Abstract
In mouse Paneth cells, α-defensins, termed cryptdins (Crps), are activated by matrix metalloproteinase-7-mediated proteolysis of inactive precursors (pro-Crps) to bactericidal forms. The activating cleavage step at Ser43 ↓ Ile44 in mouse pro-Crp4-(20–92) removes nine acidic amino acids that collectively block the membrane-disruptive behavior of the Crp4 moiety of the proform. This inhibitory mechanism has been investigated further to identify whether specific cluster(s) of electronegative amino acids in pro-Crp4-(20–43) are responsible for blocking bactericidal activity and membrane disruption. To test whether specific cluster(s) of electronegative amino acids in pro-Crp4-(20–43) have specific positional effects that block bactericidal peptide activity and membrane disruption, acidic residues positioned at the distal (Asp20, Asp26, Glu27, and Glu28), mid (Glu32 and Glu33), and proximal (Glu37, Glu38, and Asp39) clusters in pro-Crp4-(20–92) were mutagenized, and variants were assayed for differential effects of mutagenesis on bactericidal peptide activity. Substitution of the mid and proximal Asp and Glu clusters with Gly produced additive effects with respect to the induction of both bactericidal activity and membrane permeabilization of live Escherichia coli ML35 cells. In contrast, substitution of distal Glu and Asp residues with Gly or their deletion resulted in pro-Crp4-(20–92) variants with bactericidal and membrane-disruptive activities equal to or greater than that of fully mature Crp4. These findings support the conclusion that the most distal N-terminal anionic residues of pro-Crp4-(20–92) are primarily responsible for blocking Crp4-mediated membrane disruption in the precursor.
In the small bowel, Paneth cells at the base of the crypts of Lieberkühn secrete α-defensins and additional antimicrobial peptides at high levels in response to cholinergic stimulation and when exposed to bacterial antigens (1–4). Paneth cell α-defensins show broad spectrum antimicrobial activities and constitute the majority of bactericidal peptide activity in Paneth cell secretions (2, 5–7). The release of Paneth cell products into the crypt lumen is inferred to protect mitotically active crypt cells from colonization by potential pathogens and to confer protection from enteric infection (2, 8–10). The most compelling evidence for a Paneth cell role in enteric innate immunity is evident from studies of mice transgenic for a human Paneth cell α-defensin, HD-5, which are completely immune to infection and systemic disease from orally administered Salmonella enterica serovar typhimurium (11).
The biosynthesis of α-defensins requires post-translational activation by lineage-specific proteinases (12, 13). Although the enzymes that mediate pro-α-defensin processing in myeloid and epithelial cells differ, the overall processing schemes are the same. Both myeloid and Paneth cell α-defensins derive from ∼10-kDa prepropeptides that contain canonical signal sequences, electronegative proregions, and a 3.5–4-kDa mature α-defensin peptide in the C-terminal portion of the precursor (13–16). Pro-α-defensin processing in mouse Paneth cells is catalyzed by matrix metalloproteinase-7 (MMP-7)3 and takes place intracellularly and prior to secretion (17, 18). In mouse small intestinal epithelium, only Paneth cells express MMP-7 as components of dense core secretory granules (12), and the bactericidal activity of mouse Paneth cell α-defensins depends completely on activation of 8.4-kDa pro-Crps by MMP-7-catalyzed proteolysis (12, 18). MMP-7 gene disruption ablates pro-Crp processing such that mature, activated Crp peptides are absent from the small intestine, and innate immunity to oral bacterial infection is impaired in MMP-7-null mice (12).
MMP-7 produces active mouse α-defensins by cleaving precursors in vitro at conserved sites in the proregion including the junction of the propeptide and α-defensin moiety (18). Electropositive Arg side chains in Crp4 facilitate electrostatic interactions necessary for disruption of the electronegative bacterial cell envelope (3, 19, 20). In contrast, pro-Crps lack in vitro bactericidal activity, corresponding with a diminished ability to interact with model membranes, to disrupt large unilamellar vesicles (20), and to permeabilize live Escherichia coli ML35 cells. MMP-7-mediated proteolysis of pro-Crp4-(20–92) produces the specific cleavage intermediates pro-Crp4-(44–92), pro-Crp4-(54–92), and pro-Crp4-(59–92) (18). Because the Ser43 ↓ Ile44 cleavage event is sufficient to activate bactericidal activity, amino acids in the pro-Crp4-(20–43) region of the prosegment maintain the precursor in an inactive state by removal of nine acidic amino acids from proximity to the Crp4 component of the molecule (21).
To test the hypothesis that specific clusters of the nine anionic residue positions in the pro-Crp4-(20–43) region exert differential effects in blocking pro-Crp4 bactericidal activity, partial charge-neutralizing mutations made at Asp and Glu residue positions in pro-Crp4-(20–92) were prepared, and the variant pro-Crp4 peptides were tested for bactericidal activity and in cell permeabilization assays. The results show that the acidic residues nearest the pro-Crp4 N terminus are primarily responsible for inhibiting pro-Crp4 bactericidal activity.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant Peptides—Recombinant pro-Crp4-(20–92) proregion variants were prepared by site-directed PCR-based mutagenesis (18), expressed as N-terminal His6-tagged fusion proteins, and subsequently affinity-purified (3, 18, 22). After cleavage of the His6 tag, the peptides were further purified by HPLC. Peptide homogeneity was assessed by analytical reverse-phase HPLC and acid/urea (AU)-PAGE (23), and peptide masses were confirmed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (Voyager-DE) (21). The experimental details of mutagenesis and preparation of the individual recombinant peptides investigated (Fig. 1) are described in detail (see supplemental “Experimental Procedures”).
FIGURE 1.
Recombinant pro-Crp4 variant peptides prepared by site-directed mutagenesis. The primary structures of the recombinant peptides prepared and assayed in this study are aligned. Altered residues in mutant Crp4 peptides are shown in underlined boldface. Numbers above the pro-Crp4 sequence refer to residue positions in reference to the N-terminal Asp of native pro-Crp4-(20–92).
MMP-7-mediated Proteolysis of Recombinant Peptides—To test pro-Crp4 variants for proteolytic conversion by MMP-7, 10-μg peptide samples were incubated with or without 0.5 mol eq of MMP-7 in HEPES buffer (1 mm HEPES, 15 mm NaCl, 0.5 mm CaCl2, pH 7.4) for 18 h at 37 °C. Complete peptide digests were resolved by AU-PAGE and stained with Coomassie Blue.
Bactericidal Peptide Assays—The bactericidal activities of purified recombinant peptides were tested in vitro against E. coli ML35, Staphylococcus aureus 502A, Vibrio cholerae, Listeria monocytogenes 104035, and S. enterica serovar typhimurium strains as described previously (21, 24). Samples consisting of exponentially growing bacterial cells were collected by centrifugation, washed, and resuspended in 10 mm PIPES, pH 7.4, supplemented with 0.01 volume (1%, v/v) of Trypticase soy broth (PIPES/TSB). Bacteria (5 × 106 colony-forming units (CFU)/ml) were incubated with each peptide (0–6 μm) in 50 μl of PIPES/TSB. After 60 min at 37 °C, 20 μl of each incubation mixture were diluted 1:1000 with 10 mm PIPES, pH 7.4, and 50 μl of the diluted samples were plated on Trypticase soy-agar plates using an Autoplate 4000 (Spiral Biotech Inc., Bethesda, MD). Surviving bacteria were quantitated as CFU/ml on plates after incubation at 37 °C for 12–18 h, and data were analyzed and plotted using SigmaPlot (Systat Software, Inc., San Jose, CA). Bactericidal assays performed in Figs. 3, 4, and 7 are representative of experiments that are reliably reproducible (see supplemental Fig. S4).
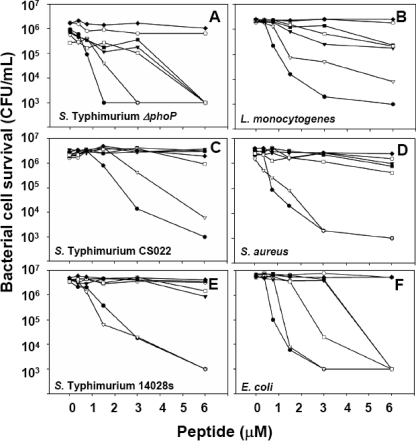
FIGURE 3.
Partial activation of pro-Crp4 bactericidal activity by mutagenesis of proximal anionic amino acid clusters. Exponentially growing S. enterica serovar typhimurium ΔphoP (A), L. monocytogenes (B), S. enterica serovar typhimurium CS022 (C), S. aureus (D), S. enterica serovar typhimurium 14028s (E), and E. coli (F) were exposed to the peptide concentrations shown at 37 °C in 50 ml of 10 mm PIPES buffer supplemented with 1% TSB for 1 h. Following peptide exposure, the bacteria were plated on TSB-agar and incubated overnight at 37 °C. Surviving bacteria were counted as CFU/ml at each peptide concentration. Values below 1 × 103 CFU/ml signify that no colonies were detected. •, Crp4; ○, pro-Crp4; ▿, (DE/G)-pro-Crp4; ▾, (DE/NQ)-pro-Crp4; □, (6-DE/G)-pro-Crp4; ▪, (3-DE/G)-pro-Crp4; ♦, (DE/G)-pro-Crp4-(20–60).
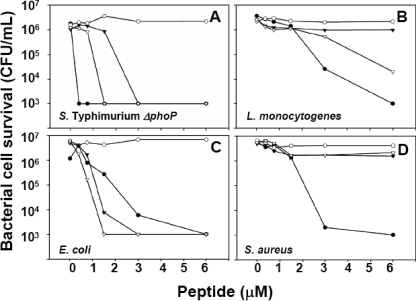
FIGURE 4.
Mutagenesis of distal proregion acidic amino acid clusters fully activates pro-Crp4. Exponentially growing S. enterica serovar typhimurium ΔphoP (A), L. monocytogenes (B), E. coli (C), and S. aureus (D) were exposed to the peptides as in Fig. 3. •, Crp4; ○, pro-Crp4; ▿, (4-DE/G)-pro-Crp4; ▾, (5-DE/G)-pro-Crp4.
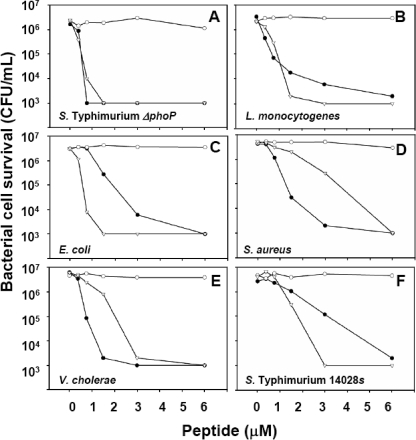
FIGURE 7.
Deletion of the distal acidic residue cluster confers full bactericidal activity on pro-Crp4. Exponentially growing S. enterica serovar typhimurium ΔphoP (A), L. monocytogenes (B), E. coli (C), S. aureus (D), V. cholerae (E), and S. enterica serovar typhimurium 14028s (F) were exposed to the peptide concentrations indicated as in Fig. 3. •, Crp4; ○, pro-Crp4; ▿, pro-Crp4-(29–92).
Peptide-mediated Permeabilization of Live E. coli—Exponentially growing E. coli ML35 cells were washed and resuspended in 10 mm PIPES/TSB as described (18, 20). Bacteria were incubated in triplicate with 0–6 μm peptide and 2.5 mm 2-ortho-nitrophenyl β-d-galactopyranoside (ONPG), a chromogenic substrate. 80 μl of E. coli ML35 cells in PIPES/TSB (∼5 × 106 CFU/ml) were added to 10 μl of ONPG and 10 μl of peptide solution and incubated at 37 °C for 2 h in a 96-well plate format. E. coli ML35 cells are β-galactosidase-constitutive and permease-negative, so ONPG diffusion into bacterial cells and ONP production are dependent on peptide-mediated membrane disruption. ONP was measured at 405 nm on a 96-well SpectraMax plate spectrophotometer (Molecular Devices, Sunnyvale, CA), and data were analyzed using SigmaPlot.
RESULTS
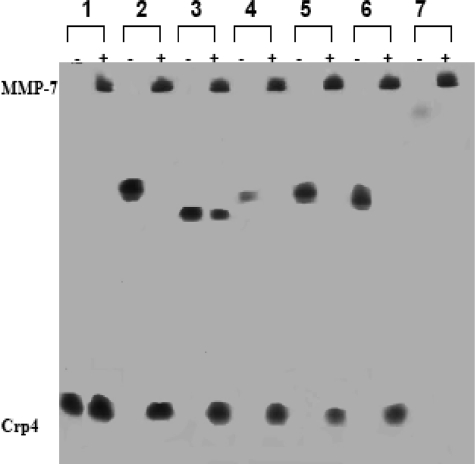
MMP-7 Processes Pro-Crp4 Anionic Proregion Variant Peptides—To test whether partial reduction of the electronegative charge in the proregion of pro-Crp4 has effects on precursor folding and processing, the native and variant pro-Crp4 peptides were incubated with MMP-7 and analyzed by AU-PAGE. Pro-Crp4 requires MMP-7-mediated proteolytic conversion to Crp4, and disruption of disulfide pairings would result in extensive peptide proteolysis (22). Because properly folded Crp4 resists proteolysis completely and Crp4 and its foldamers have distinctly different mobilities in AU-PAGE, this assay for the comigration of processing products with native Crp4 is a useful index of correct peptide folding (17, 22, 25). Pro-Crp4 and its electronegative variants were processed by MMP-7 to generate products that comigrated with Crp4, as evident from MALDI-TOF mass spectrometry and AU-PAGE mobility of all pro-Crp4 peptide variants (Fig. 2). Thus, reducing the electronegative charge of the proregion to differing extents did not destabilize the canonical α-defensin fold as judged by peptide masses and resistance to peptide proteolysis. Also, the mutagenized proregion alone, (DE/G)-pro-Crp4-(20–60), was degraded by MMP-7, indicating that mutagenesis did not modify prosegment sensitivity to proteolysis.
FIGURE 2.
MMP-7 processing of pro-Crp4 variants. The recombinant peptides (10 μg) were incubated with (+) or without (-) 0.5 mol eq of MMP-7 in 1 mm HEPES, 15 mm NaCl, 0.5 mm CaCl2, pH 7.4, for 18 h at 37 °C. The digestion products were resolved by AU-PAGE and visualized by staining with Coomassie Blue. (DE/G)-pro-Crp4 did not undergo complete cleavage due to the high ratio of peptide substrate to MMP-7. Lane 1, Crp4; lane 2, pro-Crp4; lane 3, (DE/G)-pro-Crp4; lane 4, (DE/NQ)-pro-Crp4; lane 5, (6-DE/G)-pro-Crp4; lane 6, (3-DE/G)-pro-Crp4; lane 7, (DE/G)-pro-Crp4-(20–60). The proregion and its fragments do not stain with Coomassie Blue.
Effects of Partial Electronegative Charge Neutralization on Pro-Crp4 Bactericidal Peptide Activity—To determine whether partial reduction of anionic charge in pro-Crp4-(20–43) exerts differential effects on the inhibition of pro-Crp4-(20–92) bactericidal activity, the proregion variants of pro-Crp4 were tested against S. aureus 502A, L. monocytogenes 104035, E. coli ML35, V. cholerae, and S. enterica serovar typhimurium strains including 14028s, CS022, and ΔphoP (3). Native pro-Crp4-(20–92), which lacks activity, served as a negative control peptide with bactericidal Crp4 and (DE/G)-pro-Crp4 molecules serving as positive controls (18, 21).
To test whether the bactericidal activity of (DE/G)-pro-Crp4 resulted from conferring activity on the proregion per se by mutagenesis of the electronegative residues to glycines, the mutant proregion (DE/G)-pro-Crp4-(20–60) also was assayed for microbicidal activity (see “Experimental Procedures”). In all instances, (DE/G)-pro-Crp4-(20–60) had no effect on bacterial cell survival up to 6 μm peptide concentration (Fig. 3, A–F). These findings show that the activity of (DE/G)-pro-Crp4-(20–92) is due to the proregion loss of inhibitory function and is not a consequence of (DE/G)-pro-Crp4-(20–60) being converted to a bactericidal molecule as a result of the mutagenesis.
To test the hypothesis that removal of carboxyl groups alone at Asp and Glu positions would be sufficient to eliminate proregion inhibitory effects, we prepared (DE/NQ)-pro-Crp4-(20–92) in which Asn and Gln residue positions would retain side chain length but lack anionic charge. Against the less sensitive S. aureus and S. enterica serovar typhimurium CS022 and 14028s species, even 6 μm (DE/NQ)-pro-Crp4 showed very little to no activity (Fig. 3, C–E). On the other hand, relative to Crp4 and (DE/G)-pro-Crp4, (DE/NQ)-pro-Crp4-(20–92) had attenuated activity at concentrations of 1.5 μm or less (Fig. 3, A–F) except against defensin-sensitive S. enterica serovar typhimurium ΔphoP and E. coli, where the activities were equivalent (Fig. 3, A and F). Against L. monocytogenes, (DE/NQ)-pro-Crp4 had approximately one-tenth the activity of (DE/G)-pro-Crp4 at ≥1.5 μm peptide (Fig. 3B). From these findings, we infer that anionic amino acids in the proregion appear to inhibit pro-Crp4-(20–92) bactericidal activity by a combination of charge neutralization of Crp4 cationic residues and side chain interactions that were not evident in studies of (DE/G)-pro-Crp4.
To test the hypothesis that individual clusters of anionic amino acids in pro-Crp4-(20–43) exert differential effects in blocking pro-Crp4 bactericidal activity, the peptide activities of pro-Crp4 variants with partially reduced anionic charge in the distal (Asp20, Asp26, Glu27, and Glu28), mid (Glu32 and Glu33), and proximal (Glu37, Glu38, and Asp39) anionic clusters in pro-Crp4-(20–92) were compared with pro-Crp4-(20–92) (Fig. 1). Peptides with combined charge neutralization of the distal and proximal clusters, (6-DE/G)-pro-Crp4, and of the mid and proximal clusters, (5-DE/G)-pro-Crp4, were more active than pro-Crp4 but not as active as (DE/G)-pro-Crp4 or Crp4 (Fig. 3, A–F). The proximal pro-Crp4 variant (3-DE/G)-pro-Crp4 was attenuated compared with (6-DE/G)-pro-Crp4 and (5-DE/G)-pro-Crp4, suggesting that anionic clusters contribute to blocking pro-Crp4 activity in an additive manner (Fig. 4, A–D). However, mutagenesis of the distal anionic amino acid cluster showed a markedly greater and differential effect on pro-Crp4 bactericidal activity as compared with mid and proximal cluster charge neutralization (Fig. 4).
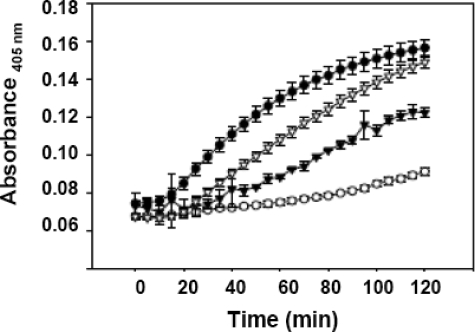
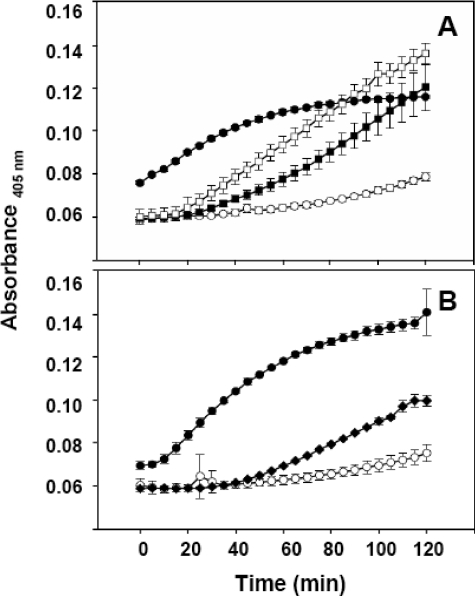
Mutagenesis of Anionic Amino Acid Residues in Proregion of Pro-Crp4 Enables Permeabilization of Live E. coli ML35 Cells—To test the hypothesis that individual clusters of anionic amino acids in pro-Crp4-(20–43) exert differential effects on defensin-microbe interactions and permeabilization of live bacteria, the pro-Crp4 variant peptides were compared with Crp4 and pro-Crp4 using the ONPG conversion assay (see “Experimental Procedures”). Consistent with their lack of bactericidal peptide activity, (DE/G)-pro-Crp4-(20–60) and pro-Crp4-(20–92) did not permeabilize E. coli (see supplemental Fig. S2). Relative to the effects of Crp4, bacterial exposure to 3 μm (DE/G)- and (DE/NQ)-pro-Crp4 resulted in a delay in measurable ONP accumulation (Fig. 5). In fact, at 3 μm, no ONP accumulation was detected as a result of exposure to pro-Crp4 variant peptides, suggesting an attenuation of membrane-disruptive activity for the partial electronegative variants (see also supplemental Fig. S3). On the other hand, at 6 μm, comparison of pro-Crp4 peptides with a combined charge neutralization of distal and proximal clusters or with only the proximal cluster neutralized supported the view that the anionic clusters provided an additive inhibitory effect on pro-Crp4-induced cell permeabilization (Fig. 6A). Deletion of the distal cluster conferred permeabilizing activity, but the induced ONPG conversion was not as robust as that caused by mature Crp4 (Fig. 6B).
FIGURE 5.
(DE/NQ)-pro-Crp4 exhibits attenuated permeabilization of live E. coli ML35 cells. Exponentially growing E. coli cells were exposed to 3 μm control peptides (•, Crp4; ○, pro-Crp4) and peptide variants in the presence of ONPG for 2 h at 37 °C. β-Galactosidase hydrolysis of ONPG was measured at 405 nm. ▿, (DE/G)-pro-Crp4; ▾, (DE/NQ)-pro-Crp4.
FIGURE 6.
The distal, mid, and proximal pro-Crp4 variants at 6 μm show differential effects on permeabilization of live E. coli ML35 cells. ONPG conversion was measured at 405 nm as described in the legend to Fig. 5. A, (6-DE/G)-pro-Crp4 (□) and (3-DE/G)-pro-Crp4 (▪); B, pro-Crp4-(29–92) (♦); A and B, Crp4 (•) and pro-Crp4 (○).
The Most N-terminal Electronegative Cluster Is Primarily Responsible for Maintaining Pro-Crp4 in an Inhibited State—Acidic amino acid residue positions in the most distal proregion cluster predominate in inhibiting pro-Crp4 bactericidal activity. For example, against S. enterica serovar typhimurium ΔphoP, L. monocytogenes, and E. coli, collective mutagenesis of the proximal and distal clusters ((6-DE/G)-pro-Crp4) increased peptide activity to a greater extent than mutagenesis at the proximal cluster alone ((3-DE/G)-pro-Crp4) (Fig. 3, A, B, and F). Mutagenesis of the distal cluster alone improved bactericidal activity much more than the combined mutagenesis of mid and proximal clusters, as evident in assays against S. enterica serovar typhimurium ΔphoP, L. monocytogenes, and E. coli (Fig. 4, A–C). For reasons that remain unclear, the recoveries of distal cluster variant (4-DE/G)-pro-Crp4 and combined proximal and mid cluster mutant (5-DE/G)-pro-Crp4 were unusually low, requiring that their bactericidal peptide activities be tested in separate experiments (Fig. 4, A–D). Remarkably, mutagenesis of the distal cluster resulted in (4-DE/G)-pro-Crp4 being as or more active than mature Crp4 at concentrations ≥1.5 μm, and the distal cluster mutant was more bactericidal than any partial charge neutralization mutants (Fig. 4A). To summarize, the relative bactericidal activities of all partial anionic pro-Crp4 variants are as follows: Crp4 = (DE/G)-pro-Crp4 = (4-DE/G)-pro-Crp4 > (6-DE/G)-pro-Crp4 = (5-DE/G)-pro-Crp4 > (3-DE/G)-pro-Crp4 > pro-Crp4. Thus, the acidic residues in the distal cluster of the proregion inhibit pro-Crp4 differentially.
Deletion of Distal Anionic Residue Cluster Confirms Its Inhibitory Role—To test the role of the distal acidic amino acid cluster further, a variant peptide, pro-Crp4-(29–92), with a deletion of the nine N-terminal amino acids of pro-Crp4-(20–92) was studied (Fig. 7, A–F). As observed for charge neutralization of the distal cluster in (4-DE/G)-pro-Crp4, deletion of the distal anionic cluster resulted in the peptide having equivalent or better activity than Crp4 (Fig. 7, A–C and F). Thus, whether the distal cluster was modified by charge neutralization or by deletion, the results confirmed that the most N-terminal acidic residue positions are mainly responsible for maintaining pro-Crp4-(20–92) in an inactive state.
DISCUSSION
This investigation tested whether specific cluster(s) of electronegative amino acids in pro-Crp4-(20–43) have specific positional effects that block bactericidal peptide activity and membrane disruption. Acidic residues positioned at the distal (Asp20, Asp26, Glu27, and Glu28), mid (Glu32 and Glu33), and proximal (Glu37, Glu38, and Asp39) clusters in pro-Crp4-(20–92) were mutagenized, and variants were assayed for differential effects of mutagenesis on bactericidal peptide activity. Certain substitutions produced additive effects on the induction of both bactericidal activity and permeabilization of live E. coli ML35 cells. In contrast, substitution of distal Glu and Asp residues with Gly or their deletion resulted in variants with bactericidal and membrane-disruptive activities equivalent to or greater than those of the fully mature Crp4 peptide. These findings support the conclusion that the distal anionic residues near the N terminus of pro-Crp4-(20–92) are primarily responsible for blocking Crp4-mediated activity in the full-length precursor.
Anionic amino acids in the proregion mediate inhibition of mouse pro-α-defensin bactericidal activity (21). α-Defensin bactericidal activity is determined by a combination of electropositive charge and amphipathicity (26–29), which enable target cell membrane disruption. Production of functional Crp4 requires specific proregion cleavage events mediated by MMP-7 (3, 12, 21, 30). Proteolysis at Ser43 ↓ Ile44 removes nine acidic residues in the proregion to produce the pro-Crp4-(44–92) intermediate, which is fully active, and findings presented here implicate the distal three acidic residues in pro-Crp4-(20–43) as having the predominant role in blocking precursor bactericidal action. Because (DE/NQ)-pro-Crp4 has intermediate bactericidal activity (Fig. 3), we cannot exclude the possibility that proregion-Crp4 side chain interactions that are not charge-neutralizing also contribute to blocking pro-Crp4 activity. Although the effect of neutralizing different anionic amino acid clusters suggested that the clusters exert additive inhibitory effects (Figs. 3 and 4), mutagenesis of the distal anionic amino acid residues alone, i.e. in (4-DE/G)-pro-Crp4 and pro-Crp4-(29–92), supports a greater role for residues positioned nearest the pro-Crp-(20–92) N terminus.
Individual pro-α-defensins may have evolved differing mechanisms to achieve the same inhibitory effect as those described for pro-Crp4. For example, in the human neutrophil α-defensin precursor pro-HNP-1, interactions between hydrophobic proregion residues with those in the HNP-1 moiety appear to be critical to maintaining pro-HNP-1 in an inactive state (30). Furthermore, in striking contrast to the findings we report, pro-HNP-1 remained inactive following deletion of nine amino acids from the pro-HNP-1 N terminus, including three acidic residues, showing that they are functionally dispensable in this respect (30). Thus, the role(s) of individual proregion residue positions in maintaining pro-α-defensin inhibition vary, possibly depending on the distribution of electropositive charge or the orientation of peptide amphipathicity along the α-defensin triple-stranded β-sheet topology. Clearly, these features vary with peptide primary structure in that all α-defensins share a common constrained tertiary structure. In this respect, Crp4 and RMAD-4 (rhesus myeloid α-defensin-4) differ markedly with regard to charge distribution and the orientation of hydrophobic side chains along the peptide fold. Still, neutralizing anionic amino acids in the proregion of pro-RMAD-4-(20–94) have the same activating effects as those described here for pro-Crp4-(20–92) (36). Possibly, the biochemistry of the HNP precursor may have a unique structural context, leaving the question open until additional molecules have been characterized.
To date, every pro-α-defensin investigated lacks bactericidal and membrane-disruptive activities (9, 18, 32), and prosegments of α-defensin precursors have a highly conserved hydrophobic motif (30) and balance the charge of their defensin partners (9, 15). Possibly, proregions are involved in attracting chaperones to facilitate trafficking of α-defensins during granulogenesis. For example, the human neutrophil defensin proregion is essential for in vitro folding (32), subcellular trafficking and sorting (31), and targeting (14). The proregion of pro-Crp4 is unstructured,4 making it difficult to visualize the actual interactions between the proregion and the mature peptide that inhibit defensin function. Also, in vitro, the pro-HNP-1 proregion, even in trans, improves the efficiency of peptide folding (32), suggesting that the proregion may confer some protection against a misfolded protein response (33–35). Conceivably, a membrane-disruptive prodefensin could alter the endoplasmic reticulum environment to induce endoplasmic reticulum stress and possibly trigger the unfolded protein response. Therefore, maintaining pro-α-defensins in an inactive state during biosynthesis and trafficking may be critical to the viability of both promyelocytes and Paneth cells, lineages that express these innate immune effector molecules at very high levels.
Supplementary Material
Acknowledgments
We thank Michael E. Selsted for useful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK044632 and AI059346. This work was also supported by the Human Frontiers Science Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S4, and Tables S1 and S2.
Footnotes
The abbreviations used are: MMP-7, matrix metalloproteinase-7; Crp4, cryptdin-4-(61–92); pro-Crp4, procryptdin-4-(20–92); (DE/G)-pro-Crp4, (D20G/D26G/E27G/E28G/E32G/E33G/E37G/E38G/D39G)-pro-Crp4-(20–92); (DE/NQ)-pro-Crp4, (D20N/D26N/E27Q/E28Q/E32Q/E33Q/E37Q/E38Q/D39N)-pro-Crp4-(20–92); (6-DE/G)-pro-Crp4, (D26G/E27G/E28G/E37G/E38G/D39G)-pro-Crp4-(20–92); (5-DE/G)-pro-Crp4, (E32G/E33G/E37G/E38G/D39G)-pro-Crp4-(20–92); (3-DE/G)-pro-Crp4, (E37G/E38G/D39G)-pro-Crp4-(20–92); (4-DE/G)-pro-Crp4, (D20G/D26G/E27G/E28G)-pro-Crp4-(20–92); (DE/G)-pro-Crp4-(20–60), (D20G/D26G/E27G/E28G/E32G/E33G/E37G/E38G/D39G)-proregion; ONP, 2-ortho-nitrophenol; ONPG, 2-ortho-nitrophenyl β-d-galactopyranoside; HPLC, high performance liquid chromatography; AU-PAGE, acid/urea-polyacrylamide gel electrophoresis; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; TSB, Trypticase soy broth; PIPES, 1,4-piperazinediethanesulfonic acid; CFU, colony-forming units.
H. J. Vogel, personal communication.
References
- 1.Selsted, M. E., and Ouellette, A. J. (2005) Nat. Immunol. 6 551-557 [DOI] [PubMed] [Google Scholar]
- 2.Ouellette, A. J. (2005) Springer Semin. Immunopathol. 27 133-146 [DOI] [PubMed] [Google Scholar]
- 3.Tanabe, H., Qu, X., Weeks, C. S., Cummings, J. E., Kolusheva, S., Walsh, K. B., Jelinek, R., Vanderlick, T. K., Selsted, M. E., and Ouellette, A. J. (2004) J. Biol. Chem. 279 11976-11983 [DOI] [PubMed] [Google Scholar]
- 4.Ouellette, A. J. (1999) Am. J. Physiol. 277 G257-G261 [DOI] [PubMed] [Google Scholar]
- 5.Ayabe, T., Satchell, D. P., Wilson, C. L., Parks, W. C., Selsted, M. E., and Ouellette, A. J. (2000) Nat. Immunol. 1 113-118 [DOI] [PubMed] [Google Scholar]
- 6.Porter, E. M., Bevins, C. L., Ghosh, D., and Ganz, T. (2002) CMLS 59 156-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zasloff, M. (2002) Nature 415 389-395 [DOI] [PubMed] [Google Scholar]
- 8.Ouellette, A. J., and Bevins, C. L. (2005) in Antimicrobial Peptides in Human Health and Disease (Gallo, R. L., ed) pp. 175-198, Horizon Scientific Press, Norfolk, UK
- 9.Ganz, T. (2003) Nat. Rev. Immunol. 3 710-720 [DOI] [PubMed] [Google Scholar]
- 10.Ganz, T. (2004) C. R. Biol. 327 539-549 [DOI] [PubMed] [Google Scholar]
- 11.Salzman, N. H., Ghosh, D., Huttner, K. M., Paterson, Y., and Bevins, C. L. (2003) Nature 422 522-526 [DOI] [PubMed] [Google Scholar]
- 12.Wilson, C. L., Ouellette, A. J., Satchell, D. P., Ayabe, T., Lopez-Boado, Y. S., Stratman, J. L., Hultgren, S. J., Matrisian, L. M., and Parks, W. C. (1999) Science 286 113-117 [DOI] [PubMed] [Google Scholar]
- 13.Ghosh, D., Porter, E., Shen, B., Lee, S. K., Wilk, D., Drazba, J., Yadav, S. P., Crabb, J. W., Ganz, T., and Bevins, C. L. (2002) Nat. Immunol. 3 583-590 [DOI] [PubMed] [Google Scholar]
- 14.Ganz, T., Liu, L., Valore, E. V., and Oren, A. (1993) Blood 82 641-650 [PubMed] [Google Scholar]
- 15.Michaelson, D., Rayner, J., Couto, M., and Ganz, T. (1992) J. Leukocyte Biol. 51 634-639 [DOI] [PubMed] [Google Scholar]
- 16.Valore, E. V., and Ganz, T. (1992) Blood 79 1538-1544 [PubMed] [Google Scholar]
- 17.Ayabe, T., Satchell, D. P., Pesendorfer, P., Tanabe, H., Wilson, C. L., Hagen, S. J., and Ouellette, A. J. (2002) J. Biol. Chem. 277 5219-5228 [DOI] [PubMed] [Google Scholar]
- 18.Shirafuji, Y., Tanabe, H., Satchell, D. P., Henschen-Edman, A., Wilson, C. L., and Ouellette, A. J. (2003) J. Biol. Chem. 278 7910-7919 [DOI] [PubMed] [Google Scholar]
- 19.Satchell, D. P., Sheynis, T., Kolusheva, S., Cummings, J., Vanderlick, T. K., Jelinek, R., Selsted, M. E., and Ouellette, A. J. (2003) Peptides (Elmsford) 24 1795-1805 [DOI] [PubMed] [Google Scholar]
- 20.Satchell, D. P., Sheynis, T., Shirafuji, Y., Kolusheva, S., Ouellette, A. J., and Jelinek, R. (2003) J. Biol. Chem. 278 13838-13846 [DOI] [PubMed] [Google Scholar]
- 21.Weeks, C. S., Tanabe, H., Cummings, J. E., Crampton, S. P., Sheynis, T., Jelinek, R., Vanderlick, T. K., Cocco, M. J., and Ouellette, A. J. (2006) J. Biol. Chem. 281 28932-28942 [DOI] [PubMed] [Google Scholar]
- 22.Maemoto, A., Qu, X., Rosengren, K. J., Tanabe, H., Henschen-Edman, A., Craik, D. J., and Ouellette, A. J. (2004) J. Biol. Chem. 279 44188-44196 [DOI] [PubMed] [Google Scholar]
- 23.Selsted, M. E. (1993) in Genetic Engineering: Principles and Methods (Setlow, J. K., ed) pp. 131-147, Plenum Press, New York
- 24.Jing, W., Hunter, H. N., Tanabe, H., Ouellette, A. J., and Vogel, H. J. (2004) Biochemistry 43 15759-15766 [DOI] [PubMed] [Google Scholar]
- 25.Ayabe, T., Wulff, H., Darmoul, D., Cahalan, M. D., Chandy, K. G., and Ouellette, A. J. (2002) J. Biol. Chem. 277 3793-3800 [DOI] [PubMed] [Google Scholar]
- 26.Matsuzaki, K., Mitani, Y., Akada, K. Y., Murase, O., Yoneyama, S., Zasloff, M., and Miyajima, K. (1998) Biochemistry 37 15144-15153 [DOI] [PubMed] [Google Scholar]
- 27.Zanetti, M., Gennaro, R., and Romeo, D. (1997) Ann. N. Y. Acad. Sci. 832 147-162 [DOI] [PubMed] [Google Scholar]
- 28.White, S. H., Wimley, W. C., and Selsted, M. E. (1995) Curr. Opin. Struct. Biol. 5 521-527 [DOI] [PubMed] [Google Scholar]
- 29.Hristova, K., Selsted, M. E., and White, S. H. (1997) J. Biol. Chem. 272 24224-24233 [DOI] [PubMed] [Google Scholar]
- 30.Zou, G., de Leeuw, E., Lubkowski, J., and Lu, W. (2008) J. Mol. Biol. 381 1281-1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, L., and Ganz, T. (1995) Blood 85 1095-1103 [PubMed] [Google Scholar]
- 32.Wu, Z., Li, X., Ericksen, B., de Leeuw, E., Zou, G., Zeng, P., Xie, C., Li, C., Lubkowski, J., Lu, W. Y., and Lu, W. (2007) J. Mol. Biol. 368 537-549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walter, P., and Johnson, A. E. (1994) Annu. Rev. Cell Biol. 10 87-119 [DOI] [PubMed] [Google Scholar]
- 34.Kuliawat, R., and Arvan, P. (1992) J. Cell Biol. 118 521-529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Traub, L. M., and Kornfeld, S. (1997) Curr. Opin. Cell Biol. 9 527-533 [DOI] [PubMed] [Google Scholar]
- 36.Kamdar, K., Maemoto, A., Qu, X., Young, S. K., and Ouellette, A. J. (2008) J. Biol. Chem. 283 32361-32368 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.