Abstract
Large networks of proteins govern embryonic stem (ES) cell pluripotency. Recent analysis of the critical pluripotency factors Oct4 and Nanog has identified their interaction with multiple transcriptional repression complexes, including members of the mSin3A-HDAC complex, suggesting that these factors could be involved in the regulation of Oct4/Nanog function. mSin3A is critical for embryonic development, but the mechanism by which the mSin3A-HDAC complex is able to regulate ES cell pluripotency is undefined. Herein we show that the mSin3A-HDAC complex positively regulates Nanog expression in ES cells through Sox2, a critical ES cell transcription factor and regulator of Nanog. We have identified the mSin3A-HDAC complex to be present at the Nanog promoter only under proliferating conditions concurrent with histone acetylation. We find that Sox2 associates with mSin3A-HDAC complex members both in vitro and in vivo, similar to the interactions found between Oct4/Nanog and the mSin3A-HDAC complex. Knockdown of mSin3A-HDAC complex members or HDAC inhibitor treatment reduces Nanog expression, and overexpression of mSin3A-HDAC complex subunits stimulates Nanog expression. Our data demonstrate that the mSin3A-HDAC complex can positively regulate Nanog expression under proliferating conditions and that this activity is complementary to mSin3A-mediated p53-dependent silencing of Nanog during differentiation.
Precise modulation of transcriptional activation and repression in ES2 cells is crucial for proper development, lineage differentiation, and genomic stability. At the core of ES cell self-renewal are the transcription factors Oct4, Sox2, and Nanog. These factors are central to maintaining pluripotency, as well as directing appropriate lineage commitment (1, 2). In addition to regulating the transcription of other genes, Oct4, Nanog, and Sox2 also activate one another's transcription. Oct4 and Sox2 together recognize and bind to a highly conserved consensus sequence, which is essential for Nanog expression in both mouse and human ES cells (3, 4). In addition to Oct4 and Sox2, many other transcription factors have been linked to regulation of Nanog expression and ES cell pluripotency. For example, Sall4 (5), FoxD3 (6), and STAT3 and T (7) transcription factors positively regulate Nanog expression, whereas p53 (8) and GCNF (9) suppress Nanog expression.
In addition to specific transcription factors, the active or inactive transcriptional state of genes is established through highly regulated modulation of underlying chromatin structure (10). This is achieved by a large family of histone-modifying and chromatin-remodeling enzymes (11). Nanog is known to interact with a variety of chromatin-modifying complexes. Analysis of the Nanog interaction network has shown that it is linked to repressor proteins (12). A recent protein interaction study identified the interaction of Oct4 and Nanog with a novel repressor complex, NODE, which contains several chromatin-associated proteins, including mSin3A, HDAC1, and HDAC2 (13). ES cells that have lost Mbd3, a component of the nucleosome-remodeling complex NuRD, show LIF-independent growth with no effect on Nanog expression or in the expression of lineage-specific genes (14), illustrating how chromatin-modifying complexes can regulate Nanog expression. In contrast, knockdown of Mta1/2-containing repression complexes led to ES cell differentiation by specifically up-regulating endoderm lineage markers (13). Collectively, these studies show that Nanog and Oct4 interact with multiple repression complexes to regulate their target genes and hence to control the fate of ES cells.
HDAC1 and HDAC2 are members of a number of deacetylase complexes, including the mSin3A-HDAC complex, the NuRD complex, the BCH10-containing complex, and the CoREST complex (15). Although they have broadly similar functions in regulating transcription, the mSin3A-HDAC complex can be distinguished from these complexes by the presence of the mSin3A protein. mSin3A has been well described as a core component of a multiprotein co-repressor complex known to silence gene expression by deacetylating histones (for review, see Ref. 16) and has been shown to play an essential role in early embryonic development (17). ES cells derived from mSin3A-/- blastocysts form significantly smaller colonies and eventually die in culture compared with wild-type or mSin3A+/- ES cells (18). Likewise, HDAC1 null ES cells also show growth defects compared with wild-type ES cells, and loss of HDAC1 contributes to embryonic lethality prior to E10.5 (19). HDAC2-/- mice display difficulty progressing through gestation (20). These observations allude to a function of the mSin3A-HDAC complex in ES cell survival and proliferation. However the mechanism by which mSin3A-HDAC complex regulates ES cells still remains unclear.
Our goal was to investigate the mechanisms underlying Nanog transcription. We sought out co-activator or co-repressor complexes (21) that could regulate Oct4 or Sox2, and consequently Nanog itself. Using chromatin immunoprecipitation (ChIP) we have identified components of the mSin3A-HDAC complex present at the Nanog promoter under proliferating conditions, despite also finding acetylated histones at this loci. This binding is lost upon differentiation, concurrent with histone deacetylation. Depletion of mSin3A-HDAC activity by siRNA or treatment with HDAC inhibitors negatively affects Nanog transcription but does not similarly affect Oct4. Overexpression of mSin3A-HDAC complex members stimulates expression from the Nanog promoter in a reporter system. In addition, mSin3A-HDAC complex members interact with Sox2 in vitro and in vivo, providing a link between a factor known to positively regulate Nanog expression and the mSin3A-HDAC complex. Our studies demonstrate that the mSin3A-HDAC complex is required for ES cell proliferation due to its role in positively regulating the expression of Nanog.
EXPERIMENTAL PROCEDURES
ES Cell Culture—Murine J1 ES cells were grown under typical ES cell conditions on gelatinized tissue culture plates in feeder-free conditions. For differentiation, ES cells were plated at 1.5 × 105 cells/ml in differentiation media (Dulbecco's modified Eagle's medium (Invitrogen, 11965-092), 10% fetal bovine serum (HyClone, SH30070), penicillin/streptomycin (Invitrogen, 15140-122), and 5 μm retinoic acid (Sigma, R2625)) for 5 days with a change in medium every other day. For drug treatments, adherent ES cells at 75% confluence were washed in phosphate-buffered saline, then incubated with normal ES cell media plus 5 μm retinoic acid, 100 ng/ml trichostatin A (Millipore, 19-138), 5 mm sodium butyrate (Millipore, 19-137), or 3 mm valproic acid (Sigma, P4543) for 3 or 6 h as noted.
Plasmid Construction—To generate a pNusA-Sox2 bacterial expression vector, a Gateway Conversion Cassette, RFA isoform (Invitrogen) was inserted into pET43.1a(+) (Novagen) downstream of the NusA coding sequence. Next, a C-terminal STREP-II purification tag sequence (IBA, Germany) was introduced downstream of the Gateway attR2 site, in-frame with cDNA sequences introduced by Gateway LR recombination (Invitrogen). Lastly, murine Sox2 cDNA was introduced into this plasmid by Gateway LR recombination with a Sox2 Gateway donor vector. To generate luciferase expression plasmids pGL3-TK-Luc and pGL3-O/S-TK-Luc, plasmids pGL3-Basic (Promega, E1751, GenBank™/EMBL accession number U47295) and pRL-TK (Promega, E2241, GenBank™/EMBL accession number AF025846) were digested with BglII/HindIII. The TK promoter from pRL-TK was subcloned into pGL3-Basic to generate pGL3-TK-Luc. pGL3-TK-Luc was digested with KpnI/BglII. Oligonucleotides containing tandem Oct4/Sox2 binding sites from the Nanog promoter were annealed together and ligated into the digested pGL3-TK to generate pGL3-O/S-TK-Luc. Forward primer: 5′-cttcttttgcattacaatgtccattcttttgcattacaatgtccattcttttgcattacaatgtccaa-3′. Reverse primer: 5′-gatcttggacattgtaatgcaaaagaatggacattgtaatgcaaaagaatggacattgtaatgcaaaagaaggtac-3′. To generate expression plasmids for Oct4, Sox2, mSin3A, and HDAC2, total mouse ES cell cDNA was amplified using reverse-transcription (Bio-Rad, 170-8891) from total RNA. Gene-specific primers for Oct4, Sox2, mSin3A, and HDAC2 were used to amplify the cDNA then recombined into a Gateway donor vector (Invitrogen), which was then sequence-verified. The cDNAs were introduced into pDEST26 (Invitrogen, 11809-019) by Gateway LR recombination.
Chromatin Immunoprecipitation—ChIP was performed using 2.0 × 107 ES cells as described in the online protocol provided by Upstate (Millipore). Purified DNA was amplified with primers spanning the Oct4-Sox2 binding site (forward primer: 5′-ggatgtctttagatcagaggatgccc-3′; reverse primer: 5′-ccacagaaagagcaagacaccaacc-3′) and p53 binding site (forward primer: 5′-tcagacttgcgttaaaaagccgcac-3′; reverse primer: 5′-gcttagggggcatcctctgatct-3′) on the Nanog promoter and Oct4-Sox2 binding site (forward primer: 5′-ggcacgcttagggctaacctg-3′; reverse primer: 5′-ctccactctgtcatgctcacctcc-3′) on the Oct4 promoter using Applied Biosystems SYBR PCR mastermix (Applied Biosystems, 4309155). Relative enrichment was calculated as 2⁁(ΔCT(control CHIP) - ΔCT(experimental CHIP)), where ΔCT is equal to the CT(immunoprecipitated sample) - CT(input). Antibody sources were: Histone H3 Ac (Millipore, 06-942), H3 K4 2Me (Millipore, 07-030), H3 K4 3Me (Millipore, 07-473), H3 K9 1Me (Millipore, 07-450), H3 K9 2Me (Millipore, 07-441), H3 K27 3Me (Millipore, 05-851), H3 K79 2Me (Millipore, 05-835), Sox2 (Abcam, ab15830), RNAPII (Abcam, ab817), HDAC1 (Abcam, ab7028), HDAC2 (Abcam, ab7029), Oct4 (R&D Biosystems, AF1759), Nanog (Cosmo Bio Co., RECRCAB0002P-F), and mSin3A (Santa Cruz Biotechnology, sc994).
RNA Isolation, qPCR, and Analysis of Transcript Levels—Total RNA was purified with an RNeasy Miniprep kit (Qiagen, 74106), DNase-treated (Promega, M6101), and reverse-transcribed (Bio-Rad, 170-8891). Quantitative PCR (qPCR) reactions were performed using 10 ng of cDNA in an ABI7500 Fast Real-Time PCR System as per the manufacturer's instructions (Applied Biosystems). TaqMan gene expression assays (all from Applied Biosystems) were are follows: Oct4 (Mm00658129_ gH), Sox2 (Mm00488369_s1), Hand1 (Mm00433931_m1), HoxA5 (Mm00439362_m1), Nestin (Mm00450205_m1), mSin3A (Mm00488255_m1), Rex1 (Mm01194090_g1), Bmp4 (Mm00432087_ m1), Gata4 (Mm00484689_m1), Sox17 (Mm00488363_m1), Cdx2 (Mm00432449_m1), Eomes (Mm01351985_m1), Fgf5 (Mm00438919_m1), p21 (Mm00432448_m1), and Mdm2 (Mm01233136_m1). A custom TaqMan assay was designed for Nanog gene expression: forward primer, 5′-TCCTCGCCCTTCCTCTGAA-3′; reverse primer, 5′-CAGGACTTGAGAGCTTTTGTTTGG-3′; and, reporter sequence, 5′-CAGCCCTGATTCTTCT-3′. To calculate changes in gene expression, average threshold values (CT) were determined for three PCR reactions and calculated as -fold change compared with control samples using the comparative ΔΔCT method.
RNA Interference—SMARTpool siRNA targeting HDAC1 (Dharmacon, M-040287-01), HDAC2 (Dharmacon, M-046158-00), mSin3A (Dharmacon, M-044653-00), or control siRNA targeting Lamin (Dharmacon, D-001050-01) were incubated with DharmaFECT4 (Dharmacon, T-2004) in Opti-MEM (Invitrogen, 319850-62) for 30 min at room temperature to allow siRNA-lipid complexes to form. 3.0 × 105 ES cells were added to the transfection reagents for final concentrations of 84 nm siRNA and 1.17 μl/ml DharmaFECT4. For siRNA combinations (targeting 2–3 genes), the total siRNA concentration remained constant (42 nm of each of two or 28 nm of each of 3 targets). Total RNA and protein were harvested from cells after incubation with siRNA for 96 h.
Protein Purification and Protein-Protein Interactions— NusA-tagged Sox2 was expressed in Escherichia coli BL21(DE3) cells (Invitrogen, C6000-03) and purified under native conditions using Streptactin resin (IBA, 2-1232-005). NusA pull-down assays were carried out using 15 μg of NusA-Sox2 and 0.5 mg of ES cell nuclear extract. Endogenous Sox2 and interacting proteins were identified using 300 μg of nuclear and cytoplasmic fractions (NE-PER, Pierce 78833) from drug-treated or untreated ES cells with 3 μg of Sox2 antibody (Abcam, ab15830). Bound complexes were washed in Nonidet P-40 buffer, resuspended in SDS sample buffer, and electrophoresed on an SDS-PAGE gel and blotted onto a polyvinylidene difluoride membrane according to standard procedures. Blots were probed with primary antibodies: HDAC1 (Santa Cruz Biotechnology, sc7872), HDAC2 (Santa Cruz Biotechnology, sc7899), mSin3A (Santa Cruz Biotechnology, sc994), Nanog (Millipore, AB5731), and Sox2 (Millipore, AB5603).
Size-exclusion Chromatography—ES cell nuclear extract was prepared with Pierce NE-PER reagents. 11 mg of extract was dialyzed against 1 liter of Nonidet P-40 buffer (50 mm Tris, 150 mm NaCl, 1% Nonidet P-40, 5 mm EDTA), then concentrated with Amicon Ultra-15 concentrator columns (Millipore, UFC901096). 1 ml (∼10 mg) of dialyzed extract was loaded onto two tandem Superose 6 10/300 GL columns (GE Life Sciences, 17-5172-01) under isotonic conditions with a flow rate of 0.2 ml/min using a Bio-Rad Duoflow FPLC. 150 0.4-ml fractions were collected, and 30 μl from every fifth fraction was run on an SDS for Western blotting for mSin3A and Sox2.
Luciferase-reporter Assays—NIH3T3 cells were seeded at a density of 2.5 × 105 cells per well of a 6-well dish and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1× penicillin/streptomycin and maintained at 37 °C and 5% CO2. DNA transfections were carried out using Lipofectamine 2000 (Invitrogen, 11668-019) as per the manufacturer's instructions. Cells were harvested after 48 h, and the luciferase activity of the lysate was measured with the Dual-Luciferase reporter assay system (Promega, E1960) using the Envision luminometer (PerkinElmer Life Sciences).
RESULTS
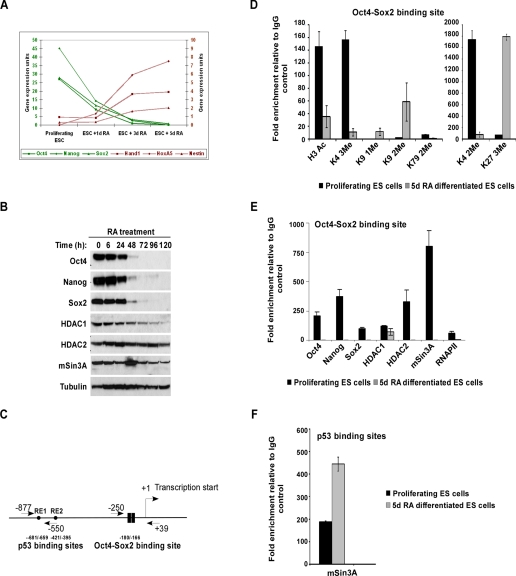
mSin3A-HDAC Complex Binds to the Nanog Promoter during ES Cell Proliferation—To gain insight into Nanog transcriptional regulation in mouse ES cells, we identified factors that occupy the Nanog promoter during ES cell proliferation and differentiation by ChIP. To accomplish this, we established 5-day retinoic acid (RA) treatment minus LIF as a benchmark for differentiated ES cells. This treatment completely differentiates the cells as marked by loss of Oct4, Sox2, and Nanog expression (Fig. 1, A and B), and increased expression of lineage markers Hand1, HoxA5, and Nestin (Fig. 1A). We chose this method of differentiation over LIF withdrawal, because RA uniformly differentiates ES cells in to neural lineage (22), whereas LIF withdrawal results in heterogeneous expression of pluripotency and lineage markers. We compared histone modifications and binding of a panel of factors at the Nanog promoter between 5-day RA differentiated and proliferating ES cells, using qPCR primers that span the Oct4-Sox2 binding site (Fig. 1C). As expected, we found active histone modifications (H3Ac, H3K4 di- and trimethyl, and H3K79 dimethyl) (Fig. 1D), transcription factors (Oct4, Nanog, and Sox2), and RNA polymerase II (Fig. 1E) present at the Nanog in the promoter only under proliferating conditions, and not after differentiation. Additionally, repressive histone modifications (H3K9 mono- and dimethyl and H3K27 trimethyl) (Fig. 1D) were present only after differentiation. Surprisingly, we observed HDAC1, HDAC2, and mSin3A, members of the mSin3A-HDAC complex, at the Oct4-Sox2 binding site on the Nanog promoter under proliferating, but not differentiated conditions (Fig. 1E). Because the mSin3A-HDAC complex has been well characterized as a transcriptional repressor, we did not expect to find it at the promoter of an actively transcribed gene. We assayed the expression of mSin3A, HDAC1, and HDAC2 over a 5-day RA treatment of ES cells, and detected all components throughout the time course (Fig. 1B) indicating that the loss of the complex from the Nanog promoter is not due to a global loss of these proteins. As previously reported (8), we also observed increased mSin3A occupancy at the p53 binding sites (diagram of Nanog promoter in Fig. 1C) on the Nanog promoter following RA-induced differentiation (Fig. 1F). These data clearly demonstrate that occupancy of mSin3A at the Oct4-Sox2 binding site is distinct from that of the p53 binding site, and it is possible that mSin3A may have varying functions under different cellular conditions.
FIGURE 1.
Recruitment of the mSin3A-HDAC complex to the Nanog promoter during ES cell proliferation. A, qPCR analysis of ES cells in the presence of 5 μm retinoic acid (RA) showing a decrease in pluripotency markers Oct4, Nanog, and Sox2 and an increase in lineage markers Hand1, HoxA5, and Nestin. B, Western blot showing levels of Oct4, Nanog, Sox2, HDAC1, HDAC2, and mSin3A upon RA treatment (5 μm). Tubulin was used as a loading control. C, schematic of the proximal Nanog promoter showing positions of Oct4-Sox2 and p53 binding sites relative to the transcription start site. Arrows denote location of primers used for chromatin immunoprecipitation (ChIP) relative to the transcription start site.D and E, ChIP assays were performed for histone modifications, transcription factors, and deacetylase complex members as indicated using protein extracts of proliferating and 5-day RA-treated ES cells, with primers amplifying the Oct4-Sox2 binding site of the Nanog promoter. Values are expressed as -fold enrichment relative to IgG control ChIP. Results shown are the average of three independent PCR reactions. F, ChIP from proliferating and 5-day RA-treated ES cells using antibodies to mSin3A with primers amplifying the p53 binding site of the Nanog promoter.
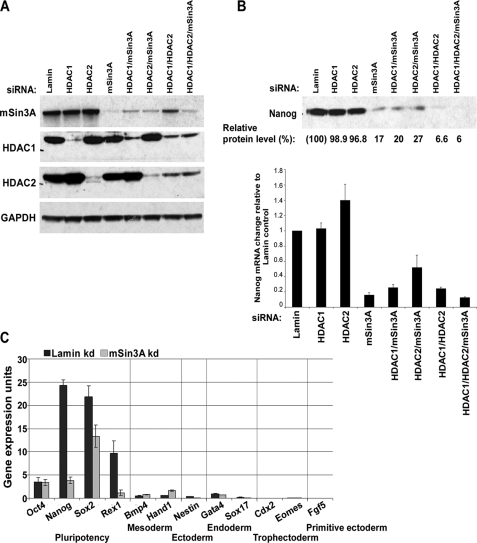
Knockdown of mSin3A Diminishes Nanog Expression—Because the mSin3A-HDAC complex plays a central role in transcriptional repression (15), it was intriguing to find it bound to the Nanog promoter during proliferation, especially because the histones at this site are heavily acetylated. To assay the possible role for this complex in Nanog regulation, we used siRNAs to inhibit the expression of mSin3A-HDAC complex subunits. If the mSin3A-HDAC complex is essential for positively regulating Nanog, loss of mSin3A-HDAC complex members would be expected to diminish Nanog expression. We knocked down HDAC1, HDAC2, and mSin3A (individually or in combination) with siRNAs for 96 h, and then analyzed Nanog transcript and protein levels. We saw efficient knockdown of target proteins (Fig. 2A) and found that levels of Nanog protein and mRNA dropped by ∼80% in the presence of mSin3A siRNA (alone or in combination with HDAC1/2) with no significant change in Nanog levels by HDAC1 or HDAC2 siRNA alone (Fig. 2B). This suggests that mSin3A is the key member of the complex regulating Nanog expression. We did see a reduction in the level of Nanog by knocking down both HDAC1 and HDAC2 together, but this knockdown also resulted in roughly 60% reduction of mSin3A protein levels (Fig. 2A), further suggesting a key role for mSin3A in Nanog regulation. We also examined earlier time points of mSin3A siRNA knockdown and observed that mSin3A levels were reduced by ∼80% within 24 h of siRNA treatment, and Nanog levels dropped by ∼50% (data not shown). To rule out the possible involvement of p53 in suppressing Nanog expression, we assayed expression of direct p53 targets p21 and Mdm2. Activated p53 increases expression of both p21 and Mdm2, but under these conditions we saw very little change in their expression (data not shown), indicating that p53 has not been activated. This suggests that mSin3A siRNA-mediated down-regulation of Nanog is independent of the p53 pathway. We next wanted to examine the expression of differentiation markers in the mSin3A knockdown ES cells, to eliminate the possibility that changes in Nanog levels are being caused by differentiation due to the loss of mSin3A. As seen previously (Fig. 2B), mSin3A knockdown resulted in decreased expression of not only Nanog but also in Sox2 and Rex1 levels (Fig. 2C). However, mSin3A knockdown had no effect on expression of Oct4 or a variety of lineage markers (Bmp4, Hand1, Gata4, Sox17, Cdx2, Eomes, and Fgf5) (Fig. 2C). Taken together, these results suggest that reduction in mSin3A levels reduces Nanog expression, and this effect is not due to an induction of differentiation either through p53 activation or mSin3A itself.
FIGURE 2.
RNA interference knockdown of mSin3A-HDAC complex members affects Nanog expression without inducing differentiation. A, Western blot showing levels of mSin3A, HDAC1, and HDAC2 proteins in siRNA-treated ES cells. Lamin siRNA was used as a negative control. Glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. B, levels of Nanog protein (top) and mRNA (bottom) in response to knockdown of mSin3A and associated HDAC members; 96 h after indicated siRNA treatment. Relative protein levels were quantified by image densitometry. Values for Nanog mRNA and protein were normalized to Lamin siRNA. C, mRNA expression levels of pluripotency-related genes (Oct4, Nanog, Sox2, and Rex1), mesodermal lineage markers (Bmp4 and Hand1), ectodermal lineage marker (Nestin), endodermal lineage markers (Gata4 and Sox17), trophectodermal lineage markers (Cdx2 and Eomes), and primitive ectoderm lineage (Fgf5) after siRNA treatment for Lamin (control) or mSin3A.
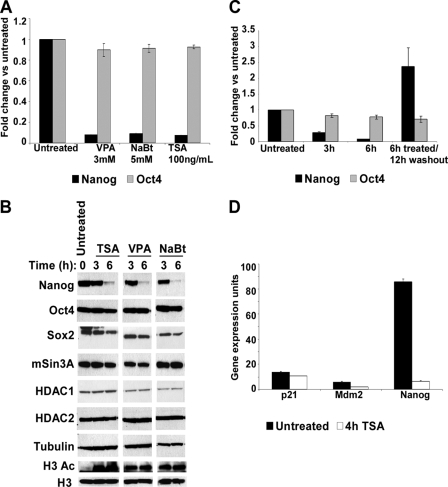
Enzymatic Activity of the mSin3A-HDAC Complex Is Vital for Nanog Expression—Because knockdown of the mSin3A-HDAC complex suggests a positive role for this co-repressor complex in Nanog regulation, we questioned if the enzymatic activity of the complex is important for regulating Nanog. To test this, we blocked deacetylase activity in ES cells by treating them with the HDAC inhibitors valproic acid, sodium butyrate, or trichostatin A (TSA). We found that, within 6 h, ES cells treated with any of these inhibitors showed significantly lower Nanog transcript (Fig. 3A) and protein levels (Fig. 3B) as measured by qPCR and Western blot analysis, respectively. Global histone acetylation was robustly increased (Fig. 3B), demonstrating the potent activity of the deacetylase inhibitors. The effect of these inhibitors did not permanently impact pluripotency, because Nanog expression could be rescued by drug withdrawal (Fig. 3C), and Oct4 levels were largely unaffected (Fig. 3, A–C). In addition, alkaline phosphatase staining and morphology analysis were similar to untreated ES cells (data not shown). To further validate that the reduction in Nanog is p53-independent, we assayed the expression of p53 target genes p21 and Mdm2. Neither of these two targets showed any change in their expression in the TSA-treated ES cells (Fig. 3D), highlighting that prevention of deacetylase activity directly leads to the reduction of Nanog, independent of p53 activity.
FIGURE 3.
HDAC inhibitors reduce Nanog gene expression. A, qPCR analysis of Nanog and Oct4 gene expression in the presence of valproic acid (VPA), sodium butyrate (NaBt), and trichostatin A (TSA) for 6 h expressed as -fold change over those untreated. B, Western blot showing levels of Nanog, Oct4, Sox2, mSin3A, HDAC1, and HDAC2 protein levels in the presence of TSA, valproic acid, and sodium butyrate for the times indicated. Total acetylated histone H3 levels were analyzed using H3Ac-specific antibodies as a positive control for HDAC inhibitor treatment. Tubulin and total histone H3 were used as loading controls. C, qPCR analysis of Nanog and Oct4 mRNA in the presence of 100 ng/ml TSA expressed as -fold change over those untreated. Duration of treatment and washout is indicated at the bottom. D, qPCR analysis of Nanog, p21, and Mdm2 mRNA in the presence of 100 ng/ml TSA for 4 h expressed as gene expression units relative to glyceraldehyde-3-phosphate dehydrogenase.
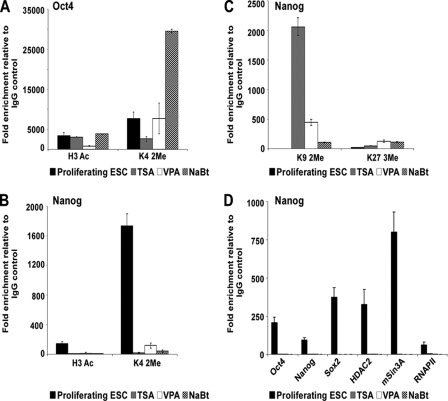
We next examined histone modifications at the Nanog and Oct4 promoters after treatment with deacetylase inhibitors. The Oct4 promoter still displayed active histone modifications in the presence of deacetylase inhibitors (Fig. 4A), consistent with a transcriptionally active locus. Following inhibitor treatment, the Nanog promoter showed loss of active histone modifications (Fig. 4B), an increase in repressive chromatin modifications (Fig. 4C), and complete loss of Oct4, Nanog, Sox2, mSin3A-HDAC complex, and RNA polymerase II (Fig. 4D). These results indicate that there is an HDAC-sensitive step in the regulation of Nanog and that the enzymatic activity of the mSin3A-HDAC complex is critical to Nanog expression.
FIGURE 4.
Analysis of Nanog and Oct4 promoters in the presence of HDAC inhibitors. A and B, ChIP analysis for active histone modifications (H3 Ac and H3 K4 2Me) at the Oct4 (A) and Nanog (B) promoters in the presence of HDAC inhibitors. C, ChIP analysis for repressive histone modifications (H3 K9 2Me and H3 K27 3Me) at the Nanog promoter in the presence of HDAC inhibitors. D, ChIP analysis for transcription and chromatin modifying factors at the Nanog promoter in the presence of HDAC inhibitors.
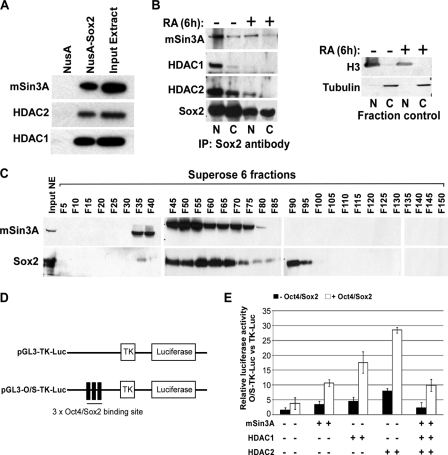
mSin3A-HDAC Complex Interacts with Sox2 to Positively Regulate Nanog Expression—Given that deacetylase activity is important for Nanog expression, we wanted to determine the target of the deacetylase activity of mSin3A-HDAC complex. We reasoned that it might be a non-histone factor present at the Nanog promoter, because the histones at the Nanog promoter remain acetylated in the presence of the mSin3A-HDAC complex under proliferating conditions. We focused our analysis on Sox2, because previous studies have shown that activity of SRY, a prototypical member of the Sox family, is regulated by HDAC3 during mammalian sex determination (23). A recent study by Liang and colleagues (13) has shown that both Nanog and Oct4 associate with multiple deacetylase-containing complexes, including NuRD, mSin3A, and Pml, but did not demonstrate the mechanism by which these complexes might regulate Nanog or Oct4 activity. Pull-down experiments using NusA-tagged Sox2 and ES cell nuclear extract showed that Sox2 interacts with the mSin3A-HDAC complex (Fig. 5A). To confirm these observations, we co-immunoprecipitated endogenous Sox2 from nuclear and cytoplasmic fractions of ES cells treated for 6 h with either RA or vehicle control. We confirmed the purity of the nuclear and cytoplasmic fractions using Western blots for histone H3 and α-tubulin. Sox2 interacted strongly with mSin3A, HDAC1, and HDAC2 in the nuclear fraction of vehicle-treated ES cells (Fig. 5B), and to a lesser extent with mSin3A and HDAC2 in the cytoplasm of vehicle-treated ES cells. Following RA treatment, there was a marked reduction in the robustness of this interaction. These results demonstrate that Sox2 interacts with the mSin3A-HDAC complex in vitro and in vivo, and this interaction is more prominent under proliferating conditions.
FIGURE 5.
The mSin3A-HDAC complex interacts with Sox2 to cooperatively stimulate Nanog transcription. A, in vitro NusA-Sox2 pulldown from ES cell nuclear extract. mSin3A-HDAC complex subunits were detected by Western blot. B, immunoprecipitation of endogenous Sox2 from nuclear and cytoplasmic fractions of ES cells treated for 6 h with 5 μm RA or vehicle. mSin3A-HDAC complex subunits were detected by Western blot using the indicated antibodies. Western blots for H3 and tubulin indicate the purity of the fractions. C, fractionation of ES cell nuclear extract. Western blots of every fifth fraction indicate that mSin3A and Sox2 are present in majority of the same fractions. D, schematic diagram of the pGL3-TK-Luc and pGL3-O/S-TK-Luc constructs. pGL3-TK-Luc has luciferase driven by the TK promoter. pGL3-O/S-TK-Luc contains three tandem copies of the Oct4-Sox2 binding site from the Nanog promoter inserted upstream of the TK promoter. E, increase in luciferase activity by the addition of Oct4/Sox2, and mSin3A-HDAC complex members. The graph represents the -fold increase in luciferase activity between pGL3-O/S-TK-Luc and pGL3-TK-Luc after adding the indicated factors.
To further confirm the interaction of Sox2 with mSin3A, we performed size-exclusion chromatography on ES cell nuclear extract. We fractionated the nuclear extract into 150 fractions, and probed every 5th fraction for mSin3A and Sox2. Most of the fractions that contained Sox2 also contained mSin3A, confirming that they are part of the large complex (Fig. 5C). We do find some fractions with only mSin3A, or only Sox2, indicating that these two proteins likely have biological functions independent of one another as well.
Because we find both the mSin3A-HDAC complex and Sox2 at the Nanog promoter during active transcription of Nanog, we wanted to determine if the mSin3A-HDAC complex cooperates with Sox2 to promote Nanog expression. To address this, we took advantage of a luciferase reporter system in NIH3T3 cells. These cells do not normally express Oct4, Nanog, or Sox2, which allows us to more finely control for which factors are affecting Nanog expression. We engineered three tandem repeats of the Oct4/Sox2 binding sites into the pGL3-TK-Luc reporter vector (Fig. 5D), where the thymidine kinase promoter drives a low level of luciferase expression. This vector, pGL3-O/S-TK-Luc, or its parental vector was transfected into NIH3T3 cells, along with expression vectors for Oct4 and Sox2. Transfection of pGL3-O/S-TK-Luc together with Oct4/Sox2 expression vectors more than doubled the luciferase activity over the parental vector with Oct4/Sox2 (Fig. 5E), verifying that addition of the binding sites increased Oct4/Sox2-mediated transcription. We next transfected the mSin3A-HDAC complex members into the assay system either with or without Oct4/Sox2. Although the absolute increase in luciferase varied between HDAC1, HDAC2, or mSin3A, the addition of any of the complex members increased luciferase activity ∼3-fold over the complex member without Oct4/Sox2. The increased luciferase activity with HDAC1 or HDAC2 compared with mSin3A may be due to interaction of these factors with endogenous mSin3A protein in 3T3 cells. We next added all three mSin3A-HDAC complex members together (±Oct4/Sox2), however we did not see any further increase in luciferase activity compared with that seen with each factor alone. Taken together, these results suggest that the mSin3A-HDAC complex positively regulates Nanog expression, and this is enhanced by the presence of Oct4 and Sox2.
DISCUSSION
Broad cohorts of proteins are involved in the regulation of Nanog (10). We reveal a novel role for the mSin3A-HDAC complex in Nanog transcriptional regulation, whereby the mSin3A-HDAC complex binds to the Nanog promoter at the Oct4/Sox2 binding sites, and its occupancy at the promoter correlates with active Nanog transcription. Luciferase assays confirm that the mSin3A-HDAC complex increases the transcriptional activity from these binding sites upon co-expression with Oct4/Sox2. This transcriptional activator role of mSin3A is different from its p53-dependent role in Nanog silencing (8). When we examined the region of p53 binding site on the Nanog promoter, we also observed increased mSin3A occupancy following RA-induced differentiation (Fig. 1F). It is not unexpected that the mSin3A-HDAC complex could operate in different capacities at these distinct sites to positively or negatively regulate Nanog expression, perhaps through interactions with specific transcription factors. However, the reduction in Nanog expression we see with mSin3A siRNA is independent of the p53 pathway.
Although core histones are the primary substrate of the mSin3A-HDAC complex, increasing evidence points to the importance of regulation of non-histone proteins by this complex (for review, see Ref. 24). Interaction of the mSin3A-HDAC complex with various factors impairs their ability to activate transcription of target genes (25–28). Recent protein interaction studies identified Nanog and Oct4 as novel interactors of transcriptional repressor complexes, including mSin3A-HDAC (12, 13). Wang and colleagues describe both Nanog and Oct4 as interactors of HDAC2 in proliferating ES cells (12), and Liang and colleagues demonstrate that Nanog and Oct4 associate with additional repressor proteins, including mSin3A-containing complexes (13). Our studies focused primarily on the mSin3A-HDAC complex and found that it interacts with Sox2. We show that this interaction can be largely disrupted by inducing differentiation in ES cells. The presence of the mSin3A-HDAC complex at the Nanog promoter can also be disrupted by inducing differentiation or by deacetylase inhibitor treatment, suggesting that acetylation may be important in the regulation of these pluripotency factors.
The activities of both histone deacetylases and acetyltransferases have been shown to play a role in ES cell differentiation. Blocking deacetylation of histones using small molecules such as TSA delays the formation of embryoid bodies, indicating that histone deacetylation is necessary for full progression through differentiation (29). However, analysis by McCool and colleagues (30) indicated that there is a global increase in acetylation over the course of differentiation of ES cells and that changes in histone acetylation during differentiation vary across different promoters. Additionally, the authors demonstrate changes in gene expression can be seen within 2 h of TSA treatment. In our study, we examine early stages of deacetylase inhibition by assaying levels of pluripotency factors after 3 or 6 h of TSA treatment. We find that one of the earliest events during deacetylase inhibition (3 h) is the down-regulation of Nanog expression without a concurrent change in Oct4 levels, and we believe that this change is tied to the activity of the deacetylase enzymes. We have also found that long term TSA treatment differentiates ES cells (data not shown). There are clearly diverse roles for deacetylases in ES cells, and it will be interesting to see how different genes are regulated by deacetylases during development.
Histone deacetylase enzymes are most commonly associated with the suppression of eukaryotic gene transcription (for review, see Ref. 31). However, there are a few reports documenting a role for deacetylases in gene activation. For example, Sin3p, the yeast homologue of mSin3A, has been shown to regulate transcriptional activation of Hog1 target genes by deacetylating target promoters and conferring resistance to osmotic stress (32). YY1 is actively acetylated and deacetylated, and acetylation of its zinc finger domains decreases its ability to bind DNA (33). We observed that the mSin3A-HDAC complex stimulates Nanog expression, and we surmise that this effect is mediated by interaction with Sox2. It will be interesting to determine if the mSin3A-HDAC complex is involved in regulating the activity of Sox2. In addition to repressor complexes, recent reports using Chip-seq technology demonstrate that the co-activator CBP/p300 is recruited to genomic sequences bound by clusters of transcription factors that include Nanog, Oct4, and Sox2 (34). Depletion of Nanog, Oct4, and Sox2 by RNA interference reduced binding of CBP/p300 to these genomic clusters in ES cells. This finding suggests that there is an active balance of HAT and HDAC activity regulating Nanog and its targets. Further analysis will be needed to determine how different families of co-activator and co-repressor complexes regulate the pluripotency transcription factor network in response to the cellular environment.
Acknowledgments
We thank B. Emerson at the Salk Institute for Biological Studies; A. Baltus, J. Doench, and H. Arthanari at Harvard University for helpful comments; and colleagues at Novartis Institute for Biomedical Research, particularly E. Li, T. Chen, F. Gaudet, and T. Gebuhr, for insightful discussions. The pCIG-HDAC1 cDNA was a kind gift from Dr. Ramesh Shivdasani (Dana Farber Cancer Institute).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ES, embryonic stem; HDAC, histone deacetylase; ChIP, chromatin immunoprecipitation; siRNA, small interference RNA; qPCR, quantitative PCR; RA, retinoic acid; LIF, leukemia inhibitory factor; TSA, trichostatin A.
References
- 1.Johnson, B. V., Rathjen, J., and Rathjen, P. D. (2006) Curr. Opin. Genet. Dev. 16 447-454 [DOI] [PubMed] [Google Scholar]
- 2.Masui, S., Nakatake, Y., Toyooka, Y., Shimosato, D., Yagi, R., Takahashi, K., Okochi, H., Okuda, A., Matoba, R., Sharov, A. A., Ko, M. S., and Niwa, H. H. (2007) Nat. Cell Biol. 9 625-635 [DOI] [PubMed] [Google Scholar]
- 3.Kuroda, T., Tada, M., Kubota, H., Kimura, H., Hatano, S. Y., Suemori, H., Nakatsuji, N., and Tada, T. (2005) Mol. Cell Biol. 25 2475-2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodda, D. J., Chew, J. L., Lim, L. H., Loh, Y. H., Wang, B., Ng, H. H., and Robson, P. (2005) J. Biol. Chem. 280 24731-24737 [DOI] [PubMed] [Google Scholar]
- 5.Wu, Q., Chen, X., Zhang, J., Loh, Y. H., Low, T. Y., Zhang, W., Zhang, W., Sze, S. K., Lim, B., and Ng, H. H. (2006) J. Biol. Chem. 281 24090-24094 [DOI] [PubMed] [Google Scholar]
- 6.Pan, G., Li, J., Zhou, Y., Zheng, H., and Pei, D. (2006) FASEB J. 20 1730-1732 [DOI] [PubMed] [Google Scholar]
- 7.Suzuki, A., Raya, A., Kawakami, Y., Morita, M., Matsui, T., Nakashima, K., Gage, F. H., Rodríguez-Esteban, C., and Izpisúa Belmonte, J. C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 10294-10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin, T., Chao, C., Saito, S., Mazur, S. J., Murphy, M. E., Appella, E., and Xu, Y. (2005) Nat. Cell Biol. 7 165-171 [DOI] [PubMed] [Google Scholar]
- 9.Gu, P., LeMenuet, D., Chung, A. C., Mancini, M., Wheeler, D. A., and Cooney, A. J. (2005) Mol. Cell Biol. 25 8507-8519 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 10.Boyer, L. A., Mathur, D., and Jaenisch, R. (2006) Curr. Opin. Genet. Dev. 16 455-462 [DOI] [PubMed] [Google Scholar]
- 11.Kaeser, M. D., and Emerson, B. M. (2006) Curr. Opin. Genet. Dev. 5 508-512 [DOI] [PubMed] [Google Scholar]
- 12.Wang, J., Rao, S., Chu, J., Shen, X., Levasseur, D. N., Theunissen, T. W., and Orkin, S. H. (2006) Nature 444 364-368 [DOI] [PubMed] [Google Scholar]
- 13.Liang, J., Wan, M., Zhang, Y., Gu, P., Xin, H., Jung, S. Y., Qin, J., Wong, J., Cooney, A. J., Liu, D., and Songyang, Z. (2008) Nat. Cell Biol. 10 731-739 [DOI] [PubMed] [Google Scholar]
- 14.Kaji, K., Caballero, I. M., MacLeod, R., Nichols, J., Wilson, V. A., and Hendrich, B. (2006) Nat. Cell Biol. 8 285-292 [DOI] [PubMed] [Google Scholar]
- 15.Verdin, E. (ed) (2006) Histone Deacetylases: Transcriptional Regulation and Other Cellular Functions, pp. 23-60, Humana Press, Totowa, NJ
- 16.Silverstein, R. A., and Ekwall, K. (2005) Curr. Genet. 47 1-17 [DOI] [PubMed] [Google Scholar]
- 17.Dannenberg, J. H., David, G., Zhong, S., van der Torre, J., Wong, W. H., and Depinho, R. A. (2005) Genes Dev. 19 1581-1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowley, S. M., Iritani, B. M., Mendrysa, S. M., Xu, T., Cheng, P. F., Yada, J., Liggitt, H. D., and Eisenman, R. N. (2005) Mol. Cell Biol. 25 6990-7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagger, G., O'Carroll, D., Rembold, M., Khier, H., Tischler, J., Weitzer, G., Schuettengruber, B., Hauser, C., Brunmeir, R., Jenuwein, T., and Seiser, C. (2002) EMBO J. 21 2672-2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trivedi, C. M., Luo, Y., Yin, Z., Zhang, M., Zhu, W., Wang, T., Floss, T., Goettlicher, M., Noppinger, P. R., Wurst, W., Ferrari, V. A., Abrams, C. S., Gruber, P. J., and Epstein, J. A. (2007) Nat. Med. 13 324-331 [DOI] [PubMed] [Google Scholar]
- 21.Narlikar, G. J., Fan, H. Y., and Kingston, R. E. (2002) Cell 108 475-487 [DOI] [PubMed] [Google Scholar]
- 22.Okada, Y., Shimazaki, T., Sobue, G., and Okano, H. (2004) Dev. Biol. 275 124-142 [DOI] [PubMed] [Google Scholar]
- 23.Thevenet, L., Mejean, C., Moniot, B., Bonneaud, N., Galeotti, N., Aldrian-Herrada, G., Poulat, F., Berta, P., Benkirane, M., and Boizet-Bonhoure, B. (2004) EMBO J. 23 3336-3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glozak, M. A., Sengupta, N., Zhang, X., and Seto, E. (2005) Gene (Amst.) 363 15-23 [DOI] [PubMed] [Google Scholar]
- 25.Rayman, J. B., Takahashi, Y., Indjeian, V. B., Dannenberg, J. H., Catchpole, S., Watson, R. J., te Riele, H., and Dynlacht, B. D. (2002) Genes and Dev. 16 933-947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy, M., Ahn, J., Walker, K. K., Hoffman W. H., Evans, R. M., Levine, A. J., and George, D. L. (1999) Genes Dev. 13 2490-2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai, Y., Kurokawa, M., Yamaguchi, Y., Izutsu, K., Nitta, E., Mitani, K., Satake, M., Noda, T., Ito, Y., and Hirai, H. (2004) Mol. Cell Biol. 24 1033-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koipally, J., Renold, A., Kim, J., and Georgopoulos, K. (1999) EMBO J. 18 3090-3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, J. H., Hart, R. L., and Skalnik, D. G. (2004) Genesis 38 32-38 [DOI] [PubMed] [Google Scholar]
- 30.McCool, K. W., Xu, X., Singer, D. B., Murdoch, F. E., and Fritsch, M. K. (2007) J. Biol. Chem. 282 6696-6706 [DOI] [PubMed] [Google Scholar]
- 31.Ng, H. H., and Bird, A. (2000) Trends Biochem. Sci. 25 121-126 [DOI] [PubMed] [Google Scholar]
- 32.de Nadal, E., Zapater, M., Alepuz, P. M., Sumoy, L., Mas, G., and Posas, F. (2004) Nature 427 370-374 [DOI] [PubMed] [Google Scholar]
- 33.Yao, Y. L., Yang, W. M., and Seto, E. (2001) Mol. Cell Biol. 21 5979-5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, Xi., Xu, H., Yuan, P., Fang, F., Huss, M., Vega, V. B., Wong, E., Orlov, Y. L., Zhang, W., Jiang, J., Loh, Y. H., Yeo, H. C., Yeo, Z. X., Narang, V., Govindarajan, K. R., Leong, B., Shahab, A., Ruan, Y., Bourque, G., Sung, W. K., Clarke, N. D., Wei, C. L., and Ng, H. H. (2008) Cell 133 1106-1117 [DOI] [PubMed] [Google Scholar]