle re-entry following MMC-induced arrest.
FIGURE 3.
Deletion of KSR1 sustains MMC-induced G2/M cell cycle arrest. KSR1–/–, KSR1+/+, or KSR1–/– MEFs expressing ectopic KSR1 were treated with MMC (0.5 μg/ml) for 6 h (A) or IR (8 Gy) (B) and were analyzed by propidium iodide staining and flow cytometry at the indicated times after treatment. The percentage of cells in G1 phase (black bars), S phase (gray bars), or G2/M phases of the cell cycle (white bars) are shown. The values are the averages ± standard deviations of three independent trials.
To ensure that KSR1 was responding specifically to ICLs, we also performed cell cycle analysis following IR. KSR1–/–, KSR1+/+, and KSR1–/– MEFs expressing ectopic KSR1 were treated with IR (at 8 Gy), fresh medium was added, and the cells were incubated for 8, 24, or 48 h. Within 8 h following IR, all three cell lines showed an increase in the number of cells at G2/M, indicating an arrest at this checkpoint (Fig. 3B). By 24 h, all of the cell lines had recovered from IR-induced G2/M arrest and demonstrated an increase in the G1 population, indicating they had re-entered the cell cycle. These data indicate that cell cycle re-entry following IR is not dependent on KSR1. ERK is activated by IR within 1 h, as compared with 8 h following MMC treatment (Fig. 1). This rapid activation of ERK may explain why cells recover more rapidly from IR rather than MMC-induced damage. Nonetheless, these data demonstrate that KSR1 specifically mediates reinitiation of the cell cycle following damage by ICLs. Therefore, we focused on the role of KSR1 in response to MMC.
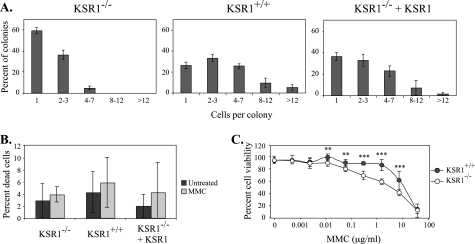
KSR1 Is Required for Proliferation Following MMC Treatment—To determine whether KSR1 is required for survival following MMC treatment, we performed a colony formation assay. KSR1–/–, KSR1+/+, or KSR1–/– MEFs expressing KSR1 were seeded at low density and were treated with MMC (0.5 μg/ml) for 2 h. MMC was removed, and the cells were incubated for 1 week in normal growth medium. Untreated cells formed large colonies within 1 week, whereas cells treated with MMC formed small colonies comprised of very few cells. The rate of cell survival could not be quantified in this assay because of the small size of the colonies in the MMC-treated cells. However, the number of cells per colony was determined to assess the proliferative potential of MMC-treated cells. Colonies of KSR1–/– MEFs treated with MMC were predominantly single cells (Fig. 4A). In contrast, the majority of colonies of KSR1-expressing MEFs were multicellular, indicating that these cells were able to proliferate following MMC treatment. These data further indicate that KSR1–/– MEFs are defective in their ability to re-enter the cell cycle and divide following the repair of ICLs.
FIGURE 4.
KSR1 is not required for survival following MMC treatment. A, proliferation following MMC treatment is decreased in KSR1–/– MEFs. KSR1–/–, KSR1+/+, and KSR1–/– MEFs expressing ectopic KSR1 were seeded at low density in a colony formation assay and were treated with 0.5 μg/ml MMC for 2 h. After 1 week, the number of cells/colony was determined. The values are the averages ± the standard deviation of three fields/dish. The results are representative of two independent experiments. B, cell viability following MMC treatment is not altered by loss of KSR1. KSR1–/–, KSR1+/+, and KSR1–/– MEFs expressing ectopic KSR1 were left untreated (dark gray bars) or treated with MMC (0.5 μg/ml, light gray bars) for 2 h. Cell survival was determined by counting with Trypan blue after 3 days of incubation in fresh medium. The values are the averages ± the standard deviations of three trials. C, KSR1–/– (open circles) and KSR1+/+ MEFs (closed circles) were treated with the indicated doses of MMC and assayed by colorimetric MTT assay to measure cell viability as described under “Experimental Procedures.” The asterisks represent statistical significance (**, p < 0.01; ***, p < 0.001).
We also performed Trypan blue staining to assess viability following MMC (0.5 μg/ml, 2 h) treatment. Three days following the removal of MMC, KSR1–/–, KSR1+/+, or KSR1–/– MEFs expressing KSR1 were treated with trypsin and counted in the presence of Trypan blue to assess cell survival. Levels of cell death were low (6% or less) and did not vary significantly among the cell lines (Fig. 4B). However, we also performed an MTT assay to assess cell viability following 5 days of continuous MMC treatment at various doses. Under these conditions, KSR1+/+ MEFs exhibited enhanced viability compared with KSR1–/– MEFs near the concentrations of MMC used in these studies (0.5 μg/ml), likely because of the impaired ability of the KSR1–/– MEFs to move through the cell cycle (Fig. 4C). These data indicate that cells expressing KSR1 retain mitotic potential following treatment with MMC.
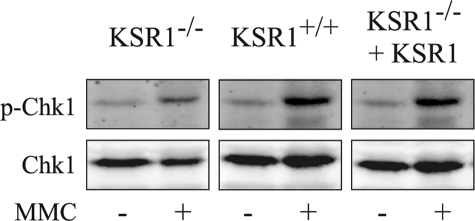
KSR1 Does Not Affect Cell Cycle Arrest or Repair of ICLs—To test whether KSR1 was required for the activation of cellular pathways leading to cell cycle arrest following MMC treatment, we examined the effects of KSR1 disruption on the activation of Chk1 (Fig. 5). Chk1 is phosphorylated and activated by ATM/ATR following DNA damage (55). In turn, Chk1 phosphorylates the phosphatase Cdc25, which promotes its cytoplasmic sequestration, prevents CDK activation, and inhibits cell cycle progression (42, 56). We treated KSR1+/+, KSR1–/–, and KSR1–/– MEFs expressing KSR1 with varying concentrations of MMC (0.5, 5, and 50 μg/ml) for 8 h and assessed Chk1 phosphorylation by Western blot. We could not detect Chk1 phosphorylation in cells treated with MMC at concentrations less than 50 μg/ml (data not shown). Chk1 was phosphorylated following treatment with 50 μg/ml MMC in KSR1–/– and KSR1–/– MEFs expressing KSR1, indicating that KSR1 is not required for Chk1 activation (Fig. 5). These data are consistent with the results of the CBPI assay (Fig. 2) and flow cytometric analysis (Fig. 3) and suggest that KSR1 is not required for the immediate sensing of DNA damage and induction of cell cycle arrest.
FIGURE 5.
KSR1 is not required for Chk1 phosphorylation. KSR1–/–, KSR1+/+, or KSR1–/– MEFs expressing ectopic KSR1 were treated with 50 μg/ml MMC for 8 h or were untreated. The lysates were analyzed by Western blot using antibodies against Chk1 or phospho-Chk1 (Ser345).
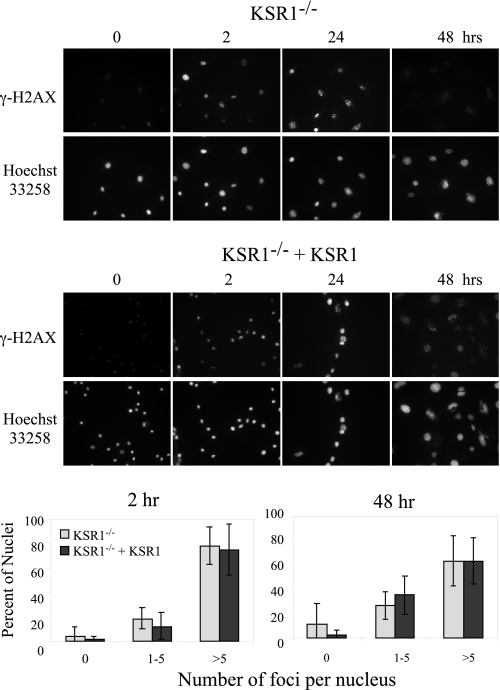
It is possible that ICLs caused by MMC are not efficiently repaired in KSR1–/– cells. As a protective mechanism against genomic instability, cells with unrepaired ICLs would not be released from cell cycle arrest. To assess DNA repair, we used immunofluorescence to detect the presence of nuclear foci containing phosphorylated histone H2AX (γ-H2AX). γ-H2AX has been used as a specific marker for the presence of DSBs. Following DNA damage, histone H2AX is phosphorylated by ATM/ATR and localizes at sites of DSBs until repair is complete (57, 58). Because DSBs are generated as an intermediate during repair of ICLs, the appearance and disappearance of γ-H2AX foci can quantitatively reflect the repair of ICLs (59–61). We treated KSR1–/– and KSR1–/– MEFs expressing KSR1 with MMC (0.5 μg/ml for 2 h) and used an anti-γ-H2AX antibody to detect nuclear foci 0, 2, 24, or 48 h after treatment. MMC treatment caused the formation of γ-H2AX foci within 2 h in both KSR1–/– and KSR1–/– MEFs expressing KSR1, demonstrating the presence of DNA damage (Fig. 6). Forty-eight hours after MMC treatment, the percentage of cells with γ-H2AX foci was significantly decreased in both KSR1–/– and KSR1-expressing cell lines, indicating that the DNA damage did not persist in the absence of KSR1. These data indicate that KSR1 is not required for the repair of ICLs.
FIGURE 6.
KSR1 is not required for repair of interstrand cross-links. KSR1–/– or KSR1–/– MEFs expressing ectopic KSR1 were treated with MMC (0.5 μg/ml) for 2 h and were fixed at the indicated time points following MMC treatment. The cells were assessed for DNA damage-containing foci by immunostaining for γ-H2AX. The nuclei were counterstained with Hoechst 33258. Photomicrographs (top) and quantification of the number of γ-H2AX foci/cell (bottom) are shown.
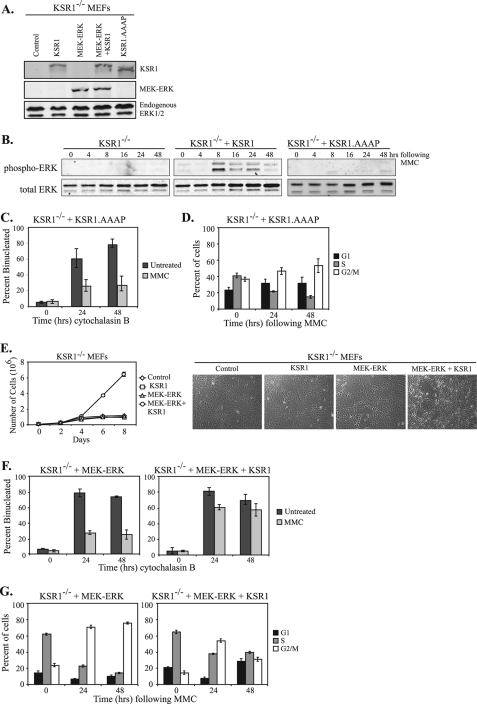
KSR1 Regulates Cell Cycle Reinitiation through ERK—ERK activation is an important step in the cellular response to genotoxic stress. KSR1 is required for maximal ERK activation following MMC-induced DNA damage (Fig. 1). Therefore, we hypothesized that KSR1 regulates cell cycle reinitiation following MMC treatment in an ERK-dependent manner. KSR1 contains a consensus ERK-binding sequence (residues 471–474, FSFP). Mutation of the ERK binding domain on KSR1 (KSR1.AAAP) results in decreased ERK activation and impaired H-RasV12-induced senescence and transformation (31). Consequently, the KSR1-ERK interaction optimizes the biological effects of ERK activation. To determine whether the KSR1-ERK interaction was required for KSR1 to promote cell cycle re-entry, KSR1.AAAP was expressed in KSR1–/– MEFs at levels comparable with the KSR1–/– MEFs stably expressing KSR1 (Fig. 7A). We assessed phospho-ERK levels in KSR1–/– MEFs expressing WT KSR1 or KSR1.AAAP. Expression of KSR1.AAAP did not restore MMC-induced ERK activation, demonstrating that this construct does not promote KSR1-scaffolded ERK activation (Fig. 7B).
FIGURE 7.
The KSR1-ERK interaction is required for cell cycle reinitiation. A, KSR1–/– or KSR1–/– MEFs expressing KSR1, MEK-ERK, MEK-ERK+KSR1, or KSR1.AAAP were lysed and subjected to Western blotting for the indicated proteins using an anti-KSR1 (top panel) or anti-ERK1/2 antibody (bottom two panels). B, KSR1–/– or KSR1–/– MEFs expressing either WT KSR1 or KSR1.AAAP were assessed for MMC-induced ERK activation. MEFs were treated with 0.5 μg/ml MMC for 2 h, washed with PBS, and incubated for the indicated times in fresh medium. Total and phospho-ERK were assessed by Western blotting. C, KSR1–/– MEFs expressing KSR1.AAAP were treated with 0.5 μg/ml MMC for 2 h (light gray bars) or left untreated (dark gray bars). Following MMC treatment, the cells were incubated with cytochalasin B for the indicated times and analyzed by CBPI assay. The percentage of binucleated cells is indicated. The values are the averages ± standard deviations of three trials. Control KSR1–/– and KSR1–/– MEFs expressing ectopic KSR1 are shown in Fig. 2. D, KSR1–/– MEFs expressing KSR1.AAAP were treated with MMC (0.5 μg/ml) for 6 h and were analyzed by propidium iodide staining and flow cytometry at the indicated times after treatment. The percent of cells in G1 phase (black bars), S phase (gray bars), or G2/M phases of the cell cycle (white bars) are shown. The values are the averages ± standard deviations of three independent trials. Control KSR1–/– and KSR1–/– MEFs expressing ectopic KSR1 are shown in Fig. 3. E, KSR1–/– MEFs (diamonds) or KSR1–/– MEFs expressing either KSR1 (squares), MEK-ERK (triangles), or MEK-ERK+KSR1 (circles) were seeded at 4 × 104 cells/35-mm-diameter dish. Triplicate dishes were assessed for cell number every 48 h on a Beckman Coulter counter. Focus formation assays were performed as described under “Experimental Procedures.” Representative photomicrographs are shown (right panel). F and G, KSR1–/– MEFs expressing MEK-ERK or MEK-ERK+KSR1 were analyzed by CBPI assay (F) or flow cytometry (G) as described above.
KSR1–/– MEFs and KSR1–/– MEFs expressing WT KSR1 or KSR1.AAAP were evaluated by CBPI assay. Treatment with MMC caused a decrease in the percentage of binucleated cells within 24 h of cytochalasin B treatment, demonstrating that the KSR1-ERK interaction is not required for cell cycle arrest in response to MMC (Fig. 7C). Similar to the results seen with the KSR1–/– MEFs (compare with Fig. 2B), the percentage of binucleated cells did not increase within 48 h of cytochalasin B treatment in cells expressing KSR1.AAAP. These data indicate that the KSR1-ERK interaction is required for cell cycle reinitiation following MMC treatment and suggest that KSR1 functions in an ERK-dependent manner to reinitiate the cell cycle following repair of ICLs.
To further examine the role of KSR1-scaffolded ERK activation in cell cycle reinitiation, cell cycle analysis was performed by flow cytometry. KSR1–/– MEFs and KSR1–/– MEFs expressing WT KSR1 or KSR1.AAAP were treated with 0.5 μg/ml MMC for 6 h. Cells were washed with PBS to remove MMC, fed with fresh medium, and incubated for 24–48 h. The cells were collected and DNA was stained with propidium iodide for flow cytometric analysis. Within 24 h following MMC treatment, KSR1–/– MEFs expressing KSR1.AAAP showed an increase in the number of cells in G2/M phase, indicating an arrest in the cell cycle at this checkpoint (Fig. 7D). In contrast to cells expressing WT KSR1, the percent of KSR1.AAAP cells arrested in G2/M phase remained high 48 h following MMC treatment (compare with Fig. 3A), These data are similar to the sustained MMC-induced G2/M arrest that was observed in KSR1–/– MEFs (Fig. 3A). Thus, disruption of the KSR1-ERK interaction (KSR1.AAAP) and blocking MMC-induced ERK phosphorylation prohibits cells from re-entering the cell cycle following MMC-induced arrest.
If the sole function of KSR1 in promoting cell cycle reinitiation is to regulate ERK activation, then expression of a constitutively active form of ERK should rescue the cell cycle defects in KSR1–/– MEFs. A form of ERK fused to its upstream activator kinase MEK is constitutively active, localizes to the nucleus, and promotes cell transformation (48). We generated KSR1–/– MEFs stably expressing this MEK-ERK fusion protein either alone or with ectopic KSR1 and performed Western blot analysis to show protein expression (Fig. 7A). We further demonstrated that the MEK-ERK fusion protein was in fact active. Constitutive activation of the Ras/Raf/MEK/ERK pathway leads to transformation of MEFs (48); therefore we examined whether KSR1 was required for the transforming potential of the MEK-ERK construct. To study transformation, KSR1–/– or KSR1–/– MEFs expressing KSR1, MEK-ERK, or MEK-ERK+KSR1 were assessed for proliferation rate and loss of contact-inhibited growth. As our laboratory has shown previously, KSR1–/– and KSR1–/– MEFs expressing KSR1 grow at similar rates and exhibit contact-inhibited growth (19). As expected, MEFs expressing MEK-ERK+KSR1 grow considerably faster than null control cells and induce focus formation (Fig. 7E). Surprisingly, KSR1–/– MEFs expressing the MEK-ERK fusion protein alone do not have increased proliferation or form foci (Fig. 7E). These data demonstrate that the MEK-ERK fusion protein is active in KSR1–/– MEFs but requires the scaffold protein KSR1 for its biological functions.
To study the role of activated ERK in cell cycle recovery following MMC, a CBPI assay was performed. KSR1–/– MEFs expressing MEK-ERK or MEK-ERK+KSR1 were treated with MMC for 2 h followed by treatment with cytochalasin B for 24–48 h. KSR1–/– MEFs expressing a MEK-ERK fusion protein were primarily mononucleated following MMC treatment, indicating they had undergone cell cycle arrest (Fig. 7F, left panel). The percentage of binucleated cells expressing the MEK-ERK fusion protein did not increase within 48 h following MMC treatment. However, expression of ectopic KSR1 in cells also expressing the constitutively active ERK construct significantly enhanced cell cycle recovery following MMC (Fig. 7F, right panel). These data demonstrate that constitutive activation of ERK alone is not sufficient to promote cell cycle re-entry after MMC treatment.
Cell cycle analysis was also performed by flow cytometry. KSR1–/– MEFs expressing MEK-ERK or MEK-ERK+KSR1 were treated with 0.5 μg/ml MMC for 6 h. The cells were washed with PBS to remove MMC, fed with fresh medium, and incubated for 24–48 h. The cells were collected, and DNA was stained with propidium iodide for flow cytometric analysis. Within 24 h following MMC treatment, KSR1–/– MEFs expressing a MEK-ERK fusion protein, both with and without KSR1, showed an increase in the number of cells at G2/M (Fig. 7G). Similar to the results seen with the KSR1–/– MEFs (Fig. 3A) and KSR1–/– MEFs expressing KSR1.AAAP (Fig. 7D), constitutive activation of ERK did not release cells from G2/M arrest (Fig. 7G, left panel). Only cells expressing both KSR1 and the MEK-ERK fusion protein were able to progress though the cell cycle and into the G1 phase (Fig. 7G). These data further demonstrate that KSR1 is not required for the initial cell cycle arrest in response to DNA damage. Instead, KSR1 is required for the release from cell cycle arrest or resumption of the cell cycle following MMC-induced damage. We show here that constitutive activation of ERK alone is not sufficient for reinitiation of the cell cycle following the cell response to ICLs. Instead, KSR1-scaffolded ERK activation is required for cell cycle recovery following MMC-induced G2/M arrest.
DISCUSSION
We demonstrate here that the molecular scaffold KSR1 is required for cell cycle recovery from MMC-induced DNA damage. Cells that lack KSR1 do not re-enter the cell cycle as efficiently as KSR1-expressing cells following MMC treatment (Figs. 2 and 3). This requirement was dependent on KSR1-mediated ERK signaling. Cells lacking KSR1 were defective in ERK phosphorylation following MMC treatment (Fig. 1). In addition, expression of a mutated form of KSR1 unable to bind ERK (KSR1.AAAP; Fig. 7) was unable to rescue the defects in ERK phosphorylation or cell cycle recovery. Overexpression of a constitutively active form of ERK was also insufficient to rescue the cell cycle reinitiation defect in KSR1–/– MEFs (Fig. 7). These data reveal a novel coupling of a mitogenic scaffold protein to the repair and recovery from a specific type of DNA damage.
UV, IR, and MMC induce different types of DNA damage, thereby activating different cellular checkpoints and damage responses. UV radiation primarily induces intrastrand DNA cross-links, IR induces DSBs, and MMC induces ICLs (52–54). The fact that KSR1 is required for cell cycle re-entry following treatment specifically with MMC, but not UV or IR, suggests that KSR1 is required to specifically regulate the cellular response to ICLs. ICLs are primarily sensed during the S phase, leading to cell cycle arrest in the late S or G2 phase of the cell cycle (1, 39, 40). KSR1 may be required for cell cycle reinitiation from this specific type of cell cycle arrest.
We have also determined that KSR1 is required for ERK activation induced by both UV and IR (Fig. 1). KSR1 was not required for the reinitiation of the cell cycle following UV or IR treatment (Figs. 2 and 3). However, this does not exclude the possibility that KSR1-mediated ERK activation is important in regulating other aspects of the cellular responses to these types of DNA damage. KSR1 enhances cell death induced by cisplatin treatment (13) but, in contrast, promotes cell propagation in cells treated with MMC by facilitating cell cycle progression (Fig. 4C). This indicates that KSR1-mediated ERK signaling may facilitate diverse cellular responses in different contexts. Although both of these chemotherapeutics cause interstrand cross-links, cisplatin has additional effects (62–64) that may account for the difference in response to cells expressing KSR1.
It is not understood how cells release from cell cycle arrest following repair of DNA damage. It may be that the magnitude of the signal indicating damage and triggering arrest falls below a threshold, and a cellular brake is released. Alternatively, the cell may actively signal that damage has been repaired. Our data support an active mechanism for the release from cell cycle arrest. ICLs are efficiently repaired in both the presence and absence of KSR1, as indicated by the resolution of γ-H2AX-containing foci (Fig. 6). However, KSR1–/– MEFs are deficient in cell cycle re-entry following resolution of the damage. These data indicate that the absence of persistent damage is not sufficient to allow the resumption of the cell cycle. Rather, the repair of DNA damage can be uncoupled from the reinitiation of the cell cycle, suggesting that an active signaling process may trigger cell cycle re-entry. Our data indicate that KSR1-mediated ERK activation is required for this release from cell cycle arrest.
We find that cells treated with MMC undergo G2/M arrest; cells expressing KSR1 are able to reinitiate the cell cycle following removal of MMC (Fig. 3). These data suggest that KSR1 may be required for the release from a G2/M checkpoint. The G2/M checkpoint is controlled by the cdc2·cyclin B complex (65). Several studies suggest that the main target of the G2/M DNA damage checkpoint is phosphorylation of cdc2 at Tyr15, which leads to its inactivation (66). Cdc2 can be phosphorylated on Tyr15 by two mitotic kinases, Wee1 and, to a lesser extent, Myt1, which prevent cells from entering into mitosis (67, 68). To progress through the cell cycle, cdc2 must be dephosphorylated by the mitotic phosphatase Cdc25C (69). The activity of Cdc25C can be either positively or negatively regulated by phosphorylation on different sites. For instance, in response to DNA damage, Chk1 phosphorylates Cdc25C on Ser216, which creates a 14-3-3 docking site that sequesters Cdc25C in the cytoplasm where it cannot activate cdc2, thus resulting in cell cycle arrest at G2/M (56). Recently, it has been shown that ERK can bind to and phosphorylate Cdc25C on a number of sites that are required for maximal phosphatase activity of Cdc25C, further enhancing mitotic progression (70). It is possible that KSR1-mediated ERK activation may be required for Cdc25C activation.
p42 MAPK activation is also required for release of G2/M arrest in clam oocytes (71). Delayed p42 MAPK activation results in inhibition of cell cycle re-entry following meiotic stimulation with KCl and the phosphatase inhibitor molybdate (71). These studies, as well as the data presented here, suggest that KSR1-mediated ERK activation is required for the reinitiation of the cell cycle.
KSR1 may also mediate cell cycle recovery following MMC arrest through its interaction with other proteins. KSR1 interacts with proteins that regulate DNA repair and cell cycle arrest. For instance, KSR1 interacts with the E3 ubiquitin ligase IMP (72). IMP, also known as Brap2, was originally identified as a BRCA1-interacting protein (73). Although the role of BRCA1 is not completely clear, it appears to be important in regulating several steps of the DNA damage response, ranging from sensing and signaling DNA damage to arresting and repairing damage in cells (74). Recently, Yan et al. (75) have shown an in vivo association between BRCA1 and ERK1/2, and this association may be important in regulating the cellular response to IR-induced DNA damage in MCF-7 cells. KSR1 is also a substrate for the kinase C-TAK1 (49). C-TAK1 was originally identified as a Cdc25C-associated kinase, and it phosphorylates Cdc25C on the same residue as Chk1 to inhibit cell cycle progression (76). The interaction of KSR1 with C-TAK1 may regulate the ability of C-TAK1 to phosphorylate Cdc25C in response to DNA damage. Consequently, KSR1 function may be linked directly or indirectly to these different proteins.
These data reveal that the molecular scaffold KSR1 is required for cell cycle reinitiation and recovery following arrest caused by ICLs. Because DNA cross-linking agents are used as chemotherapeutic agents for certain cancers, alterations in KSR1 function may affect cell sensitivity to chemotherapeutic agents. The function of KSR1 or KSR1-regulated signaling may alter the efficacy of chemotherapy in specific patients. In summary, this study provides a novel link between the MAPK signaling cascade and its involvement in the DNA damage response.
Acknowledgments
We gratefully acknowledge Mika Bessho and Laura Zietlow for assistance with the UV irradiation. Dr. Ying Yan, Jessica Nearman, and Mario Fernandez performed the gamma irradiation of cells. Linda Wilkie, Victoria Smith, and Dr. Charles Kuszynski of the University of Nebraska Medical Center Cell Analysis Facility performed the flow cytometric analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant CA90400 (to R. E. L.). This work was also supported by a grant from the Nebraska Dept. of Health and Human Services (to R. E. L.) and Cancer Center Support Grant CA036727. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MAPK, mitogen-activated protein kinase; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; MMC, mitomycin C; IR, ionizing radiation; ICL, interstrand cross-link; DSB, double strand break; GFP, green fluorescent protein; MEF, mouse embryo fibroblast; CBPI, cytokinesis block proliferation index; YFP, yellow fluorescent protein; PBS, phosphate-buffered saline; SAPK, stress-activated protein kinase; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Gy, grays; WT, wild type; γ-H2AX, phosphorylated histone H2AX; MSCV, murine stem cell virus.
References
- 1.Sancar, A., Lindsey-Boltz, L. A., Unsal-Kacmaz, K., and Linn, S. (2004) Annu. Rev. Biochem. 73 39–85 [DOI] [PubMed] [Google Scholar]
- 2.Zhou, B. B., and Elledge, S. J. (2000) Nature 408 433–439 [DOI] [PubMed] [Google Scholar]
- 3.Yang, J., Yu, Y., and Duerksen-Hughes, P. J. (2003) Mutat. Res. 543 31–58 [DOI] [PubMed] [Google Scholar]
- 4.Shields, J. M., Pruitt, K., McFall, A., Shaub, A., and Der, C. J. (2000) Trends Cell Biol. 10 147–154 [DOI] [PubMed] [Google Scholar]
- 5.Wittinghofer, A. (1998) Biol. Chem. 379 933–937 [PubMed] [Google Scholar]
- 6.Marshall, C. J. (1996) Curr Opin Cell Biol. 8 197–204 [DOI] [PubMed] [Google Scholar]
- 7.Boulton, T. G., Nye, S. H., Robbins, D. J., Ip, N. Y., Radziejewska, E., Morgenbesser, S. D., DePinho, R. A., Panayotatos, N., Cobb, M. H., and Yancopoulos, G. D. (1991) Cell 65 663–675 [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto, T., Yokote, K., Tamura, K., Takemoto, M., Ueno, H., Saito, Y., and Mori, S. (1999) J. Biol. Chem. 274 13954–13960 [DOI] [PubMed] [Google Scholar]
- 9.Minden, A., Lin, A., McMahon, M., Lange-Carter, C., Derijard, B., Davis, R. J., Johnson, G. L., and Karin, M. (1994) Science 266 1719–1723 [DOI] [PubMed] [Google Scholar]
- 10.Pandey, P., Raingeaud, J., Kaneki, M., Weichselbaum, R., Davis, R. J., Kufe, D., and Kharbanda, S. (1996) J. Biol. Chem. 271 23775–23779 [DOI] [PubMed] [Google Scholar]
- 11.Liu, Z. G., Baskaran, R., Lea-Chou, E. T., Wood, L. D., Chen, Y., Karin, M., and Wang, J. Y. (1996) Nature 384 273–276 [DOI] [PubMed] [Google Scholar]
- 12.Bode, A. M., and Dong, Z. (2003) Sci STKE 2003, RE2. [DOI] [PubMed]
- 13.Kim, M., Yan, Y., Kortum, R. L., Stoeger, S. M., Sgagias, M. K., Lee, K., Lewis, R. E., and Cowan, K. H. (2005) Cancer Res. 65 3986–3992 [DOI] [PubMed] [Google Scholar]
- 14.Chou, S. F., Chang, S. W., and Chuang, J. L. (2007) Investig. Ophthalmol. Vis. Sci. 48 2009–2016 [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. W., Fang, L., Igarashi, M., Ouchi, T., Lu, K. P., and Aaronson, S. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 8302–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dent, P., Yacoub, A., Fisher, P. B., Hagan, M. P., and Grant, S. (2003) Oncogene 22 5885–5896 [DOI] [PubMed] [Google Scholar]
- 17.Wu, D., Chen, B., Parihar, K., He, L., Fan, C., Zhang, J., Liu, L., Gillis, A., Bruce, A., Kapoor, A., and Tang, D. (2006) Oncogene 25 1153–1164 [DOI] [PubMed] [Google Scholar]
- 18.Tang, D., Wu, D., Hirao, A., Lahti, J. M., Liu, L., Mazza, B., Kidd, V. J., Mak, T. W., and Ingram, A. J. (2002) J. Biol. Chem. 277 12710–12717 [DOI] [PubMed] [Google Scholar]
- 19.Kortum, R. L., and Lewis, R. E. (2004) Mol. Cell. Biol. 24 4407–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kortum, R. L., Costanzo, D. L., Haferbier, J., Schreiner, S. J., Razidlo, G. L., Wu, M. H., Volle, D. J., Mori, T., Sakaue, H., Chaika, N. V., Chaika, O. V., and Lewis, R. E. (2005) Mol. Cell. Biol. 25 7592–7604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson, G., Robinson, F., Beers Gibson, T., Xu, B. E., Karandikar, M., Berman, K., and Cobb, M. H. (2001) Endocr. Rev. 22 153–183 [DOI] [PubMed] [Google Scholar]
- 22.Pages, G., Lenormand, P., L'Allemain, G., Chambard, J. C., Meloche, S., and Pouyssegur, J. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 8319–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meloche, S., and Pouyssegur, J. (2007) Oncogene 26 3227–3239 [DOI] [PubMed] [Google Scholar]
- 24.Yan, Y., Black, C. P., and Cowan, K. H. (2007) Oncogene 26 4689–4698 [DOI] [PubMed] [Google Scholar]
- 25.Abbott, D. W., and Holt, J. T. (1999) J. Biol. Chem. 274 2732–2742 [DOI] [PubMed] [Google Scholar]
- 26.Nguyen, A., Burack, W. R., Stock, J. L., Kortum, R., Chaika, O. V., Afkarian, M., Muller, W. J., Murphy, K. M., Morrison, D. K., Lewis, R. E., McNeish, J., and Shaw, A. S. (2002) Mol. Cell. Biol. 22 3035–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Therrien, M., Michaud, N. R., Rubin, G. M., and Morrison, D. K. (1996) Genes Dev. 10 2684–2695 [DOI] [PubMed] [Google Scholar]
- 28.Stewart, S., Sundaram, M., Zhang, Y., Lee, J., Han, M., and Guan, K. L. (1999) Mol. Cell. Biol. 19 5523–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu, W., Fantl, W. J., Harrowe, G., and Williams, L. T. (1998) Curr. Biol. 8 56–64 [DOI] [PubMed] [Google Scholar]
- 30.Muller, J., Cacace, A. M., Lyons, W. E., McGill, C. B., and Morrison, D. K. (2000) Mol. Cell. Biol. 20 5529–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kortum, R. L., Johnson, H. J., Costanzo, D. L., Volle, D. J., Razidlo, G. L., Fusello, A. M., Shaw, A. S., and Lewis, R. E. (2006) Mol. Cell. Biol. 26 2202–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozano, J., Xing, R., Cai, Z., Jensen, H. L., Trempus, C., Mark, W., Cannon, R., and Kolesnick, R. (2003) Cancer Res. 63 4232–4238 [PubMed] [Google Scholar]
- 33.Yan, F., John, S. K., and Polk, D. B. (2001) Cancer Res. 61 8668–8675 [PubMed] [Google Scholar]
- 34.Yan, F., John, S. K., Wilson, G., Jones, D. S., Washington, M. K., and Polk, D. B. (2004) J. Clin. Investig. 114 1272–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szatmari, E., Kalita, K. B., Kharebava, G., and Hetman, M. (2007) J. Neurosci. 27 11389–11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKay, M. M., and Morrison, D. K. (2007) J. Biol. Chem. 282 26225–26234 [DOI] [PubMed] [Google Scholar]
- 37.Dronkert, M. L., and Kanaar, R. (2001) Mutat. Res. 486 217–247 [DOI] [PubMed] [Google Scholar]
- 38.Noll, D. M., Mason, T. M., and Miller, P. S. (2006) Chem. Rev. 106 277–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreassen, P. R., D'Andrea, A. D., and Taniguchi, T. (2004) Genes Dev. 18 1958–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pichierri, P., and Rosselli, F. (2004) EMBO J. 23 1178–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng, Y., Forbes, K. C., Wu, Z., Moreno, S., Piwnica-Worms, H., and Enoch, T. (1998) Nature 395 507–510 [DOI] [PubMed] [Google Scholar]
- 42.Peng, C. Y., Graves, P. R., Thoma, R. S., Wu, Z., Shaw, A. S., and Piwnica-Worms, H. (1997) Science 277 1501–1505 [DOI] [PubMed] [Google Scholar]
- 43.Takizawa, C. G., and Morgan, D. O. (2000) Curr. Opin. Cell Biol. 12 658–665 [DOI] [PubMed] [Google Scholar]
- 44.Kuraoka, I., Kobertz, W. R., Ariza, R. R., Biggerstaff, M., Essigmann, J. M., and Wood, R. D. (2000) J. Biol. Chem. 275 26632–26636 [DOI] [PubMed] [Google Scholar]
- 45.Fisher, L. A., Bessho, M., and Bessho, T. (2008) J. Biol. Chem. 283 1275–1281 [DOI] [PubMed] [Google Scholar]
- 46.Cipak, L., Watanabe, N., and Bessho, T. (2006) Nat. Struct. Mol. Biol. 13 729–733 [DOI] [PubMed] [Google Scholar]
- 47.Niedernhofer, L. J., Odijk, H., Budzowska, M., van Drunen, E., Maas, A., Theil, A. F., de Wit, J., Jaspers, N. G., Beverloo, H. B., Hoeijmakers, J. H., and Kanaar, R. (2004) Mol. Cell. Biol. 24 5776–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson, M. J., Stippec, S. A., Goldsmith, E., White, M. A., and Cobb, M. H. (1998) Curr. Biol. 8 1141–1150 [DOI] [PubMed] [Google Scholar]
- 49.Muller, J., Ory, S., Copeland, T., Piwnica-Worms, H., and Morrison, D. K. (2001) Mol. Cell 8 983–993 [DOI] [PubMed] [Google Scholar]
- 50.Fenech, M., and Morley, A. A. (1985) Mutat. Res. 147 29–36 [DOI] [PubMed] [Google Scholar]
- 51.Stiff, T., Reis, C., Alderton, G. K., Woodbine, L., O'Driscoll, M., and Jeggo, P. A. (2005) EMBO J. 24 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Setlow, R. B. (1966) Science 153 379–386 [DOI] [PubMed] [Google Scholar]
- 53.Roots, R., Kraft, G., and Gosschalk, E. (1985) Int. J. Radiat. Oncol. Biol. Phys. 11 259–265 [DOI] [PubMed] [Google Scholar]
- 54.Kano, Y., and Fujiwara, Y. (1981) Mutat. Res. 81 365–375 [DOI] [PubMed] [Google Scholar]
- 55.Zhao, H., and Piwnica-Worms, H. (2001) Mol. Cell. Biol. 21 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanchez, Y., Wong, C., Thoma, R. S., Richman, R., Wu, Z., Piwnica-Worms, H., and Elledge, S. J. (1997) Science 277 1497–1501 [DOI] [PubMed] [Google Scholar]
- 57.Redon, C., Pilch, D., Rogakou, E., Sedelnikova, O., Newrock, K., and Bonner, W. (2002) Curr. Opin. Genet. Dev. 12 162–169 [DOI] [PubMed] [Google Scholar]
- 58.Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S., and Bonner, W. M. (1998) J. Biol. Chem. 273 5858–5868 [DOI] [PubMed] [Google Scholar]
- 59.Akkari, Y. M., Bateman, R. L., Reifsteck, C. A., Olson, S. B., and Grompe, M. (2000) Mol. Cell. Biol. 20 8283–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Silva, I. U., McHugh, P. J., Clingen, P. H., and Hartley, J. A. (2000) Mol. Cell. Biol. 20 7980–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rothfuss, A., and Grompe, M. (2004) Mol. Cell. Biol. 24 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siddik, Z. H. (2003) Oncogene 22 7265–7279 [DOI] [PubMed] [Google Scholar]
- 63.Tomasz, M. (1995) Chem. Biol. 2 575–579 [DOI] [PubMed] [Google Scholar]
- 64.Friedberg, E. C., Graham, G. C., Siede, W., Wood, R. D., Schultz, R. A., and Ellenberger, T. (2005) DNA Repair and Mutagenesis, ASM Press, Washington, D.C.
- 65.Smits, V. A., and Medema, R. H. (2001) Biochim. Biophys. Acta 1519 1–12 [DOI] [PubMed] [Google Scholar]
- 66.Atherton-Fessler, S., Parker, L. L., Geahlen, R. L., and Piwnica-Worms, H. (1993) Mol. Cell. Biol. 13 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lundgren, K., Walworth, N., Booher, R., Dembski, M., Kirschner, M., and Beach, D. (1991) Cell 64 1111–1122 [DOI] [PubMed] [Google Scholar]
- 68.Parker, L. L., Atherton-Fessler, S., and Piwnica-Worms, H. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 2917–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dunphy, W. G. (1994) Trends Cell Biol. 4 202–207 [DOI] [PubMed] [Google Scholar]
- 70.Wang, R., He, G., Nelman-Gonzalez, M., Ashorn, C. L., Gallick, G. E., Stukenberg, P. T., Kirschner, M. W., and Kuang, J. (2007) Cell 128 1119–1132 [DOI] [PubMed] [Google Scholar]
- 71.Shibuya, E. K., Boulton, T. G., Cobb, M. H., and Ruderman, J. V. (1992) EMBO J. 11 3963–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matheny, S. A., Chen, C., Kortum, R. L., Razidlo, G. L., Lewis, R. E., and White, M. A. (2004) Nature 427 256–260 [DOI] [PubMed] [Google Scholar]
- 73.Li, S., Ku, C. Y., Farmer, A. A., Cong, Y. S., Chen, C. F., and Lee, W. H. (1998) J. Biol. Chem. 273 6183–6189 [DOI] [PubMed] [Google Scholar]
- 74.Venkitaraman, A. R. (2002) Cell 108 171–182 [DOI] [PubMed] [Google Scholar]
- 75.Yan, Y., Black, C. P., Cao, P. T., Haferbier, J. L., Kolb, R. H., Spieker, R. S., Ristow, A. M., and Cowan, K. H. (2008) Cancer Res. 68 5113–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peng, C. Y., Graves, P. R., Ogg, S., Thoma, R. S., Byrnes, M. J., 3rd, Wu, Z., Stephenson, M. T., and Piwnica-Worms, H. (1998) Cell Growth Differ. 9 197–208 [PubMed] [Google Scholar]