Abstract
The ergot fungus Claviceps purpurea produces both ergopeptines and simple d-lysergic acid alkylamides. In the ergopeptines, such as ergotamine, d-lysergic acid is linked to a bicyclic tripeptide in amide-like fashion, whereas in the d-lysergylalkanolamides it is linked to an amino alcohol derived from alanine. We show here that these compound classes are synthesized by a set of three non-ribosomal lysergyl peptide synthetases (LPSs), which interact in a combinatorial fashion for synthesis of the relevant product. The trimodular LPS1 assembles with LPS2, the d-lysergic acid recruiting module, to synthesize the d-lysergyltripeptide precursors of ergopeptines from d-lysergic acid and the three amino acids of the peptide chain. Alternatively, LPS2 can assemble with a distinct monomodular non-ribosomal peptide synthetase (NRPS) subunit (ergometrine synthetase) to synthesize the d-lysergic acid alkanolamide ergometrine from d-lysergic acid and alanine. The synthesis proceeds via covalently bound d-lysergyl alanine and release of dipeptide as alcohol with consumption of NADPH. Enzymatic and immunochemical analyses showed that ergometrine synthetase is most probably the enzyme LPS3 whose gene had been identified previously as part of the ergot alkaloid biosynthesis gene cluster in C. purpurea. Inspections of all LPS sequences showed no recognizable peptide linkers for their protein-protein interactions as in NRPS subunits of bacteria. Instead, they all carry conserved N-terminal domains (C0-domains) with similarity to the C-terminal halves of NRPS condensation domains pointing to an alternative mechanism of subunit-subunit interactions in fungal NRPS systems. Phylogenetic analysis of LPS modules and the C0-domains suggests that these enzyme systems most probably evolved by module duplications and rearrangements from a bimodular ancestor.
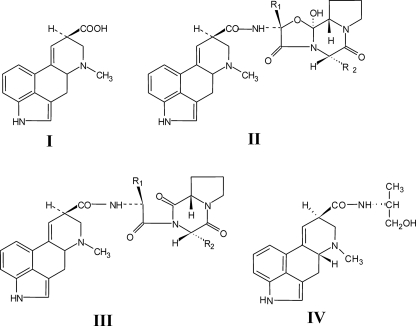
d-Lysergic acid (see Fig. 1, I) is the pharmacophore of the various amide/peptide-type ergot alkaloids. In these compounds, the carboxyl group of d-lysergic acid is amidated with simple amino alcohols or small peptide chains, which, depending on their structures, confer the tetracylic methylergolene skeleton of d-lysergic acid similarity to different neurotransmitters such as dopamine, serotonine, or adrenaline (1). Several d-lysergic acid alkaloids or synthetic derivatives are used in the treatment of a number of disorders in the vascular and central nervous systems (2, 3). The natural d-lysergic acid amides are produced by a wide variety of ascomycete fungi, mostly belonging to the family Clavicipitaceae (4). Most prominent among these is Claviceps purpurea, which grows on cereals and forms there the sclerotia known as ergot, which has long been the main source of these compounds (5). Remarkably, d-lysergic acid rarely occurs in free form in these fungi and, like its biosynthetic precursors, the clavine alkaloids agroclavine and elymoclavine, has little biological activity (6).
FIGURE 1.
Structures of d-lysergic acid peptide alkaloids. I, d-lysergic acid; II, general structure of ergopeptines; III, general structure of ergopeptams; and IV, structure of ergometrine (synonymous with ergonovine and ergobasine).
In the ergopeptines (see Fig. 1, II) d-lysergic acid is amidated with bicyclic tripeptide chains. The first two amino acid positions of the tripeptide chains are normally occupied by non-polar amino acids. The third amino acid is nearly always proline. Biosynthetically, the ergopeptines are derived from the l-ergopeptams (d-lysergyl peptide lactams, see Fig. 1, III), structurally identical to ergopeptines except that the cyclol bridge between the α-C of the first amino acid and the carboxyl group of proline is missing (7). l-Ergopeptams have, like the ergopeptines, l-proline, whereas the d-ergopeptams, which in ergot fungi sometimes occur as co-products with ergopeptines, have d-proline. d-Ergopeptams are considered as dead-end products of the alkaloid peptide pathway arising from spontaneous isomerization of l-ergopeptams prior to their conversion to ergopeptines, the regular end products of the alkaloid pathway (7).
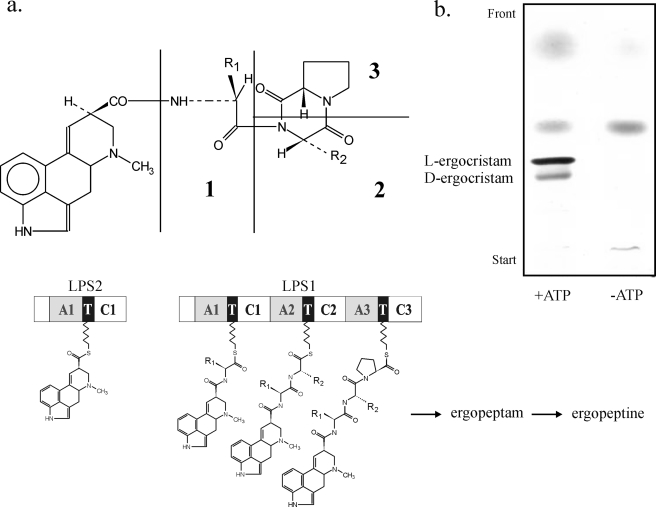
The l-ergopeptams are assembled non-ribosomally from d-lysergic acid and the amino acids of the peptide chain by the lysergyl peptide synthetases (LPS1 and LPS2)2 (8–10). The monomodular LPS2 recruits d-lysergic acid, the formal N terminus of the chain, as a thioester. d-Lysergic acid is then progressively elongated to the d-lysergyl-mono-, -di-, and -tripeptides by the trimodular LPS1 (see Fig. 2a). Enzyme-bound d-lysergyl tripeptide is finally released as l-ergopeptam. LPS1 and LPS2 have sizes of 370 and 141 kDa, respectively, and are a rare example of fungal NRPS systems composed of subunits (9). Their genes have been sequenced and analyzed (11, 12). Inspection of other ergot fungi-producing ergopeptines by DNA sequencing has revealed both LPS1 and LPS2 gene orthologues (designated lpsA and lpsB) in the respective genomes and also that in C. purpurea the lpsA gene may occur in duplicate (13–16). Both lpsAs and lpsBs have been found in the ergot fungi so far analyzed to be co-localized with most if not all genes encoding the steps of clavine alkaloid biosynthesis, including the conversion of elymoclavine to d-lysergic acid forming an ergot alkaloid biosynthesis cluster (11, 15, 17, 18).
FIGURE 2.
a, NRPS assembly line of the l-ergopeptams (ergopeptines). Ergopeptams have the general structure given in the structural formula above. In ergotamam, R1 is methyl and R2 is benzyl; in ergocristam R1 is isopropyl and R2 is benzyl; in ergocryptam R1 is isopropyl and R2 is isobutyl. Amino acid positions 1, 2, and 3 in the ergopeptam structure correspond to modules A1TC1 (M1), A2TC2 (M2), and A3TC3 (M3) of LPS1. The free-standing d-lysergic acid module is LPS2. b, ergocristam synthesis catalyzed by LPS1/LPS2 in crude extracts from C. purpurea strain Ecc93. The lanes show TLC separation on silica gel of extracts from reaction mixtures containing d-lysergic acid, [U-14C]valine, phenylalanine, and proline, with and without MgATP. Authenticity of reaction products was checked by radiochemical analysis as described previously (9). The solvent system was solvent system I.
In the simple d-lysergic acid amides such as ergometrine (synonymous with ergonovine and ergobasine) (Fig. 1, IV) and d-lysergic acid hydroxyethylamide, d-lysergic acid is amidated with alaninol or ethanolamine, respectively. Ergometrine has a pronounced uterotonic effect compared with ergotamine as revealed by the finding that pure ergotamine acted more slowly than when aqueous extracts of ergot were administered (19). In field sclerotia of C. purpurea, the non-polar ergopeptines are always accompanied by these simple water-soluble d-lysergic acid alkylamides, indicating that the fungus augments the toxic effects by producing two different classes of d-lysergic acid alkaloids characterized by short and longer acting effects on the central nervous and vascular system (20). In contrast to the ergopeptines, however, it was unknown how d-lysergic acid is assembled into that second class of ergot alkaloids represented by ergometrine nor which enzymes are involved.
EXPERIMENTAL PROCEDURES
Strains and Cultures—Cultivation of strain Ecc93 of C. purpurea was described previously (8, 9). It produces mainly ergocristine with minor amounts of ergosine, ergotamine, and ergometrine (together with other low molecular weight alkaloids). Strain Ecc93 was routinely selected for maintenance of alkaloid high production as described previously (21).
Chemicals and Radiochemicals—d-Lysergic acid was prepared by alkaline hydrolysis of ergotamine. Ergometrine was kindly donated by Dr. J. J. Sanglier, Novartis AG, Basel (Switzerland). Dihydrolysergic acid and dihydroergometrine were obtained by catalytic hydrogenation over palladium on charcoal of d-lysergic acid and ergometrine, respectively. Dihydrolysergylalanine was synthesized as described (10). [9,10-3H]Dihydroergocryptine (17.5 Ci/mmol) was from Hartmann Analytics, Braunschweig (Germany). l-[U-14C]Alanine (148 mCi/mmol), l-[U-14C]valine (256 mCi/mmol) and l-[U-14C]phenylalanine (460 mCi/mmol) were from Amersham Biosciences. [9,10-3H]Dihydrolysergic acid was prepared from [9,10-3H]dihydroergocryptine as described previously (8).
Nucleic Acid Analysis—Extraction of chromosomal DNA of C. purpurea was with the PeqGold Fungal DNA Mini Kit according to the manufacturer's instructions (Peqlab, Erlangen, Germany). For PCR analysis, TaqDNA polymerase was used (Peqlab). Plasmid for subcloning of PCR fragments for sequencing was pCR4-TOPO (Invitrogen) and for expression of lpsCred was pQE30UA (Qiagen). Oligonucleotides for PCR amplification of portions of lpsC from C. purpurea Ecc93 were: IO_lps3red_for, 5′-GTGTTTGTGACAGGCGCCAGCGGATT; IO_lps3red_rev, 5′-CGGTTAAAGGATCCCCTATTACGAGGCC; IO_lps3C_for, 5′-CGAACACGCATCATTGTCTGCGC; IO_lps3C_rev, 5′-CAAGTGACCACACGAATACCCAGCC; IO_lps3A_for, 5′-GGATAACCCCTATGAATGATCGACCG; IO_lps3A_rev, 5′-CCTTTGAGAGAATGCTATGGAGGTTGG; IO_lps3Nterm_for, 5′-GTTTTATATGAGAACCAAGGTCCC; and IO_lps3Nterm_rev, 5′-GCAGACCTTCCCAGATAACACG. DNA sequencing was performed by AGOWA (Berlin, Germany). Sequences and multiple alignments were computed and arranged with ClustalW (22) and Genedoc (23).
Cloning and Expression of lpsCred—The PCR fragment encoding a 192-amino acid-long region of the reductase domain of lpsC (accession number AJ884677) was obtained by amplifying a DNA fragment between nucleotides 3785 and 4361 of the lpsC gene using primers LPS3red_for/rev and genomic DNA from C. purpurea Ecc93 as template. The fragment was ligated by TOPO-cloning into vector pQE30UA (Qiagen), resulting in LpsCred_pQE30UA. Heterologous expression of plasmid LpsCred_pQE30UA in Escherichia coli M15 was in 1 liter of LB medium, 100 μg/ml ampicillin, and 25 μg/ml kanamycin at 37 °C. At an A600 of 0.6, induction was with 1 mm isopropyl-1-thio-β-d-galactopyranoside. Cells were harvested after further 4 h of incubation at 37 °C. Expression of LpsCred_pQE30UA resulted in the formation of inclusion bodies. For purification of LpsCred from E. coli M15 transformed with plasmid cpps3red_pQE30UA, 6 g of cells was resuspended in 0.05 m phosphate buffer (pH 8.0) containing 0.5 mg/ml lysozyme and passed through a French Press cell at 8,000 p.s.i. After incubation in the presence of 10 mm MgCl2 and 20 μg ml–1 DNase I (grade II, Sigma) for 30 min at room temperature, the suspension was centrifuged at 1500 × g for 30 min to separate the insoluble cellular fraction containing inclusion bodies. The collected fraction of inclusion bodies was resuspended in 0.05 m phosphate buffer (pH 8.0) containing 1% Triton X-100, disintegrated by ultrasonification (3 × 30 s, 50 watts) and centrifuged as before. The inclusion bodies were precipitated, and the procedure was repeated two times with buffers containing 0.5% Triton X-100 and finally buffer without Triton X-100. The inclusion bodies were dissolved in 0.05 m phosphate buffer (pH 8.0) containing 8 m urea and 4 mm dithiothreitol, and the resultant solution was centrifuged at 20,000 × g for 30 min. The clear supernatant containing denatured protein was dialyzed against 0.05 m phosphate buffer (pH 8.0) at 4 °C for 24 h, with two changes of buffer. The resulting suspension was centrifuged at 20,000 × g for 30 min. The supernatant containing the renatured protein was concentrated by ultrafiltration with Centricon YM10 membranes (Millipore, Schwalbach, Germany) and further purified by preparative SDS-PAGE (15%, 3-mm gel thickness) by the Laemmli method. Protein bands containing the recombinant protein were cut out from the gel and minced into small pieces for electroelution of protein overnight using a Biotrap BT 1000 (Schleicher & Schüll, Dassel, Germany) in 0.1 m Tris-HCl, 0.1% SDS, pH 8.0. Eluted protein was concentrated in a Centricon 30 microconcentrator (Amicon) to the desired concentration.
Methods of Analysis—Techniques for amino acid analysis, radiomeasurements, and radiochemical analysis of enzymatically formed peptides have been described elsewhere (8, 9). TLC solvent systems for amino acid analysis on Silica Gel 60 plates (Merck, Darmstadt, Germany) were isopropanol:acetic acid:water (7:3:2, by vol.) or butanol:acetic acid:water (4:2:2, by vol.). For separation of d-lysergyl peptides on silica gel TLC plates solvent systems were: I, ethyl acetate:methanol:water:dimethylformamide (90:5:5:0.5, by vol.); II, ethyl acetate:methanol:water:dimethylformamide (84:10:5:1, by vol.); III, ethyl acetate:propanol:methanol:water (2:2:2:1, by vol.); and IV, ethanol:water (75:25, by vol.). Protein blotting was performed by using the Fast Blot B34 semi-dry system (Biometra Göttingen, Germany) and Immobilon-P transfer membrane (Millipore) with a current density of 2.5 A/cm2 for 60 min. Washing and incubation procedures were according to standard protocols. Serum against the Red domain of LPS3 was obtained by immunization of a rabbit with LpsCRed (performed at Biogenes GmbH, Berlin). For Western blotting analysis, serum was used in 1:500 dilution. Secondary antibody was monoclonal phosphatase-conjugated anti-rabbit antibody (Sigma) at 1:20,000 dilution. Visualization of immunoreactive bands was with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate treatment at concentrations of 0.25 mg ml–1 and 0.15 mg ml–1, respectively.
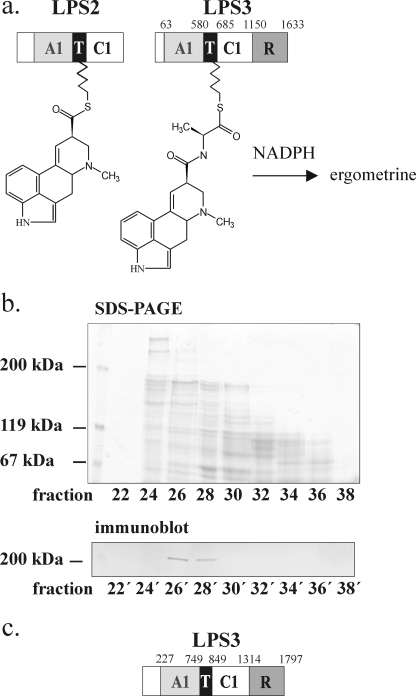
Enzyme Purification—All operations were performed at 4 °C. Generally, freshly harvested mycelium from 8-day-old cultures of strain Ecc93 was suspended in a 3- to 4-fold volume of buffer A (9) and passed twice through a French Press cell at 10,000 p.s.i. The homogenate was centrifuged at 20,000 × g for 15 min. The supernatant was cleared by the addition of polymin P (0.1% final concentration). After centrifugation as above, to the supernatant was added ammonium sulfate (55% final concentration) and left on ice overnight. The precipitate was collected by centrifugation at 20,000 × g for 30 min, and pellets were taken up in buffer B (9) at final protein concentration between 15 to 20 mg ml–1. Prior to use, the protein extract was desalted into buffer B by using PD10 columns (Amersham Biosciences). For further fractionation, enzyme was concentrated by adsorption to DEAE-cellulose pre-equilibrated with buffer B and subsequent elution with 0.15 m NaCl in buffer B. 2-ml portions of such concentrated enzyme (50 mg ml–1 protein) were passed through a Superdex TM200 gel filtration column (Amersham Biosciences) previously equilibrated with buffer B at a flow rate of 1 ml min–1. 1.5-ml fractions were collected. Enzymes were localized by their ability to form thioester with radioactive substrates as [3H]dihydrolysergic acid (LPS2), [14C]alanine (LPS3), or [14C]phenylalanine (LPS1).
Enzyme Assays—Enzyme thioester formations of dihydrolysergic acid or amino acids were measured as described (9). For in vitro synthesis of ergometrine or ergopeptams, ammonium sulfate-precipitated (55% saturation) fractions desalted into buffer B on Sephadex PD10 gel filtration columns (Amersham Biosciences) were routinely used at final concentrations of 10–12 mg ml–1 protein. Standard assays contained 200 μl of extract with 2.0 μCi of [14C]alanine, 15 mm ATP, 2 mm NADPH, 20 mm MgCl2, and 0.25 mm d-lysergic acid or dihydrolysergic acid in a total volume of 200 μl (substrate mixture for each assay had been freeze-dried before and kept at –80 °C prior to use). In case of kinetic analyses, alanine concentrations were adjusted by addition of labeled or non-labeled alanine when necessary. For in vitro synthesis of ergopeptines, 200 μl (unless stated otherwise) of enzyme solution was incubated for the indicated times at 26 °C with 2 μCi of the [14C]-amino acid chosen as radiolabel, 2.5 mm of the other substrate amino acids, 15 mm ATP, 20 mm MgCl2, and 0.25 mm d-lysergic acid or dihydrolysergic acid. In case of labeling with [3H]dihydrolysergic acid, 5 μCi was used. Reactions were stopped by adding 1 ml of water and extracted twice with 2.5 ml of ethyl acetate. Combined extracts were evaporated and applied to TLC plates and developed in appropriate solvent systems (solvent system I for ergopeptams; II for ergometrine). Radioactive products were detected by radioscanning or autoradiography. For quantitation, radioactive zones were cut out from plates and counted in a liquid scintillation counter. LPS2- and LPS3-associated activities were quantitated by titration with [14C]phenylalanine or [14C]alanine, respectively, and subsequent determination of acid stable radioactivity as described (10). One unit of LPS is the amount of enzyme that binds 1 pmol of the relevant substrate in a 30-min reaction at 30 °C (in buffer B). For the analysis of enzyme-bound reaction intermediates of ergometrine synthesis, reaction incubations (in each case performed in triplicate and combined after incubation for 20 min) were precipitated with 3 ml of trichloroacetic acid (10% w/v) and left on ice for 1 h. After centrifugation and repeated washings with trichloroacetic acid and finally with EtOH, the protein pellet was dissolved in 1 ml of 1 m NaOH and left for 20 min at 37 °C. After neutralization to pH 7 with 1 m HCl, protein was removed by centrifugation, and the aqueous phase was subjected to solid-phase extraction on IFSORB as described previously (10). Radioactivity was eluted from the solid phase with methanol and after concentration subjected to TLC on silica gel using solvent systems II, III, and IV.
RESULTS AND DISCUSSION
A Cell-free System of Ergopeptam and Ergometrine Biosynthesis—To analyze the enzymatic basis in C. purpurea for the production of the two diverse d-lysergic acid alkaloid classes, a cell-free system catalyzing both ergopeptine and ergometrine synthesis was established. C. purpurea strain Ecc93 produces ergopeptines (ergocristine and ergotamine) as main components and low molecular weight ergot alkaloids, including ergometrine, as minor components of the alkaloid mixture. Ergometrine was present at 2–3% (w/w) of the total alkaloid (300 mg/liter) after 10 days of cultivation. Partially purified extracts from broken cells of strain Ecc93 showed LPS1- and LPS2-associated activity as revealed by the in vitro formation of l-ergocristam (d-lysergylvalylcyclo(phenylalanylproline) together with minor amounts of d-ergocristam (Fig. 2b) at a kcat of ∼1.2–1.4 min–1 (8–11 units/mg of protein). comparable to that of l-ergotamam synthesis catalyzed by LPSs from C. purpurea strain D1 (10). The quantitation of total LPS1-type enzyme was by measurement of capacity of enzyme fraction to catalyze formation of acid-stable [14C]phenylalanine-enzyme thioester from enzyme and [14C]phenylalanine in the presence of MgATP. Ergotamine is a minor component (15–25%) of the ergopeptine mixture elaborated by strain Ecc93. Accordingly, cell-free l-ergotamam (d-lysergylalanylcyclo(phenylalanylproline)) formation was also detectable in the protein extract, albeit at considerably lower kcat (∼0.12–0.2 min–1; not shown), which reflects either side specificity of the enzyme-catalyzing ergocristam synthesis or indicates a second LPS1 species catalyzing l-ergotamam formation present at a much lower level than that catalyzing l-ergocristam.
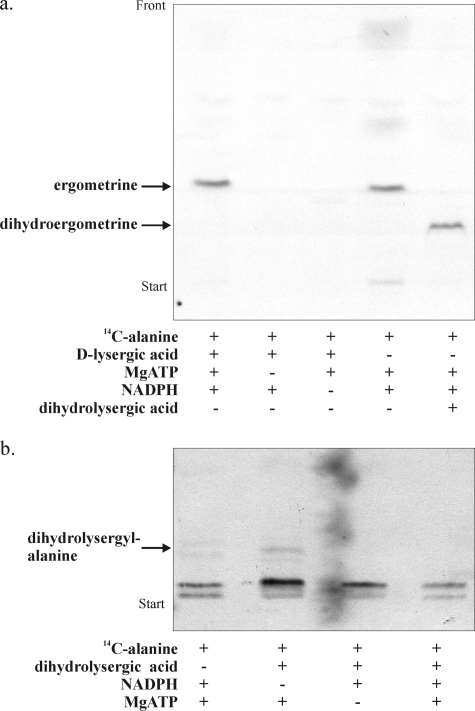
The same enzyme extract from strain Ecc93 catalyzing ergopeptam synthesis was incubated with d-lysergic acid, MgATP, and radioactive alanine in the presence of reducing cofactors such as NADH or NADPH. Earlier work (24–26) had suggested alanine as the precursor of the alaninol moiety of ergometrine (Fig. 1, IV), which pointed to a step requiring reducing cofactors for conversion of the carboxyl group to a hydroxymethyl group. The analysis of reaction products by TLC indeed revealed formation of a radioactive compound dependent on MgATP and NADPH (Fig. 3a). NADH was not a substrate (not shown). Because partial purified enzyme fractions from C. purpurea usually contain traces of d-lysergic acid from the fungal cells, which sticks to protein, the dependence of compound formation on d-lysergic acid initially was equivocal. However, when the same incubations contained dihydrolysergic acid instead of d-lysergic acid (i.e. in excess over endogenous d-lysergic acid traces), a compound with lower Rf value was detected, in agreement with the lower Rf value of dihydrolysergic acid in TLCs compared with d-lysergic acid (7). Acid hydrolysis of the two compounds yielded radioactive alaninol in each case. To confirm the lower migrating compound as dihydroergometrine, [3H]dihydrolysergic acid was incubated in the presence of non-labeled alanine (not shown). Alkaline hydrolysis of the compound in this case gave radioactive dihydrolysergic acid. Moreover, TLC cochromatography of the two sister compounds with authentic ergometrine or dihydroergometrine in solvent systems I, II, and III showed that they had the same Rf values as these standards. The enzyme activity in the extract catalyzing ergometrine synthesis was termed ergometrine synthetase.
FIGURE 3.
a, ergometrine synthesis catalyzed by ergometrine synthetase in crude extract from C. purpurea strain Ecc93. The lanes show TLC separation on silica gel of extracts from reaction mixtures containing d-lysergic acid (or dihydrolysergic acid), [U-14C]alanine, MgATP, and NADPH. Each lane represents an experiment with the addition or omission of the indicated substrate. The solvent system was solvent system II. b, analysis of enzyme-bound reaction intermediates of ergometrine synthesis bound to ergometrine synthetase. The lanes show TLC separation on a silica gel of radioactive material that was split off ergometrine synthetase when incubated with different reaction mixtures. These mixtures contained dihydrolysergic acid, alanine, MgATP, and NADPH. Each lane represents an experiment with the addition or omission of the indicated substrate. [U-14C]Alanine was used as radiolabel in each experiment. The solvent system was solvent system III. Note: covalently bound reaction product is only visible in the case of NADPH. Authentic dihydrolysergylalanine (10) was used as the chromatographic standard. The shadow on the autoradiogram is from a radioactive contamination in the screen.
Mechanism of Ergometrine Synthesis—Reaction velocity measurements showed that the reaction catalyzed by the ergometrine synthetase proceeded for up to 60 min, with linearity for the first 10 min after which it gradually declined. Half-life time of enzyme activity on ice was 6 h. The reaction rate depended on protein concentration in a strictly linear manner over a concentration range of more than one order of magnitude (0.5–10 mg ml–1). The Km values were 123 ± 10 μm for alanine and 2–3 μm for dihydrolysergic acid and thus comparable to the values obtained previously in the case of ergotamam synthesis (9). Km for NADPH was 8.7 ± 1 μm. kcat of the ergometrine producing reaction was 1.3–1.4 min–1 calculated from enzyme units obtained from measurements of [14C]alanine-enzyme thioester formation (ranging between 0.8 and 1.6 unit/mg of protein, dependent on the quality of enzyme preparation) under the assumption that the NRPS responsible for ergometrine synthesis has one binding site for alanine. Thus, ergometrine synthetase units usually were present about one order of magnitude less than LPS1 units in agreement with the fact that ergometrine is a minor component of the alkaloid mixture elaborated by C. purpurea Ecc93.
To address the formation of covalently bound intermediates during ergometrine synthesis, enzyme-substrate complexes obtained from different incubations with [14C]alanine in which dihydrolysergic acid, NADPH, or ATP each that had been omitted were saponified with NaOH, and radioactive material released from the enzyme was analyzed by TLC. As was to be expected, no radioactive compounds were found when dihydrolysergic acid or ATP was omitted from incubation mixtures (Fig. 3b). By contrast, in case when NADPH was absent, a compound was split off the enzyme suggesting that NADPH plays a role in release of a covalently bound intermediate during ergometrine synthesis. Accordingly, when all substrates of the ergometrine reaction were present again no covalent intermediate was obtained, whereas ergometrine was formed (the latter not shown). This confirms the essential role of NADPH in the turnover of enzyme. The radioactive compound that accumulated in the absence of NADPH on the enzyme was identified as dihydrolysergylalanine by TLC comparison with chemically synthesized dihydrolysergylalanine in solvent systems II, III, and IV. The intermediacy of d-lysergylalanine as a covalently bound intermediate in ergometrine synthesis was also confirmed by the observation that addition of non-labeled alaninol in up to 30-fold molar excess over radiolabeled alanine did not suppress the incorporation of radiolabel into ergometrine, whereas non-labeled alanine did (not shown). This finding clearly excludes that free alaninol is involved in the formation of ergometrine. Moreover, the presence of dihydrolysergylalanine as a covalently bound intermediate explains previous reports in which it was noted that free d-lysergyl-alanine was not incorporated into ergometrine in vivo and in vitro (26).
Substrate Specificity of Ergometrine Synthetase—The substrate specificity of ergometrine synthetase was strict for alanine. Attempts to synthesize enzymatically structural analogs of ergometrine, e.g. dihydromethylergometrine from [3H]dihydrolysergic acid and aminobutyric acid, failed. Other amino acids structurally related to alanine, some of which in vitro can occupy the first modules of LPS1-type enzymes such as serine, valine, norvaline, leucine, norleucine, or 2-amino-n-octanoic acid (Walzel (47)), were also not incorporated into new dihydrolysergic acid alkanolamides. Similarly, none of these amino acids could significantly suppress incorporation of radiolabeled alanine into ergometrine at a 30-fold molar excess. Non-labeled alanine, of course, suppressed [14C]alanine incorporation by 95% in the same molar ratio. These data indicate a rather strict substrate specificity of ergometrine synthetase for its substrate alanine and possibly explain why ergometrine besides the closely related d-lysergic acid α-hydroxyethylamide is the sole member of this alkaloid class.
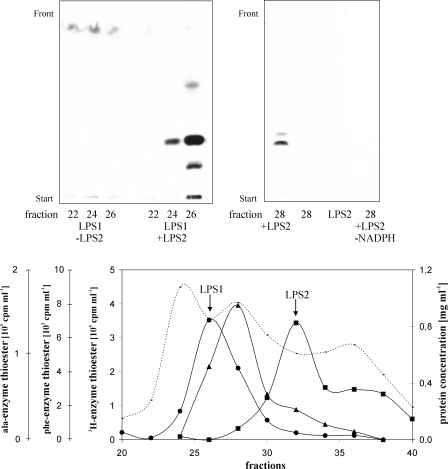
LPS2 Is the d-Lysergic Acid-activating Subunit of Ergometrine Synthetase—To identify the enzyme components of ergometrine formation, the protein extract catalyzing synthesis of both ergometrine and ergopeptam was fractionated on a Superdex TM200 gel filtration column (see Fig. 4). Resulting fractions were assayed for the presence of NRPS subunits by enzyme thioester formation with radioactive phenylalanine (for LPS1), radioactive dihydrolysergic acid (for LPS2 or the hypothetical d-lysergic acid-activating component of ergometrine synthetase) and radioactive alanine (alanine activation for ergometrine synthetase). Three distinct activity peaks appeared at molecular weight ranges of the column of ∼400 kDa (phenylalanine), 200 kDa (alanine), and 150 kDa (d-lysergic acid). Enzyme assays for ergotamam and ergometrine synthesis were performed by combining fractions from each peak with fractions from the other two peaks. Clearly, as expected, the peak in the 400-kDa fraction range representing LPS1 complemented with the 150-kDa (LPS2) peak fraction in synthesizing d-ergotamam (Fig. 4, upper left panel). Strikingly, the fractions containing LPS2 also efficiently complemented the alanine-activating enzyme peak in synthesizing ergometrine (Fig. 4, upper right panel), indicating that the alanine peak (200 kDa) obviously contained all functions for adenylation, thioesterification of alanine, and, importantly, reduction of d-lysergylalanine to ergometrine (ergometrine synthetase). All other combinations between activity peaks failed to catalyze any formation of d-lysergic acid alkaloids. This clearly showed that LPS2 is the d-lysergic acid-activating component not only for ergopeptam synthesis but also for the synthesis of simple d-lysergic acid alkaloids. These data also unequivocally reveal that the trimodular LPS1 plays no significant role in the synthesis of d-lysergic acid alkylamides, in contrast to previous reports (18).
FIGURE 4.
Fractionation and complementation analysis of LPS enzymes of C. purpurea strain Ecc93 by gel filtration on Superdex TM200. Ammonium sulfate- and DEAE-cellulose-fractionated enzyme extract was applied onto a Superdex TM200 (Amersham Biosciences) column, and fractions were assayed for enzyme-thioester formation of LPS1 (with [U-14C]phenylalanine (•)), LPS2 (with [3H]dihydrolysergic acid (▪)), and ergometrine synthetase (with [U-14C]alanine (▴)). Each peak was tested for complementation with the others by incubating 150 μl of each peak fractions of LPS1, LPS2, and ergometrine synthetase in standard conditions, except that the volume was 350 μl instead of 200 μl. Label for ergotamam synthesis was [U-14C]phenylalanine, for ergometrine synthesis [U-14C]alanine. All reaction products were chromatographed on silica gel plates in solvent system II (upper panels). Note, that in the conditions used only d-ergotamam is formed due to the presence of 6 mm dithiocrythritol in the flow buffer, which was not desalted prior to incubation.
LPS3, Encoded by lpsC, Is the Alanine-activating Subunit of Ergometrine Synthetase—Recently, continued sequencing (16) of the ergot alkaloid biosynthesis gene cluster in C. purpurea strain P1 revealed, besides a second lpsA copy (cpps4), also the additional NRPS gene lpsC encoding a protein of a calculated mass of 178.5 kDa. This protein, named LPS3, encodes a free-standing module composed of adenylation (A-), thiolation (T-), condensation (C-), and, interestingly enough, a C-terminal Red-domain (R-domain, oxidoreductase or reduction domain) with homology to the C-terminal Red-domains of other NRPS systems (27, 28) catalyzing reductive release of their products (Fig. 5a). The Red-domain was seen for the first time as a release domain in the aminoadipate reductase Lys2 of yeast and other fungi (29, 30). The amino acid sequence of the LPS3 Red-domain has highest identity (40%) to the Red-domain of the peramin synthetase PerA of the endophytes Epichloe and Neotyphodium (31). These fungi are also d-lysergic acid alkaloid producers (13). As in LPS3 in PerA the Red-domain lies at the C-terminal end of the enzyme and is claimed to catalyze release of the product peramin (31). The amino acid sequence of the NRPS portion of LPS3 (A-, T-, and C-domain), has highest similarity to the sequences of the first and third modules of the known LPS1-type enzymes. The identities were 47 and 31%, respectively, in the case of Neotyphoideum and in the case of LPS4 from C. purpurea they were 42 and 38%, respectively. By contrast, the second modules of LPS1-type enzymes were less similar to LPS3 (see below). In view of its calculated size of 178.5 kDa and of the presence of the Red-domain at its C-terminal end, LPS3 could be a good candidate for catalyzing ergometrine synthesis in C. purpurea Ecc93 as shown in Fig. 5a. However, C. purpurea strain P1 and its relatives were not found to produce ergometrine, which left the role of the protein questionable. To determine whether the ergometrine-synthesizing C. purpurea strain Ecc93 contained the LPS3 gene homologue (lpsC), PCR analysis with various primer pairs derived from the lpsC sequence of strain C. purpurea P1 was performed using C. purpurea Ecc93 chromosomal DNA as template. Sequence analysis of amplified portions of lpsC encoding the Red-domain and the A-, C-, and T-domains (deposited as part of this work under GenBank™ EU730584) indicated that lpsC of strain Ecc93 is ∼99% identical to the lpsC sequence from strain P1. To address the existence of LPS3 as a protein in C. purpurea strain Ecc93, antibodies against the Red-domain were raised in rabbits with antigen obtained by expression of part of the Red-domain encoding region of the lpsC homologue from Ecc93, named lpsCred, in E. coli (see “Experimental Procedures”). Immunoblot analysis of the enzyme fraction of the gel filtration separations from Fig. 4 containing the LPS enzymes with the serum obtained showed a single band of 195–200 kDa present only in the main fractions of the ergometrine synthetase peak, leaving little doubt that the alanine-activating subunit of ergometrine synthetase is LPS3 (Fig. 5b).
FIGURE 5.
Identification of LPS3 from C. purpurea Ecc93 and proof of its structure. a, structure derived from the lpsC sequence in the ergot alkaloid biosynthesis gene cluster of C. purpurea strain P 1 (16) (derivative of C. purpurea ATCC 20102). A, adenylation; T, thiolation; C, condensation; R, reductase; calculated Mr: 178,200. The proposed reaction mechanism was in conjunction with LPS2. b, immunoblot analysis of ergometrine synthetase fractions of the gel filtration separation in Fig. 4 with antibody raised against the Red-domain of LPS3 encoded by the lpsC sequence of Ecc93. The analysis shows that LPS3 is present only in those fractions catalyzing alanine thioester formation and complementing with LPS2 in the biosynthesis of ergometrine (shown below). c, corrected domain arrangement of LPS3 showing the C0-domain, which contributes 19 kDa to the originally calculated molecular mass of the enzyme in accordance with the experimentally determined value.
Attempts to detect ergometrine synthesis in protein extracts of C. purpurea strains P1 and D1 failed, which may be based on more than 10 years of constant selection of their parents for high production of ergopeptines (8, 21). In fact, they produce only traces of low molecular weight alkaloids in contrast to C. purpurea strain Ecc93 and they have elevated levels of LPS1-type enzymes, which may outcompete LPS3 from productive contacts with LPS2, the d-lysergic acid-recruiting enzyme. Immunoblot analysis of protein extracts of P1 and D1 with anti-Red antibodies did neither reveal a plausible band attributable to LPS3 in crude extracts nor in fractionated protein preparations suggesting too little or no formation of LPS3. Nevertheless, reverse transcription-PCR analysis of total RNA with primers derived from the lpsC sequence revealed the presence of the lpsC transcript in strains P1 and D1 as well as in Ecc93.3 Whether ergometrine synthesis occurs in C. purpurea strain P1 can therefore eventually be clarified when both lpsA genes in this strain will be disrupted and ergometrine synthesis will be studied in the absence of interfering LPS1-type enzyme.
The knowledge of the LPS3 amino acid sequence permitted one to determine the specificity determining amino acid residues in the substrate binding pocket of the LPS3 A-domain using Ravel's prediction algorithm NRPS BLAST server (32). The analysis revealed the signature sequence DIFLAGII, which did not match the alanine-specific consensus DLFFCGGP, for which precedent is the A-domain of the first module of LPS1 (reported as alanine-activating) of C. purpurea (11). The reason for the deviation of the LPS3 signature may lie in the fact that NRPS BLAST server does not take into account the known side specificity of that module in in vitro conditions (10). By contrast, NRPS predictor (33), another algorithm that considers not only the 8 amino acids in the A-domain binding pocket, but a total of 34 amino acids surrounding the amino acid substrate at 8-Å minimal distance predicted specificity of LPS3 for small amino acids glycine/alanine as it did for the first modules of LPS1 and its orthologue LPSA on a small cluster basis. On a large cluster basis (considering side specificity) it correctly categorized the LPS1 and LPSA A1-domains as side-specific for aliphatic amino acid substrates but erroneously categorized the LPS3-A-domain into a class of A-domains that have aliphatic and aromatic hydroxyamino acids as substrates, which is totally inconsistent with the experimental data. This shows that the NRPS predictor (33), like the BLAST server (32), classifies the LPS3 substrate binding pocket as unique. From this, we infer that the LPS3 signature represents a true consensus for alanine.
All LPS Enzymes Have a Conserved N-terminal Domain—In the bacterial NRPSs and polyketide synthases interactions between modular enzyme subunits were found to be mediated by interpeptide linkers. In the case of NRPS they are part of the N-terminal C-domain of acceptor subunits and of the C-terminal epimerization (E-) domain of donor subunits as in the case of gramicidin, tyrocidin, and surfactin peptide synthetases (34–36). Such sequences (communication-mediating domain-A and communication-mediating domain-D, respectively) were not seen in the protein sequences of the LPS enzymes, which was not unexpected, because the donor subunits LPS2 (or LPSB) do not have C-terminal E-domains. Instead, they have the C-domain-catalyzing condensation of d-lysergic acid in cis at their C-terminal ends, whereas the acceptor subunits (i.e. LPS1 or LPS3) do not (Fig. 2). This raises the question how molecular interaction between the C-domain of LPS2 and its downstream subunits takes place. Remarkably, LPS1, LPS4 from C. purpurea, and LPSA from Neotyphodium lolii all have conserved stretches of 200–250 amino acid residues in front of their first A-domains. However, from its deduced amino acid sequence, LPS3 from C. purpurea did not possess such an N-terminal end, which raised concerns about the significance of these sequences as possible sites of protein interactions.
The clue to this conundrum was provided by reconsidering the immunoblot analyses of SDS-PAGE gels of the LPS3 preparations from gel filtration separations with anti-Red antibodies. As mentioned above, in such blots LPS3 showed up as a 200-kDa band (Fig. 5b) that in effect contrasted the calculated molecular mass of 178.5 kDa derived from the published LPS3 sequence. Upon inspection of the 5′-region of sequences of both lpsC of C. purpurea strain P1 (14) and Ecc93, surprisingly, a 29-nucleotide intron separating a 489-bp upstream exon from the original annotated 5′-end (16) was detected, the position of which is nearly the same as in the gene sequences of all other LPSs. The resultant upstream exon encoded a stretch of 163 amino acids, and its deduced amino acid sequence had high similarity to the N-terminal ends of the other LPS enzymes (Fig. 6). Moreover, the deduced amino acid sequence neatly added an extra 19 kDa to the previously calculated 179.5 kDa of LPS3 (yielding a total of 198.5 kDa), which was then in perfect agreement with the observed size of ∼200 kDa in immunoblots (Fig. 5c).
FIGURE 6.
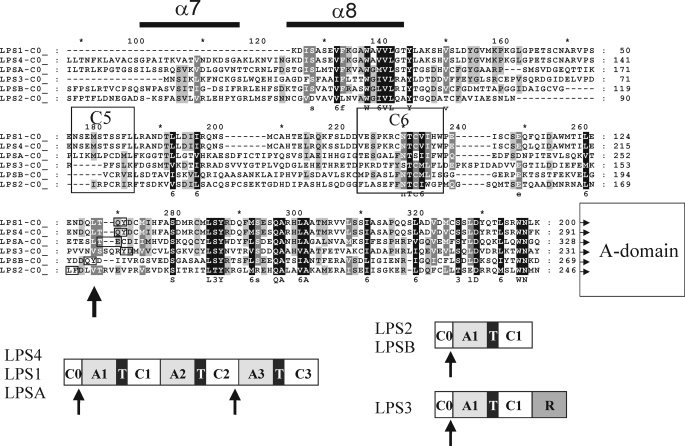
Alignment of N-terminal regions of LPS enzymes. The beginning of the corresponding A-domains is denoted by the conserved WN at the end of each sequence. C5 and C6 denote the signature consensus of C5- and C6-motifs of C-domains of NRPSs. The helical region α7/8 is defined by comparison with the VibH sequence and by modeling analysis using the PHYRE fold recognition server (40). The arrows denote the locations of the introns in the C0- and the C2-domains in the alignment and in the domain organizations of LPS enzymes given below. Intron boundaries are boxed.
Structure of the N-terminal Region of LPS Enzymes—Analysis of BLASTP hits of the N-terminal sequences of the elongating LPS enzyme subunits indicated that they all have similarity to the C-terminal halves of C-domains of NRPSs. Structure determination of VibH, a free-standing C-domain of the vibriobactin assembly line (37) or of TycC6, a peptidyl carrier protein (T)-C-didomain from the tyrocidin assembly line (38), has shown that C-domains are pseudodimers of two subdomains each having αβα sandwich structure (37). The longest versions of these N-terminal domains, present in LPS4 and LPSA, begin at the C4-motif of C-domains, whereas the shorter versions in LPS1, LPS2, LPSB, and LPS3 begin between the C4- and C5-motifs (Fig. 6). Clearly, these half-domains cannot catalyze condensations, because they lack the catalytically important C3-motif (HHXXXDG) (39). The alignment in Fig. 6 shows that they are most conserved in the region between C4 and C5 (Fig. 6), which corresponds to helices α7 and α8 of the C-terminal subdomain of VibH and TycC6. Secondary structure predictions of the C-half domains at the N-terminal regions of the LPS enzymes using the PHYRE protein fold recognition server (40) show that these helices form part of the αβα sandwich structure in the C-terminal half of VibH and are exposed to the surface of the domain. According to the previous numbering of C-domains in the LPS enzyme subunits the C-half domains were denoted C0-domains.
C0-domains in NRPS Systems—BLASTP searches revealed besides the LPSs a considerable number of other fungal and some bacterial NRPS enzymes with N-terminal C0-domains. In bacterial NRPS enzyme systems with C0-domains such as the mycosubtilin or iturin synthetases in Bacillus subtilis (41), the C0-domains are at the N termini of elongating acceptor subunits, e.g. MycB and MycC. Thus they differ from the gramicidin or tyrocidin systems by replacement of the N-terminal C-domain by a C0-domain (41). The mechanism of interaction between Myc subunits is not known. In the case of VibF, an NRPS subunit involved in vibriobactin synthesis, dimerization via a non-functional C-domain in VibF has been described (42). However, the domain's contact site leading to dimerization remained unknown. Therefore, we must await future investigations on the role of the C0-domains in the various NRPS systems. Interestingly enough, we also noted the presence of C0-domains in a number of fungal NRPS enzymes consisting of a single polypeptide chain, e.g. aminoadipylcysteinylvaline synthetase from Penicillium chrysogenum (GenBank™ accession number P26046.1) or HC-toxin synthetase from Cochliobolus carbonum (HC-toxin synthetase, GenBank™ accession number Q01886.2). They lie directly in front of the first modules in these enzymes, a position reminiscent of the location of N-terminal C-domains in cyclosporin synthetase or enniatin synthetase (43). It is not known whether these N-terminal C-domains are functionally necessary or whether they are required for structural reasons (43).
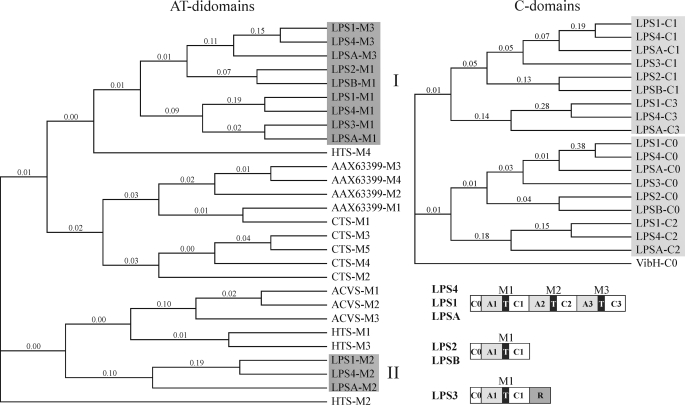
Phylogeny of C0-domains of the LPS Enzymes—The conservation of intron position in orthologous and paralogous genes has been proposed as a strong indicator for phylogenetic relationship due to gene duplication (44). The conservation of intron position in the C0-domains of all LPS proteins and also of all second C-domains (C2) of the LPS1-type enzymes therefore raises questions about their phylogenetic relatedness (Fig. 6). Because the sequential order of domains in the LPS is A-T-C, which contrasts the canonical C-A-T organization of catalytic domains in the NRPS (45, 46), neighbor joining analyses and multiple alignments were performed with amino acid sequences of all AT-didomains of LPS enzymes and of a number of fungal NRPS unrelated to alkaloid biosynthesis. On the other hand, the amino acid sequences of all C-domains of LPS, including the C0-domains, were compared. The phylogenetic tree obtained in the case of the AT-didomains depicted in Fig. 7 (left panel) shows that LPS AT-didomains can be grouped into two clades (I and II) distant from each other and from didomains of other fungal NRPS, including the didomains of CTS, a pentamodular NRPS from C. purpurea unrelated to alkaloid peptide biosynthesis. The gene of CTS has been cloned previously by our group (accession number ABR23346). Clade I (shaded in Fig. 7) of LPS AT-didomains contains the AT-didomain of all first modules (M1s, domain order A-T-C) of LPS enzymes, i.e. from LPS1-type, LPS2-type, and LPS3. This homology suggests that all of these AT-didomains probably arose by gene duplication of an ancestor module leading to a paralogous set of modules each with a different function (donor subunit or acceptor subunit) in the two alkaloid peptide assembly systems. Remarkably, clade I also harbors the AT-didomains of all third modules (M3s) of the LPS1-type proteins, which strengthens the assumption that the first and third AT-didomains of LPS1-type enzymes may have evolved from a common ancestor module as postulated previously (14) or that the M3s arose from M1s by gene duplication. An additional argument (14) for gene duplication was that on the gene level the A-domains of first (M1) and third (M3) modules each are preceded by an intron at nearly the same position (Fig. 6). By contrast, clade II harbors only the didomains (M2) of the LPS1-type enzymes (shaded II in Fig. 7). They are phylogenetically distant from clade I suggesting that M2s evolved in a way different from that of M1s and M3s.
FIGURE 7.
Phylogenetic analysis of AT-didomains and C-domains of the various LPSs from ergot fungi. AT-didomains: Analysis was done using ClustalW using sequences of AT-didomains of LPS2 and from several other fungal NRPS and of an NRPS (CTS) from C. purpurea strain D1 (ATCC 20102) unrelated to d-lysergic acid alkaloid biosynthesis. The shading under the clades contains AT-didomains from the first modules (M1) and the third modules (M3) (clade I), and the second modules (M2) of LPS enzymes (clade II). The other AT-didomains from other fungi or from C. purpurea (CTS) are unrelated to alkaloid peptide synthesis. The accession numbers are as follows: LPS1 (C. purpurea) (CAB39315); LPS4 (C. purpurea) (CAI59268); LPSA (N. lolii) (AAL26315); LPS2 (C. purpurea) (CAD28788); LPSB (N. lolii) (ABM91454); LPS3 (C. purpurea) (CAI59267); HC-toxin synthetase (C. carboneum) (Q01886); CTS (C. purpurea) (ABR23346); AAX63399 (Hypocrea virens) (AAX63399); aminoadipylcysteinylvaline synthetase (Penicillim chrysogenum) (P26046); and VibH (Vibrio cholerae) (AAD48879). C-domains: phylogenetic tree of all C-domains of LPS enzymes along with VibH-C-terminal half domain. The phylogenetic trees were generated with ClustalW using the amino acid sequences of C-terminal halves of the regular C-domains and of C0-domains (for detail see supplemental material). The phylogenetic tree was essentially the same when full-length C-domain sequences were used.
A picture congruent with these analyses emerged when the sequences of all C-domains of the LPSs were analyzed (shown in Fig. 7, right panel). Their resultant phylogenetic tree consists of two clades one comprising all first C-domains (C1) and third C-domains (C3), whereas the other comprises the C2-domains together with the C0-domains. This indicates phylogenetic relatedness between the C1- and C3-domains in analogy to the AT-didomains of M1 and M3 and suggests that the modules were duplicated as A-T-C units but not C-A-T units. On the other hand, the relatedness between the amino acid sequences of C0- and C2-domains is in clear accordance to the conserved intron position in their gene sequences and suggests a common ancestor of these domains different from that of C1- and C3-domains.
It may be speculated that the C0-domains are remnants of an ancestor module that once lay in front of the putative M1 ancestor both possibly encoded by a single transcript. During evolution of LPS enzymes, the DNA of this ancient module may have been duplicated and finally lost except for that portion encoding the C0 portion. This eventually gave rise to the formation of LPS2 (which still possesses the C0-domain) and LPS3 and the first modules of LPS1-type enzymes. Next, insertion of an M2 into its present position between modules 1 (M1) and 3 (M3) of LPS1-type may have occurred. In view of the significant similarity between M2s and the C0s in their C-domain portion it may be speculated that M2 evolved from the putative ancestor module. Furthermore, when one considers that fungal modules are arranged in the domain order A-T-C (but not C-A-T) it is easy to understand that the second intron of the LPS1-type genes arose by acquisition of the hypothetical M2-module (i.e. duplication of ancient M0) and not by duplication of M1 as suggested previously (14). However, whether the C0-domains of LPSs are molecular fossils or play a functional role in the interactions of LPS2 with LPS3 (simple d-lysergic acid amides) and the first modules of LPS1-type enzymes (complex d-lysergic acid amides) must be clarified in the future.
Supplementary Material
Acknowledgments
We thank David Hopwood for critical reading of the manuscript.
Dedicated to the memory of Dr. Rainer Zocher, the pioneer in fungal peptide synthetase research (09.11.1948-18.03.2008).
This work was supported by the Deutsche Forschungsgemeinschaft SPP 1152 (Grant Ke 452/11-4). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental references and Figs. S1 and S2.
Footnotes
The abbreviations used are: LPS, d-lysergylpeptide synthetase; NRPS, nonribosomal peptide synthetase; C0-domains, modified N-terminal C-domains; lpsCred, gene fragment encoding a portion of LPS3 Red-domain; CTS, C. purpurea putative toxin synthetase; M, module; AT, adenylationthiolation didomain.
Havemann, J. and Keller, U., unpublished data.
References
- 1.Stadler, P. A., and Giger, R. (1984) in Natural Products and Drug Development (Krosgard-Larson, P., and Kofod, H., eds) pp. 463–485, Munksgaard, Copenhagen, Denmark
- 2.Burkhalter, A., Julius, D. J., and Katzung, B. G. (1998) in Basic and Clinical Pharmacology (Katzung, B. G., ed) pp. 261–286, Appleton-Lange, New York
- 3.Berde, B., and Stuermer, E. (1978) in Introduction to the Pharmacology of Ergot Alkaloids and Related Compounds (Aellig, W. H., Berde, B., and Schild, O. H., eds) pp. 1–28, Springer, Berlin
- 4.Flieger, M., Wurst, M., and Shelby, R. (1997) Folia Microbiol. 42 3–29 [DOI] [PubMed] [Google Scholar]
- 5.Kobel, H., and Sanglier, J. J. (1986) in Biotechnology (Rehm, H. J., and Reeds, G., eds) pp. 569–609, VCH Verlagsgesellschaft, Weinheim, Germany
- 6.Klotz, J. L., Bush, L. P., Smith, D. L., Schafer, W. D., Smith, L. L., Vevoda, A. O., Craig, A. M., Arrington, B. C., and Strickland, J. R. (2006) J. Animal Sci. 84 3167–3175 [DOI] [PubMed] [Google Scholar]
- 7.Stadler, P. A. (1982) Planta Med. 46 131–144 [DOI] [PubMed] [Google Scholar]
- 8.Keller, U., Han, M., and Stoeffler-Meilicke, M. (1988) Biochemistry 27 6164–6170 [DOI] [PubMed] [Google Scholar]
- 9.Riederer, B., Han, M., and Keller, U. (1996) J. Biol. Chem. 271 27524–27530 [DOI] [PubMed] [Google Scholar]
- 10.Walzel, B., Riederer, B., and Keller, U. (1997) Chem. Biol. 4 223–230 [DOI] [PubMed] [Google Scholar]
- 11.Tudzynski, P., Hölter, K., Correia, T., Arntz, C., Grammel, N., and Keller, U. (1999) Mol. Gen. Genet. 261 133–141 [DOI] [PubMed] [Google Scholar]
- 12.Correia, T., Grammel, N., Ortel, I., Keller, U., and Tudzynski, P. (2003) Chem. Biol. 10 1281–1292 [DOI] [PubMed] [Google Scholar]
- 13.Panaccione, D. G., Johnson, R. D., Wang, J., Young, C. A., Damrongkool, P., Scott, B., and Schardl, C. L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12820–12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Damrongkool, P., Sedlock, A. B., Young, C. A., Johnson, R. D., Goetz, K. E., Scott, B., Schardl, C. L., and Panaccione, D. G. (2005) DNA Seq. 16 379–385 [DOI] [PubMed] [Google Scholar]
- 15.Fleetwood, D. J., Scott, B., Lane, G. A., Tanaka, A., and Johnson, R. D. (2007) Appl. Environ. Microbiol. 73 2571–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haarmann, T., Machado, C., Lübbe, Y., Correia, T., Schardl, C. L., Panaccione, D. G., and Tudzynski, P. (2005) Phytochemistry 66 1312–1320 [DOI] [PubMed] [Google Scholar]
- 17.Haarmann, T., Ortel, I., Tudzynski, P., and Keller, U. (2006) Chembiochem 7 645–652 [DOI] [PubMed] [Google Scholar]
- 18.Panaccione, D. G., Tapper, B. A., Lane, G. A., Davies, E., and Fraser, K. J. (2003) Agric. Food Chem. 51 6429–6437 [DOI] [PubMed] [Google Scholar]
- 19.De Costa, C. (2002) Lancet 359 1768–1770 [DOI] [PubMed] [Google Scholar]
- 20.Hofmann, A. (1964) Die Mutterkornalkaloide, Enke Verlag, Stuttgart, Germany
- 21.Keller, U. (1982) Appl. Environ. Microbiol. 46 580–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997) Nucleic Acids Res. 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholas, K. B., Nicholas, H. B., Jr., and Deerfield, D. W. (1997) EMBNEW NEWS 4 14 [Google Scholar]
- 24.Agurell, S. (1966) Acta Pharm. Suec. 3 71–100 [PubMed] [Google Scholar]
- 25.Castagnoli, N., Corbett, K., Chain, E. B., and Thomas, R. (1970) Biochem. J. 117 451–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basmadjian, G. P., Floss, H. G., Gröger, D., and Erge, D. (1969) Chem. Comm. 1 418–419 [Google Scholar]
- 27.Gaitatzis, N., Kunze, B., and Müller, R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11136–11141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pospiech, A., Cluzel, B., Bietenhader, J., and Schupp, T. (1995) Microbiology 141 1793–1803 [DOI] [PubMed] [Google Scholar]
- 29.Morris, M. E., and Jinks-Robertson, S. (1991) Gene (Amst.) 98 141–145 [DOI] [PubMed] [Google Scholar]
- 30.Casqueiro, J., Gutiérrez, S., Bañuelos, O., Fierro, F., Velasco, J., and Martín, J. F. (1998) Mol. Gen. Genet. 259 549–556 [DOI] [PubMed] [Google Scholar]
- 31.Tanaka, A., Tapper, B. A., Popay, A., Parker, E. J., and Scott, B. A. (2005) Mol. Microbiol. 57 1036–1050 [DOI] [PubMed] [Google Scholar]
- 32.Challis, G. L., Ravel, J., and Townsend, C. A. (2000) Chem. Biol. 3 211–224 [DOI] [PubMed] [Google Scholar]
- 33.Rausch, C., Weber, T., Kohlbacher, O., Wohlleben, W., and Huson, D. H. (2005) Nucleic Acids Res. 33 5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gokhale, R. S., and Khosla, C. (2000) Curr. Opin. Chem. Biol. 4 22–27 [DOI] [PubMed] [Google Scholar]
- 35.Hahn, M., and Stachelhaus, T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15585–15590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanovic, A., Samel, S. A., Essen, L. O., and Marahiel, M. A. (2008) Science 321 659–663 [DOI] [PubMed] [Google Scholar]
- 37.Keating, T. A., Marshall, C. G., Walsh, C. T., and Keating, A. E. (2002) Nat. Struct. Biol. 9 522–526 [DOI] [PubMed] [Google Scholar]
- 38.Samel, S. A., Schoenafinger, G., Knappe, T. A., Marahiel, M. A., and Essen, L. O. (2007) Structure 15 781–792 [DOI] [PubMed] [Google Scholar]
- 39.Bergendahl, V., Linne, U., and Marahiel, M. A. (2002) Eur. J. Biochem. 269 620–629 [DOI] [PubMed] [Google Scholar]
- 40.Kelley, L. M., MacCallum, R. M., and Sternberg, M. J. (2000) J. Mol. Biol. 299 499–520 [DOI] [PubMed] [Google Scholar]
- 41.Duitman, E. H., Hamoen, L. W., Rembold, M., Venema, G., Seitz, H., Saenger, W., Bernhard, F., Reinhardt, R., Schmidt, M., Ullrich, C., Stein, T., Leenders, F., and Vater, J. (1999) Proc. Natl. Acad. Sci. U. S. A. 9 13294–13299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillson, N. J., Balibar, C. J., and Walsh, C. T. (2004) Biochemistry 43 11344–11351 [DOI] [PubMed] [Google Scholar]
- 43.Hornbogen, T., and Zocher, R. (2005) A Handbook of Industrial Mycology, Mycology Series, (An, Z., ed.) Marcel Dekker, New York, Vol. 22, pp. 449–478 [Google Scholar]
- 44.Fedorov, A., Merican, A. F., and Gilbert, W. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 16128–16133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linne, U., and Marahiel, M. A. (2000) Biochemistry 39 10439–10447 [DOI] [PubMed] [Google Scholar]
- 46.Sieber, S. A., and Marahiel, M. A. (2005) Chem. Rev. 105 715–738 [DOI] [PubMed] [Google Scholar]
- 47.Walzel, B. (1997) Characterization of D-lysergic Acid Activating Enzymes from The Ergot Fungus Claviceps Purpurea. M.Sc. thesis, Technische Universität Berlin, Germany
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.