Abstract
Cellular stress such as endoplasmic reticulum stress, hypoxia, and viral infection activates an integrated stress response, which includes the phosphorylation of the eukaryotic initiation factor 2α (eIF2α) to inhibit overall protein synthesis. Paradoxically, this leads to translation of a subset of mRNAs, like transcription factor ATF4, which in turn induces transcription of downstream stress-induced genes such as growth arrest DNA-inducible gene 34 (GADD34). GADD34 interacts with protein phosphatase 1 to dephosphorylate eIF2α, resulting in a negative feedback loop to recover protein synthesis and allow translation of stress-induced transcripts. Here, we show that GADD34 is not only transcriptionally induced but also translationally regulated to ensure maximal expression during eIF2α phosphorylation. GADD34 mRNAs are preferentially associated with polysomes during eIF2α phosphorylation, which is mediated by its 5′-untranslated region (5′UTR). The human GADD34 5′UTR contains two non-overlapping upstream open reading frames (uORFs), whereas the mouse version contains two overlapping and out of frame uORFs. Using 5′UTR GADD34 reporter constructs, we show that the downstream uORF mediates repression of basal translation and directs translation during eIF2α phosphorylation. Furthermore, we show that the upstream uORF is poorly translated and that a proportion of scanning ribosomes bypasses the upstream uORF to recognize the downstream uORF. These findings suggest that GADD34 translation is regulated by a unique 5′UTR uORF mechanism to ensure proper GADD34 expression during eIF2α phosphorylation. This mechanism may serve as a model for understanding how other 5′UTR uORF-containing mRNAs are regulated during cellular stress.
Phosphorylation of Ser51 in eIF2α is a key cellular response to environmental stresses such as hypoxia, endoplasmic reticulum (ER)2 stress and viral infection. The stress-induced phosphorylation of eIF2α represses general protein synthesis, which induces the expression of specific genes involved in the stress response (1, 2). Reprogramming of gene expression is vital for cellular survival and can trigger apoptosis, if the stress is severe and prolonged.
In mammals, four distinct eIF2α kinases have been identified (2). These include protein kinase R, which is activated upon binding to double-stranded RNAs or through the antiviral interferon response (3), the heme-regulated inhibitor, which senses heme availability and responds to oxidative stress (4, 5), the general control nonderepressible-2, which is regulated by amino acid availability (6), and the protein kinase R-like ER kinase, PERK, which is activated in response to an accumulation of unfolded proteins in the ER (7). Although protein kinase R, heme-regulated inhibitor, general control nonderepressible-2, and PERK can all catalyze the phosphorylation of eIF2α to halt protein synthesis, they do so in response to distinct environmental cues. For instance, the accumulation of unfolded proteins in the ER activates PERK to repress translation to ease the load of unfolded proteins in the ER, whereas translational repression during amino acid depletion is achieved through general control nonderepressible-2 activation, which provides cells sufficient time for recovery from nutrient starvation. The biological importance of each eIF2α kinase signaling pathway is reflected in their association with several diseases such as diabetes, cancer, neurodegenerative diseases, and viral infections, including hepatitis C virus and cytomegalovirus infections (8–12).
The ternary complex, composed of initiator Met-tRNAi, GTP, and the heterotrimeric initiation factor eIF2, mediates recognition of the AUG codon by scanning 40 S ribosomal subunits (2, 13). Upon correct basepairing of the codon-anticodon of the Met-tRNAi, eIF2-GTP is hydrolyzed to eIF2-GDP and subsequently released. Free eIF2-GDP is recycled to the green fluorescent protein-bound form by the guanine nucleotide exchange factor, eIF2B. Phosphorylation of the α subunit of eIF2α on Ser51 prevents the exchange of GDP for GTP by sequestering eIF2B, thus lowering the available pool of eIF2-GTP and repressing protein synthesis (14, 15).
Although general protein synthesis is repressed when eIF2α is phosphorylated, a subset of mRNAs remains actively translated under these conditions. Such mRNAs include mammalian ATF4, ATF5, and yeast GCN4 mRNAs (6, 16–20). Translation of ATF4, a member of the bZIP family of transcription factors responsible for inducing transcription of downstream stress-inducible genes, is governed by two uORFs within its 5′UTR (17, 20). Following translation of the upstream uORF, under basal conditions, ribosomes resume scanning and reinitiate translation at the downstream uORF, which leads to ribosome disassembly and prevents ATF4 translation. In contrast, under conditions where eIF2α is phosphorylated, reinitiating ribosomes have a higher probability of recruiting another eIF2 ternary complex downstream after the AUG codon of the downstream uORF and thereby initiate translation at the ATF4 ORF. This mechanism is reminiscent of the classic model of reinitiation exemplified by the translational regulation of the yeast GCN4 transcription factor (21). The expression of ATF5, another bZIP family member, is also regulated at the translational level through a similar mechanism involving uORFs in its 5′UTR, indicating that such a mechanism is conserved and likely important for regulating many mRNAs (18, 19).
The accumulation of unfolded proteins in the ER activates a multitude of intracellular signaling pathways, collectively referred to as the unfolded protein response (UPR) (22). One arm of the UPR switches on the ER-resident unfolded protein sensor, PERK, to repress global protein synthesis via eIF2α phosphorylation, which in turn induces ATF4 translation (16). ATF4 activates the transcription of downstream stress-induced genes, including CHOP and GADD34 (23). GADD34 interacts with protein phosphatase 1 to dephosphorylate eIF2α, which relieves the inhibition of translation (24, 25). This negative feedback loop is critical for translation of stress-induced genes and cell adaptation to ER stress (24–27). Although it is clear that GADD34 transcriptional induction is important, the mechanism by which the GADD34 transcript is translated during eIF2α phosphorylation remains obscure.
In this study, we demonstrate that translation of the human and mouse GADD34 mRNAs are governed through an uORF within its 5′UTR, which is responsible for translational repression during unstressed conditions and directs translation of GADD34 during eIF2α phosphorylation. Our results suggest that the human and mouse 5′UTRs use a distinct mechanism to ensure GADD34 expression during cellular stresses that induce eIF2α phosphorylation.
EXPERIMENTAL PROCEDURES
5′RACE and Plasmid Construction—The human 5′UTR GADD34-YFP, mouse 5′UTR GADD34-YFP, and human 5′UTR ATF4-YFP reporter plasmids were engineered using a two-part ligation strategy in the pcDNA3 vector. The human GADD34 5′UTR cDNA was synthesized by 5′RACE from total HepG2 RNA using the FirstChoice RLM-RACE system (Ambion) and PCR-amplified using a gene-specific primer, PrEJ18, and the 5′RACE-nested inner primer. The PCR product was TA-cloned (Invitrogen) and sequence-verified. The human GADD34 5′UTR fragment was digested with EcoRI and NcoI. The mouse GADD34 5′UTR cDNA was PCR-amplified using primers, PrEJ481 and PrEJ482, which contain a 5′ EcoRI anda3′ NcoI site. The human ATF4 5′UTR was PCR-amplified using primers, PrEJ279 and PrEJ420RR, and subsequently TA-cloned. ATF4 5′UTR was digested with a 5′ EcoRI and a 3′ BbsI site, which contains complementary ends with NcoI. The reporter enhanced YFP, eYFP (Clontech), was PCR-amplified using primers containing a 5′ NcoI and a 3′ XbaI site. The digested 5′UTR and amplified eYFP were cloned into EcoRI and XbaI sites of pcDNA3, resulting in fusion of the 5′UTR with YFP via an NcoI site. The NcoI site contains the AUG codon of eYFP. These ligations yielded the human GADD34 5′UTR-YFP (hGADD34-YFP), the mouse GADD34 5′UTR-YFP (mGADD34-YFP), and the human ATF4-YFP (hATF4-YFP) constructs. All mutant 5′UTRs, including mutations that either knocked out the AUG codon of each uORF, inserted codons between the start codon and stop codon of the mouse uORFs, or fused each uORF with the YFP ORF, were mutated using the QuikChange site-directed mutagenesis kit (Stratagene). All mutations were sequenced and verified.
The sequence of the mGADD34 uORFs overlap is 5′-..cgacAUGAacc..-3′. The AUG start codon of uORF2 overlaps with the UGA of uORF1. To create the 1 and 2 codon insertion mutants, a codon (underlined) was inserted to produce 5′-... cgacAUGUUGAacc... -3′ (1cod) and 5′-... cgacAUGUUGUUGAacc...-3′ (2cod).
Cell Culture and Stable Cell Lines—Mouse Hepa (1–6C) cells were generously provided by Maria Hatzoglou (Case Western University, Cleveland). HepG2 and Hepa cells were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (v/v), 2 mm glutamine, and 100 units of penicillin/streptomycin. Plasmid transfections in HepG2 cells were performed using Lipofectamine (Invitrogen) at 60% confluency. Stable cell lines were selected using Geneticin, and cells were passaged as pooled cell lines. Cells were treated either with 1 μm thapsigargin (Sigma), 2 mm DTT, or 100 μg/ml arsenite (Riedel de Haen) to activate eIF2α kinases and induce eIF2α phosphorylation.
Western Blot Analysis—Cells were washed two times with phosphate-buffered saline and scraped into lysis buffer containing 20 mm Hepes, pH 7.5, 150 mm NaCl, 1% Triton X-100 (v/v), 10% glycerol, 1 mm EDTA, 10 mm NaF, 17.5 mm β-glycerophosphate, and a protease inhibitor mixture (Roche Applied Science). The lysates were freeze-thawed three times and centrifuged to clear cell debris and nuclei. Protein concentration was determined using Bradford reagent (Bio-Rad). Proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride Immobilon-P or Immobilon-FL membrane (Millipore). Total eIF2α was detected using a polyclonal antibody to the C terminus of eIF2α (Cell Signaling, #9722), and phosphorylated eIF2α was detected with an epitope-specific antiserum (Cell Signaling, #9721). Immunoblots were probed with GADD34 (Santa Cruz Biotechnology, H-193), CHOP (Affinity Bioreagents MAI-250), ATF4 (Santa Cruz Biotechnology, SC-200), and green fluorescent protein (Roche Applied Science) antibodies. The anti-green fluorescent protein antibody cross-reacts with the YFP reporter protein.
Sucrose Gradient Centrifugation and Polysome Analysis—Sucrose gradient centrifugation and polysome analysis was prepared as described (28). Briefly, prior to drug treatment and cell lysis, fresh media was added to cells 4–6 h earlier. Cells were incubated with 100 μg/ml cycloheximide for 3 min at 37 °C, washed three times with 1× phosphate-buffered saline, and harvested directly on the plate using lysis buffer (15 mm Tris-HCl, pH 7.4, 15 mm MgCl2, 200 mm NaCl, 1% Triton X-100 (v/v), 100 μg/ml cycloheximide, 1 mg/ml heparin). Nuclei and cell debris were cleared by centrifugation at 12,000 × g for 10 min at 4 °C, and the resulting supernatant was loaded onto a 10–50% (w/v) sucrose gradient composed of the lysis buffer minus the Triton X-100. The gradient was centrifuged at 35,000 rpm for 3 h in an SW41 rotor at 4 °C. Fractions were collected from the top using an ISCO fraction collector and a Brandel syringe pump system. To collect RNA, 3 ml of 8 m guanidine-HCl and 5 ml of EtOH were added to each fraction, precipitated, and resuspended in water. Equal volumes of fractionated RNA were subjected to Northern blot analysis.
Radiolabel Incorporation and Immunoprecipitation of Newly Synthesized Proteins—To examine newly synthesized proteins, cells were grown in Dulbecco's modified Eagle's medium minus methionine and cysteine supplemented with 10% (v/v) dialyzed fetal bovine serum for 25 min and metabolically labeled with 100 μCi/ml [35S]methionine-cysteine for 20 min. To induce stress, 1 μm thapsigargin, 2 mm DTT, or 100 μg/ml arsenite were incubated at the same time when cells were grown in Dulbecco's modified Eagle's medium minus methionine and cysteine. Radiolabeled cells were washed two times with 1× phosphate-buffered saline and lysed in buffer containing 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm DTT, 1 mm EDTA, 0.5% (v/v) Nonidet P-40, and protease inhibitor mixture. Equal amounts of proteins, determined by Bradford reagent, were incubated with anti-green fluorescent protein antibody (Roche Applied Science) and agarose-protein G beads according to the protocol recommended by Roche Applied Science. Proteins attached to the agarose-beads were recovered by boiling in Laemmli sample buffer and separated by SDS-PAGE. The gel was dried, subjected to PhosphorImager analysis, and quantified by ImageQuant (Storm, Amersham Biosciences).
Northern Blot Analysis—Total RNA (TRIzol, Invitrogen) or RNA purified from sucrose gradient centrifugation fractions were separated on a denaturing agarose gel and transferred to Zeta-probe membrane (Bio-Rad). Radiolabeled DNA hybridization probes were generated using the Radprime kit (Invitrogen). The amount of radiolabeled probe was quantitated by PhosphorImager analysis (Storm, Amersham Biosciences).
Oligonucleotide Sequences—The sequences were PrEJ18 (5′-CTACCCATGGGTCTGGGCGGCTGGGGGC-3′), PrEJ279 (5′-TTTCTACTTTGCCCGCCCACAG-3′), PrEJ420 (RR-5′-CTAGGAAGACCCCATGGTTTCTTCAGCCCC-3′), PrEJ481 (5′-CTAGGAATTCGCTCTGAGTTTGTGGAAGATT-3′), and PrEJ482 (5′-CTAGCCATGGGTCTGGGCGGCGGGCTGCAC-3′).
RESULTS
Translational Control during ER Stress in Liver Cells—To investigate the mechanisms of translational control during ER stress, human hepatoma HepG2 cells and mouse hepatoma Hepa cells were treated with UPR-inducing agents, thapsigargin or DTT. These cells were chosen because they are highly secretory and are sensitive to drugs that accumulate unfolded proteins in the ER. Protein synthesis was measured by pulsed labeling with [35S]methionine for the indicated times followed by quantitation of the total radioactive incorporation into newly synthesized polypeptides. Both thapsigargin and DTT treatment inhibited general protein synthesis as early as 15 min after incubation and was sustained for 30 min (Fig. 1, A and B). Notably, inhibition correlated with increased phosphorylation of eIF2α observed at both time points (Fig. 1, A and B). A partial recovery in protein synthesis was observed upon prolonged incubation with thapsigargin or DTT (60 min), which correlated with reduced phosphorylation of eIF2α (Fig. 1A). Incubation with thapsigargin or DTT for extended periods also elicited an increase in GADD34 protein levels, consistent with the fact that increased GADD34 protein levels mediate eIF2α dephosphorylation (Fig. 2C) (26).
FIGURE 1.
Polysomal association of GADD34 mRNA during eIF2α phosphorylation in human HepG2 and mouse Hepa cells. HepG2 cells (A) or Hepa (1–6C) cells (B) were treated with 1 μm thapsigargin or 2 mm DTT for the indicated times and subjected to [35S]methionine/cysteine pulse-labeling. Radiolabel incorporation into newly synthesized protein was measured by trichloroacetic acid precipitation and normalized to that in untreated cells (100%) (A) or by autoradiogram (B) of SDS-PAGE analysis. The phosphorylation status of eIF2α and the total eIF2α in cell lysates were monitored by Western blot analysis using a phospho-specific antibody to phosphorylated eIF2α (top) and an antibody that recognizes the C-terminal region of eIF2α (bottom), respectively. C, sedimentation profiles at absorbance 254 nm of HepG2 lysates untreated (left) or treated with 1 μm thapsigargin for 30 min (right). Cell lysates were fractionated by a 10–50% (w/v) sucrose gradient centrifugation. The top to bottom of the gradient is represented from left to right, respectively. The sedimentation of the 40 S, 60 S, 80 S fractions and polysomes are indicated. D and E, polysomal Northern blot analysis of RNA in fractions from HepG2 (D) and Hepa (E) cell lysates after 1 μm 30 min thapsigargin (thap), 2 mm 30 min DTT, or untreated (unt) treatments as indicated to the right. The distribution of mRNAs is indicated to the left by Northern blotting of polysomal RNA. Fractions from top to bottom of the gradient are represented from left to right, respectively. Fractions containing 80 S and polysomes are indicated at the bottom. Quantitation of the Northern blots in D and E are shown in F and G, respectively. The amount of radioactive probe specific to the indicated mRNA in each fraction is indicated as a percentage of total radioactivity in all fractions within each sucrose gradient (% radioactivity).
FIGURE 2.
GADD34 expression in HepG2 cells during thapsigargin and actinomycin D treatment. A, Northern blots of RNA and (C) immunoblots of lysates from HepG2 cells alone (unt) or treated with 5 μg/ml actinomycin D (actD), 1 μm thapsigargin (thap), or thapsigargin/actinomycin D (actD/thap) for the indicated times. A, radiolabeled probes specific to GADD34, ATF4, CHOP, and GAPDH mRNAs (indicated to the right) were quantitated (B) using phosphorimaging analysis and normalized to the amount of RNA at the 3-h untreated time point. C, GADD34, ATF4, phosphorylated eIF2α, total eIF2, and CHOP were detected by antibodies as indicated to the right. Representative Western blots of at least three independent experiments are shown. For GADD34, two independent experiments are shown, which shows reproducible induction of GADD34 protein levels under thapsigargin alone and thapsigargin and actinomycin D treatment. The arrow indicates the migration of GADD34 protein, and the asterisk indicates nonspecific proteins recognized by the GADD34 antibody. D, quantitation of a representative GADD34 immunoblot. The amount of GADD34 protein was normalized to the amount of GADD34 at the 3-h untreated time point (Odyssey-Licor). E, polysomal Northern blot analysis of RNA in fractions from untreated HepG2 cells (unt) or from cells after a 30-min treatment with 1 μm thapsigargin (thap), 5 μg/ml actinomycin D (actD), or 1 μm thapsigargin (actD/thap) as indicated to the right. The distribution of GADD34 and GAPDH mRNAs is shown by Northern blot analysis using radiolabeled DNA probes. Fractions from top to bottom of the gradient are represented from left to right, respectively. Locations of 80 S and polysomes across the gradient are indicated at the bottom. Quantitation of the amount of radioactivity within each fraction is shown in supplemental Fig. S1.
To examine the translation of specific and total RNAs during ER stress, lysates of cells untreated or treated with thapsigargin were separated by sucrose gradient centrifugation. In untreated cells, the majority of ribosomes sedimented to heavier molecular weight polysomes, indicating that most ribosomes are actively engaged in translation (Fig. 1C, left panel). In contrast, treatment of cells with thapsigargin resulted in dissociation of ribosomes from mRNAs and an increase in free 40 S and 60 S subunits (Fig. 1C, right panel). The loss of polysomes is in agreement with the significant decrease in protein synthesis during thapsigargin treatment.
Polysome Association of GADD34 in Human and Mouse Liver Cells during ER Stress—We next asked whether GADD34 mRNAs are associated with polysomes during ER stress. To monitor the association of specific mRNAs with ribosomes, individual mRNAs within the sucrose gradient fractions were probed by Northern blot analysis. In untreated human HepG2 and mouse Hepa cells, actin and GAPDH mRNAs sedimented primarily to heavy molecular weight fractions 9 and 10, indicating that these mRNAs associated with many ribosomes and are actively translated (Fig. 1, D–G). Thapsigargin and DTT treatment resulted in a dramatic shift of actin and GAPDH mRNAs to lower molecular weight polysomes, consistent with these mRNAs being translationally repressed relative to those in untreated cells (Fig. 1, D–G). Conversely, ATF4 mRNA remained associated with polysomes during thapsigargin and DTT treatment in both cell lines as reported previously (Fig. 1, D and E) (16, 17). In HepG2 cells, ATF4 mRNA sedimented primarily with more ribosomes (fractions 7–9) under ER stress than under untreated conditions (fractions 6–8), indicating that this mRNA is translationally induced during eIF2α phosphorylation (Fig. 1D). Similar to ATF4, human and mouse GADD34 mRNAs shifted to higher molecular weight fractions during thapsigargin and DTT treatments compared with those during unstressed conditions (Fig. 1, D–G). Therefore, like ATF4, both human and mouse GADD34 mRNAs possess mechanisms to direct translation during ER stress when eIF2α is phosphorylated.
GADD34 Is Regulated at the Transcriptional and Translational Level during ER Stress—Given that the GADD34 mRNA was associated with more ribosomes (Fig. 1) and that GADD34 has been shown to be transcriptionally up-regulated during ER stress (24, 25), the increase in GADD34 protein expression is likely due to increases in both transcription and translation rates. To determine the contribution of translational control of GADD34 during eIF2α phosphorylation, we treated cells with the transcription inhibitor, actinomycin D, and thapsigargin. Incubation with actinomycin D inhibited the thapsigargin-induced transcription of GADD34, ATF4, and CHOP, consistent with previous reports that these genes are transcriptionally induced during ER stress (Fig. 2, A and B) (16, 24). Treatment of cells with thapsigargin alone or a combination of thapsigargin and actinomycin D induced a small but reproducible increase in GADD34 protein levels, indicating that GADD34 is translationally up-regulated (Fig. 2, C and D). Treatment with actinomycin D alone did not induce GADD34 expression. Similarly, ATF4 protein expression was increased during thapsigargin and thapsigargin/actinomycin D treatments, whereas CHOP expression was only induced during thapsigargin treatment, in agreement that transcriptional induction is required for CHOP expression during ER stress (16). The induction of GADD34 and ATF4 protein during thapsigargin/actinomycin D treatment was not as prominent as during thapsigargin treatment alone (Fig. 2, C and D), suggesting that transcriptional induction of GADD34 and ATF4 is required for maximal expression during eIF2α phosphorylation.
The increase in GADD34 protein during thapsigargin/actinomycin D treatment is likely due to the translational up-regulation of existing basal GADD34 mRNAs. To confirm this, we analyzed the polysomal distribution of GADD34 mRNAs in cells treated with thapsigargin alone or with thapsigargin/actinomycin D. Both treatments elicited a shift of GADD34 mRNAs to higher molecular weight polysomal fractions as compared with that in untreated cells (Fig. 2E and supplemental Fig. S1). In contrast, GAPDH mRNAs shifted to lower molecular weight fractions during both treatments (Fig. 2E and supplemental Fig. S1). Together with the Western blot analyses in Fig. 2C, these results demonstrated that GADD34 mRNAs are translationally activated under thapsigargin-induced ER stress.
Interestingly, a 3-h thapsigargin treatment induced eIF2α phosphorylation but by 6 h post-treatment, eIF2α phosphorylation levels had decreased, likely due to maximal induction of GADD34 protein (Fig. 2C). In contrast, during thapsigargin/actinomycin D incubation, eIF2α remained phosphorylated at 6 h post-treatment, suggesting that the moderate induction of GADD34 protein during this treatment was insufficient to reduce eIF2α phosphorylation (Fig. 2C). In summary, the optimal induction of GADD34 protein during eIF2α phosphorylation is mediated through both transcriptional and translational controls of GADD34.
The Human and Mouse GADD34 5′UTRs Mediate Polysome Association during eIF2α Phosphorylation—To begin elucidating the mechanism of GADD34 translation, we first focused on its 5′UTR. An analysis of annotated mammalian GADD34 mRNAs revealed two upstream open reading frames (uORFs) within the 5′UTR. The human (hGADD34), chimp and rat GADD34 5′UTRs contain two non-overlapping uORFs separated by 7–30 nucleotides, whereas the two uORFs of mouse (mGADD34), hamster, and bovine 5′UTRs are overlapping and out of frame by a single nucleotide (Fig. 3). Specifically, the uORFs overlap within the stop codon of the upstream uORF (uORF1) and the start AUG codon of the downstream uORF (uORF2) (Fig. 3).
FIGURE 3.
Schematic of uORFs within select mammalian GADD34 5′UTRs. Representative cDNAs encoding GADD34-related sequences in GenBank™ are shown, including human (NM_014330), rat (NM_133546), hamster (L28147), and mouse (NM_008654) GADD34 5′UTRs. The AUG codon of the GADD34 ORF is shown. The length of the 5′UTR and the space between uORFs are indicated above (nt, nucleotides). Each box represents uORF1 and uORF2 and the size of the predicted translated uORF (aa, amino acid length). A close-up view of the overlap of the mouse GADD34 uORFs is shown below. The upstream uORF (uORF1) overlaps with the downstream uORF (uORF2) by 1 nucleotide. The stop codon of uORF1 is underlined, and the AUG of uORF2 is in gray.
To determine whether the GADD34 5′UTR is sufficient to direct translation during eIF2α phosphorylation, we designed reporter RNAs containing the human or mouse GADD34 5′UTR fused upstream of an enhanced yellow fluorescent protein (YFP) ORF (Fig. 4). Stable HepG2 cell lines were generated that express different 5′UTR-YFP reporter RNAs, which are transcribed by the mammalian cytomegalovirus promoter. We first tested whether the GADD34 5′UTR could direct polysome association of the reporter RNA during thapsigargin-induced ER stress. Cell lysates were subjected to sucrose gradient centrifugation, and then fractionated. Following fractionation, the polysomal distributions of endogenous GADD34, ATF4, actin, GAPDH, and reporter RNAs were determined by Northern blot analysis. Quantitations of the Northern blots are shown in supplemental Fig. S2. For all stable cell lines, the endogenous actin, GAPDH, GADD34, and ATF4 mRNAs had similar polysomal distributions to those observed with the parental HepG2 cell line (compare Figs. 1 (D–G) and 4 and supplemental Fig. S2), indicating that these cell lines were responsive to ER stress. Briefly, actin and GAPDH mRNAs shifted to lower molecular weight polysomal fractions during thapsigargin-induced stress, whereas GADD34 and ATF4 mRNAs associated with heavier polysomal fractions under the same conditions (Fig. 4 and supplemental Fig. S2). The reporter RNA containing the human or mouse GADD34 5′UTRs (hGADD34-YFP or mGADD34-YFP, respectively) shifted to higher molecular weight polysomes during thapsigargin treatment (Fig. 4 (A and C) and supplemental Fig. S2), which is similar to the polysome distribution of the endogenous GADD34 mRNA. This suggested that the hGADD34 and mGADD34 5′UTRs were sufficient to direct translation during eIF2α phosphorylation. As predicted, a reporter RNA containing the human ATF4 5′UTR (hATF4-YFP) also associated with more ribosomes during ER stress (Fig. 4D), whereas a minimal YFP reporter RNA associated with fewer ribosomes during thapsigargin treatment (data not shown). Thus, the 5′UTRs of hGADD34, mGADD34, and hATF4 can confer resistance to the effects of eIF2α phosphorylation-dependent translational repression during ER stress.
FIGURE 4.
The 5′UTR of human and mouse GADD34 mediates polysome association during thapsigargin treatment in HepG2 cells. Sucrose gradient density centrifugation and fractionation of lysates from stable cell lines expressing reporter YFP RNA fused with wild-type human GADD34 5′UTR (hGADD34-YFP) (A), mutant human GADD34 5′UTR (1&2KO hGADD34-YFP) (B), mouse GADD34 5′UTR (mGADD34-YFP) (C), or human ATF4 5′UTR (hATF4-YFP) (D) that were incubated in the absence (unt) or presence of 1 μm thapsigargin for 30 min (thap) were performed. The reporter RNAs were transcribed by the cytomegalovirus promoter (CMV). The distribution of specific RNAs across the gradient was detected by Northern blot analysis as indicated to the left. Fractions from top to bottom of the gradient are represented from left to right, respectively. Fractions that contain the 80 S and polysomes are indicated below the fractions. Quantitations of the amount of radioactivity within each fraction are shown in supplemental Fig. S2.
To test whether the uORFs of the hGADD34 5′UTR are important for translational regulation, the AUG codons of the upstream (uORF1) and downstream (uORF2) uORFs of hGADD34 5′UTRs were mutated to AUU (1&2KO hGADD34-YFP) (Fig. 4B). In untreated cells, the 1&2KO hGADD34-YFP RNA associated with more ribosomes (fractions 7 and 8) as compared with the wild-type hGADD34-YFP reporter RNAs (fractions 6 and 7) (Fig. 4 (A and B) and supplemental Fig. S2), indicating that intact uORFs were important for keeping basal translation repressed. Under thapsigargin treatment, the mutant reporter RNA shifted to lower molecular weight polysomes similar to that observed of the endogenous actin mRNA (Fig. 4 (A and B) and supplemental Fig. S2), demonstrating that mutating the uORFs sensitized the RNA to the inhibitory effects of eIF2α phosphorylation. Thus, the hGADD34 5′UTR uORFs are important for repressing basal translation and for regulating translation during eIF2α phosphorylation.
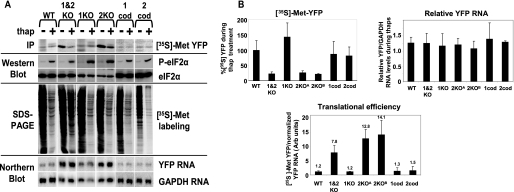
The Downstream uORF of Human GADD34 Regulates Translation during eIF2α Phosphorylation—To confirm that the hGADD34 5′UTR can direct translation during ER stress, the incorporation of [35S]methionine/cysteine into newly synthesized YFP was measured by pulse labeling followed by immunoprecipitation. We first compared the translation efficiency of wild-type and mutant 1&2KO hGADD34-YFP RNA by measuring the amount of radioactive incorporation into newly synthesized YFP normalized to the amount of reporter YFP RNA (Fig. 5B, translational efficiency = radiolabel incorporation into YFP protein divided by YFP/GAPDH RNA levels). Translation of 1&2KO hGADD34-YFP RNAs was ∼29-fold higher than that of the wild-type hGADD34-YFP RNA, which is in agreement with the polysome distributions of the reporter RNAs (Fig. 4, A and B). Thus, the uORFs are important to repress basal translation during unstressed conditions.
FIGURE 5.
The uORFs of hGADD34 5′UTR mediate translation regulation during eIF2α phosphorylation. In A: Top row, immunoprecipitates of [35S]methionine-labeled YFP from lysates of cells expressing IGR IRES-YFP, hATF4-YFP, wild-type hGADD34-YFP, YFP alone, or mutant human GADD34 5′UTR fused with YFP where the AUG codon of the upstream uORF (1KO), downstream uORF (2KO), or both uORFs were mutated (1KO, 2KO, and 1&2KO, respectively) were either left untreated (U) or treated with 1 μm thapsigargin (T), 2 mm DTT (D), or 100 μg/ml arsenite for 45 min (A). Cells were pulse-labeled with [35S]methionine for 20 min prior to harvesting. Immunoprecipitates were separated by SDS-PAGE and exposed by phosphorimaging analysis. Second row, immunoblots of lysates using antibodies that recognize phospho-eIF2α or total eIF2α. Third row, lysates were subjected to SDS-PAGE analysis and exposed to autoradiography. Bottom row, in parallel, RNA from treated cells was subjected to Northern blot analysis using probes specific for YFP or GAPDH. B, quantitation of newly synthesized [35S]methionine-labeled YFP immunoprecipitates (top left) and levels of YFP RNA normalized to GAPDH (top right) as described in A. Each bar represents the percent of [35S]methionine-labeled YFP expressed or the YFP RNA levels during the indicated drug treatments as compared with that in untreated cells, which is set at 100%. For Northern blot analysis, YFP mRNA levels were normalized to GAPDH mRNA levels. Bottom, translational efficiency of reporter YFP RNA in unstressed cells was calculated by the amount of newly synthesized [35S]methionine YFP protein normalized to YFP/GAPDH mRNA. Above each bar shows the average translational efficiency normalized to the wild-type hGADD34-YFP RNA as 1. Shown are averages ± S.D. from at least three independent experiments.
Mutating hGADD34 uORF1 and uORF2 decreases the number of polysomes associated with the YFP mRNA in thapsigargin-treated cells (Fig. 4, A and B), suggesting that, in addition to repressing translation during basal conditions, the GADD34 5′UTR is required for translation during stress conditions. To test whether the GADD34 5′UTR can direct translation during stress conditions, we measured the translation of YFP under the control of GADD34 5′UTR in the presence of thapsigargin or DTT, both of which induce ER stress and activate PERK. Treatment with either drug resulted in an overall inhibition of protein synthesis and induction of eIF2α phosphorylation in all stable cell lines (Fig. 5A). Translation of YFP alone or mutant 1&2KO hGADD34-YFP was significantly inhibited by these drugs (Fig. 5, A and B). In contrast, translation of the wild-type hGADD34-YFP RNA was largely insensitive to the effects of eIF2α phosphorylation as compared with 1&2KO hGADD34-YFP translation under the same treatments (Fig. 5, A and B). Arsenite, which induces oxidative stress and activates heme-regulated inhibitor, also leads to translation arrest. Like thapsigargin and DTT, arsenite inhibited the translation of YFP alone or mutant 1&2KO hGADD34-YFP but not that of wild-type hGADD34, indicating that the GADD34 5′UTR is required for translation during UPR as well as oxidative-stress response. As expected, the cricket paralysis virus intergenic region internal ribosome entry site (IGR IRES-YFP) and hATF4-YFP RNAs were also translated during eIF2α phosphorylation (Fig. 5A). The cricket paralysis virus intergenic region IRES can recruit the ribosome independently of initiation factors and has been shown to be resistant to eIF2α phosphorylation-dependent translational repression (29–31). Similarly, the mechanism via the ATF4 5′UTR uORFs promotes translation during eIF2α phosphorylation (17, 20). Levels of reporter RNAs were similar during eIF2α phosphorylation, thus the differences in YFP expression were due to alterations in translational efficiencies (Fig. 5, A and B). Therefore, polysome association of the wild-type hGADD34-YFP RNA during ER stress reflected protein synthesis by ribosomes.
The two uORFs (uORF1 and uORF2) within the hGADD34 5′UTR are reminiscent of the non-overlapping uORFs found within the yeast GCN4 and mouse ATF4 5′UTRs. But they differ in that hGADD34 uORFs are separated by a shorter (30 nucleotide) spacer. Previous reports have found that an intercistronic space of only 30 nucleotides limited the time for the scanning ribosomes to re-acquire another ternary complex before reaching the start codon of the downstream uORF, even during basal conditions where ternary complex is abundant (32–34). This argues against a model where reinitiation represses hGADD34 translation. However, it remains possible that the intergenic distance between the two uORFs within the hGADD34 5′UTR is not limiting, in which case reinitiation may account for the repression under basal conditions. To investigate this possibility that reinitiation takes place at the downstream uORF, we first determined whether the individual uORFs of hGADD34 5′UTR are inhibitory. To address this, we mutated the AUG codon of the upstream (uORF1) or downstream (uORF2) uORFs to AUU, separately. Mutating the uORF1 or uORF2 (1KO and 2KO, respectively) repressed basal translation to different extents. An intact uORF1 moderately inhibited translation (4.5-fold), whereas an intact uORF2 inhibited translation significantly (0.5-fold) as compared with the reporter RNA in which both uORFs are mutated (29.4 fold) (1&2KO hGADD34-YFP) (Fig. 5, A and B). This is in contrast to the upstream uORF of ATF4, which stimulates efficient reinitiation at the downstream AUG codon (17, 20). Although the arrangement of the hGADD34 uORFs is similar to that of ATF4, both hGADD34 uORFs were inhibitory thus suggesting that the mechanism of translational repression of hGADD34 is distinct from that of ATF4 and may not involve reinitiation at the downstream uORF.
We next asked whether hGADD34 5′UTR lacking either uORF1 or uORF2 could still direct translation during thapsigargin treatment. Compared with basal translation, an intact uORF2 within the GADD34 5′UTR directed translation during thapsigargin-induced ER stress (Fig. 5, A and B). In contrast, translation of the mutant 2KO hGADD34-YFP was significantly repressed similar to the reporter RNA lacking both uORFs. In all cases, the reporter RNA levels were unaltered during this treatment (Fig. 5, A and B). Thus, the downstream hGADD34 uORF2 was inhibitory during unstressed conditions and directed translation of GADD34 during eIF2α phosphorylation.
The Overlapping uORFs of the Mouse GADD34 5′UTR Control Translation during eIF2α Phosphorylation—Unlike the human version, the mouse GADD34 5′UTR contains two overlapping uORFs (Fig. 3). To investigate whether the mGADD34 uORFs control translation, we engineered mutant mGADD34-YFP expression constructs in which the AUG codons of each or both uORFs were mutated. Because the AUG codon of the downstream uORF2 overlaps with the UGA stop codon of the upstream uORF1, we engineered two mutations that mutate the AUG codon but restored the UGA stop codon. The first mutation altered the AUG codon to an UUG and the second to a CUG. We produced these two mutants to confirm that altering the AUG codon did not have an effect on the translation of the upstream uORF. In all experiments, both of these mutations produced similar results and will be referred to as 2KO mGADD34 5′UTR.
Stable HepG2 cell lines expressing the wild-type and mutant mGADD34-YFP RNAs were selected, and YFP expression was quantified by immunoprecipitation as described above. As compared with unstressed conditions, translation of wild-type mGADD34-YFP RNA was maintained during thapsigargin treatment, which is in agreement with the polysome distribution of this reporter RNA (Figs. 4C, 6A, and 6B). Mutating both mGADD34 uORFs or just uORF2 (2KO) directed higher basal YFP expression, indicating that the mGADD34 uORF2 was inhibitory (Fig. 6, A and B). Thapsigargin treatment inhibited translation of 1&2 KO mGADD34-YFP and 2KO mGADD34-YFP RNAs similar to that observed of total protein synthesis in the cell (∼20%) (Fig. 6B). In contrast, an intact uORF2 (1KO) did not significantly affect basal YFP expression and, like the wild-type version, the 1KO mGADD34-YFP RNA remained translated during thapsigargin treatment. For all stable cell lines, the level of reporter RNA was unaltered under basal and stressed conditions (Fig. 6, A and B). In summary, like the human hGADD34, the uORF2 of mGADD34 5′UTR was inhibitory under basal conditions and mediates translation during ER stress when eIF2α was phosphorylated. To confirm that the mouse uORF2 was sufficient to direct translation during ER stress, we monitored the polysome distribution of 1KO mGADD34-YFP RNA in untreated and thapsigargin-treated cells. The reporter RNA shifted to heavier polysomes during ER stress, similar to the endogenous GADD34 mRNA (data not shown).
FIGURE 6.
The overlapping uORFs of the mouse 5′UTR mGADD34 mediate translational control during eIF2α phosphorylation. Top row, immunoprecipitates of [35S]methionine-labeled YFP from lysates of cells stably expressing wild-type or mutant mouse 5′UTR mGADD34-YFP reporter RNAs as indicated above under untreated or 1μm thapsigargin treatment for 45 min. 1&2KO, 1KO, and 2KO represent mutant mouse mGADD34 5′UTR reporter RNAs in which the AUG start codon of uORF1 (1KO), uORF2 (2KO), or both (1&2KO) uORFs was mutated. Mutations that inserted one (1cod) or two (2cod) codons between the AUG codon of the uORF2 and the stop codon of the upstream uORF1 were engineered into the 5′UTR mGADD34-YFP reporter RNA (see “Experimental Procedures” for sequence). Second row, immunoblots of lysates using antibodies that recognize phospho-eIF2α or total eIF2α. Third row, cells were pulse-labeled with [35S]methionine for 20 min prior to harvesting. Lysates were subjected to SDS-PAGE analysis and exposed to autoradiograph. Bottom row, in parallel, RNA from treated cells was subjected to Northern blot analysis using probes specific for YFP or GAPDH. B, quantitation of [35S]methionine-labeled YFP immunoprecipitates (top left) and of YFP RNA levels normalized to GAPDH (top right). Bottom, each bar represents the percentage of [35S]methionine-labeled YFP expressed or YFP RNA levels during the thapsigargin treatment as compared with that in untreated cells. Translational efficiency of reporter YFP RNA in unstressed cells was calculated by the amount of newly synthesized [35S]methionine precipitates normalized to YFP/GAPDH mRNA levels. Above each bar shows the average translational efficiency normalized to the wild-type mGADD34-YFP RNA. Shown are averages ± S.D. from at least three independent experiments.
In the case of the ATF4 5′UTR, ribosomes resume scanning after translation of the upstream uORF and re-acquire another ternary complex for reinitiation at the downstream AUG. The key feature of this mechanism relies on sufficient spacing between the uORFs to give time for scanning ribosomes to re-acquire eIF2-GTP/Met-tRNAi complex (32). In contrast, the overlap of the mGADD34 uORFs suggests that translational control via the mGADD34 5′UTR is distinct from the ATF4 reinitiation mechanism. The UGA of the upstream uORF overlaps with the AUG of the downstream uORF in the –1 frame. To determine whether spacing between the UGA and AUG codon is important for regulation, we engineered mutant mGADD34 5′UTR-YFP reporters with either one or two extra codons inserted between the UGA of the upstream uORF and the AUG of the downstream uORF (see “Experimental Procedures” for specific details on the insertion sequence). Adding one or two extra codons maintained basal translation at a low level similar to the wild-type mGADD34-YFP RNAs (Fig. 6, A and B). Interestingly, the reporter RNAs containing the extra codons were still translated during thapsigargin treatment, indicating that increasing the overlap of uORFs by two codons does not significantly affect translational control mediated by the mGADD34 5′UTR during eIF2α phosphorylation (Fig. 6, A and B).
Although translation is relatively resistant to eIF2α phosphorylation, the hGADD34-YFP, mGADD34-YFP, and hATF4-YFP RNAs do not incorporate more radiolabel during ER stress when compared with basal conditions, which is contrary to its polysome distribution where these reporter RNAs shifted to higher molecular weight polysomes during thapsigargin treatment (compare Figs. 5 and 6 with Fig. 4, A, C, and D). From the polysome distribution, we would predict that translation of these reporter RNAs would be increased during stress. This may reflect differences in protocols between the two experiments. In the immunoprecipitation experiments, cells were amino acid-starved prior to labeling, whereas the cells were not in the polysome analysis protocol. To test this hypothesis, we analyzed the polysomal distribution of mGADD34-YFP in cells that were amino acid-starved. Unlike the shift to higher molecular weight polysomes during thapsigargin treatment (Fig. 4C and supplemental Fig. S2), the mGADD34-YFP RNAs did not shift in cells that were starved and treated with thapsigargin (supplemental Fig. S3). Similarly, the endogenous GADD34 mRNA also did not shift to higher molecular weight polysomes during starvation/thapsigargin treatment (compare Fig. 1F and supplemental Fig. S3). Therefore, amino acid starvation inhibited the 5′UTR-dependent translational activation of GADD34 during ER stress. The inhibitory effect of amino acid starvation on thapsigargin-induced translation of GADD34 remains to be investigated.
Leaky Scanning Permits Translation of uORF2 within the hGADD34 and mGADD34 5′UTRs—The results indicate that the 5′UTR uORF2 represses translation of hGADD34 and mGADD34 under unstressed conditions. Mutating uORF2 led to moderately higher basal levels of translation, suggesting that uORF1 does not inhibit scanning ribosomes. However, our data do not formally exclude that ribosome reinitiate after translation of uORF1, a scenario similar to the ATF4 5′UTR. Alternatively, ribosomes may skip the first AUG codon and continue scanning, an event called leaky scanning (35). The major determinants for AUG recognition by scanning ribosomes are the nucleotides surrounding the AUG codon called the Kozak consensus (13, 36, 37). In general, an optimal Kozak consensus contains both an A/G at –3 and a G at +4 given that A of AUG is +1. An AUG codon that contains either of these determinants is considered to be in strong AUG context, whereas an AUG lacking both determinants is considered a weak context. The AUG codons for both uORFs in the hGADD34 and mGADD34 5′UTRs are in poor context, thus predicting that leaky scanning occurs within the 5′UTR.
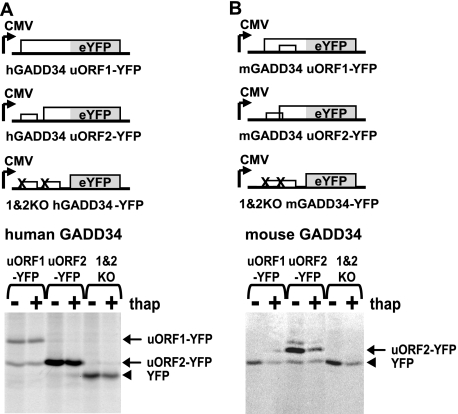
To determine whether the uORFs are recognized by scanning ribosomes, we engineered mutant hGADD34-YFP and mGADD34-YFP reporter RNAs in which the stop codon for each uORF was mutated and the uORF fused in-frame with the reporter YFP ORF (Fig. 7). The design of hGADD34 uORF1-YFP was such that the AUG codon of uORF2 is in the same frame, thus we can determine whether scanning ribosomes skip the first AUG codon. For the mGADD34 uORF1-YFP, the AUG codon of uORF2 was mutated to fuse uORF1 in frame with the YFP ORF.
FIGURE 7.
The downstream uORF2 of hGADD34 and mGADD34 5′UTRs are translated efficiently in HepG2 cells. Schematics of human (A) or mouse (B) GADD34 5′UTR-YFP reporter RNAs stably expressed in HepG2 cells. The upstream (uORF1) or downstream (uORF2) uORFs were engineered such that the reading frames of the uORF were fused with the reporter YFP ORF. For the human hGADD34 uORF1-YFP RNA, the uORF2 is in-frame with uORF1 and the YFP ORF, thus both uORF1-YFP and uORF2-YFP fusion protein products can be monitored. Below, immunoprecipitates of newly synthesized 35S-labeled YFP protein from lysates of cells in the presence or absence of 1 μm thapsigargin. Reactions were resolved on an SDS-PAGE and exposed by phosphorimaging analysis. uORF1-YFP, uORF2-YFP, and YFP ORF proteins are indicated to the right.
Under basal conditions, translation of uORF2-YFP was readily observed from the human and mouse versions of GADD34 5′UTR constructs (Fig. 7). Thapsigargin treatment moderately inhibited uORF2-YFP translation. In contrast, we observed different results with uORF1-YFP. The hGADD34 uORF1-YFP RNA produced two protein products representing translation from the AUG codon of both uORF1 and uORF2 (Fig. 7A). Thus, a proportion of scanning ribosomes skips the first AUG codon and directs translation from the downstream uORF2 AUG codon, indicating that leaky scanning was involved. Interestingly, thapsigargin treatment did not significantly affect translation of hGADD34 uORF1-YFP or uORF2-YFP or the extent of leaky scanning. Surprisingly, the mGADD34 uORF1-YFP did not produce the expected fusion uORF1-YFP protein and only resulted in translation of the YFP ORF (which is fused in-frame) (Fig. 7B). Therefore, scanning ribosomes most likely skip uORF1 to initiate translation at the downstream YFP ORF AUG codon. Levels of reporter uORF1-YFP and uORF2-YFP RNA were similar (data not shown). In summary, the 5′UTRs of human and mouse GADD34 permitted a significant amount of leaky scanning of ribosomes to bypass the AUG codon of uORF1 and directed translation of uORF2, which is a strong barrier of scanning ribosomes.
DISCUSSION
eIF2α phosphorylation elicits a cascade of signaling events that reprograms gene expression in order for the cell to adapt to environmental stress. Paradoxically, translational inhibition via eIF2α phosphorylation activates the translation of a subset of mRNAs (e.g. ATF4 and yeast GCN4), which encode transcription factors that activate downstream stress-response genes. A key component of this cellular response is the induction of GADD34 protein expression. In this report, we demonstrate that GADD34 induction is in part mediated through a 5′UTR mechanism that increases translation during ER stress. GADD34 mRNAs associate with and shift to heavier polysomes during thapsigargin- and DTT-induced ER stress (Fig. 1, D–G), indicating that GADD34 is translated under conditions of general translational arrest when eIF2α is phosphorylated. Specifically, the 5′UTR of both human and mouse GADD34 mRNAs is essential and sufficient to direct efficient translation during ER stress. Through reporter constructs, we showed that the GADD34 5′UTR induced re-distribution of reporter mRNAs to heavier polysomes during eIF2α phosphorylation, which is similar to the distribution of endogenous GADD34 mRNAs (Fig. 4 and supplemental Fig. S2). Our data, together with earlier reports (38), support the notion that induction of GADD34 translation during eIF2α phosphorylation plays an important role in the general stress response, which alleviates cellular stress, including ER stress, oxidative stress, and hypoxia.
GADD34 is an essential component of the UPR (24, 26). GADD34 interacts with protein phosphatase 1 via its C-terminal region to dephosphorylate eIF2α, leading to translational recovery (24, 26, 39, 40). Ectopic expression of a truncated GADD34 protein lacking the catalytic C-terminal region in mouse embryo fibroblasts prevents dephosphorylation eIF2α and blocks translational recovery during ER stress, resulting in premature apoptosis (24). The heightened sensitization to cell death is likely due to translational repression of stress-induced transcripts, such as the chaperone BiP, which are normally expressed during ER stress (26). Thus, the increased expression of GADD34 results in a negative feedback loop to enhance the translation of stress-induced mRNAs during eIF2α phosphorylation, which are essential for cellular survival and adaptation to environmental stress. GADD34 is also regulated at the transcriptional level, which is induced in part by ATF4 and CHOP, which are expressed during ER stress (25, 27). However, it was unclear whether the increase in GADD34 mRNA alone yielded sufficient GADD34 protein during eIF2α phosphorylation to promote the negative feedback loop. Our data demonstrates that GADD34 is translationally induced to ensure maximal expression during ER stress. We showed that GADD34 mRNAs associated with more ribosomes during ER stress, suggesting that eIF2α phosphorylation leads to increased GADD34 translation (Fig. 1, D–G). In addition, GADD34 protein increased under thapsigargin treatment in the presence of the transcription inhibitor actinomycin D (Fig. 2, C and D). This indicated that ongoing transcription was not required for GADD34 induction and that existing basal GADD34 mRNAs can be translated during ER stress. In support of this, we showed that GADD34 mRNAs associated with higher molecular weight polysomes in cells treated with thapsigargin and actinomycin D (Fig. 2E and supplemental Fig. S2). However, the induction of GADD34 protein was moderate in thapsigargin/actinomycin D-treated cells compared with that of cells treated with thapsigargin alone and was not sufficient to reduce eIF2α phosphorylation at later time points (Fig. 2D). These results are in agreement with previous observations that impaired expression of GADD34 in CHOP–/– mice embryonic fibroblasts leads to persistent eIF2α phosphorylation and loss of protein synthesis recovery (27). Therefore, the coordinated transcriptional and translational controls of GADD34 are necessary for optimal expression to dephosphorylate eIF2α and for translational recovery.
Our results revealed that the uORFs of the human and mouse GADD34 5′UTR played a significant role under basal and stress conditions. 5′UTR uORFs have been shown previously to be important for translational control of several mRNAs (41). In general, uORFs act as barriers to scanning ribosomes thereby modulating translation of the authentic ORF. Under certain cellular conditions, ribosomes can bypass the uORFs and reinitiate translation at the main ORF. The best studied mechanism of translational reinitiation is the one governing ATF4, ATF5, and yeast GCN4 translation. The major premise of this mechanism is that the translation of the upstream uORF stimulates translation or reinitiation at a downstream AUG, whereas translation of the downstream uORF leads to translation termination and dissociation of ribosomes. Following translation of the upstream uORF, if eIF2 levels become limiting (i.e. eIF2α is phosphorylated), the recruitment of the ternary complex by the ribosome is markedly reduced and as a result ribosomes have a higher probability of reinitiating translation after the downstream uORF and thereby reinitiate translation at the authentic ORF. Major determinants that control the extent of reinitiation are the intercistronic space between the uORFs and the length of the upstream uORF. After translation of uORF1, the probability of reinitiating at the downstream uORF2 depends on the length of the intercistronic space to allow for scanning ribosomes to re-acquire the ternary complex (32–34). The shorter the intercistronic space, the greater chance that scanning ribosomes will bypass uORF2. Another feature of this mechanism is that the extent of reinitiation decreases the longer the uORF, whereas efficient reinitiation occurs with translation of very short uORFs (2–3 amino acids long) (21, 42). ATF4, ATF5, and GCN4 all contain short uORFs. It has been proposed that ribosomes that translate a short uORF are still bound to certain initiation factors (i.e. eIF4G), which promotes reinitiation (43). However, when longer uORFs are translated, these factors fall off the ribosome, thus preventing reinitiation (43).
Our data indicate that the two uORFs within the human and mouse GADD34 5′UTRs control translation through a mechanism that is distinct from the well studied reinitiation mechanism. First, neither the human GADD34 uORF1 nor the mouse GADD34 5′UTR can promote reinitiation (Fig. 5 and 6). In fact, the human uORF1 moderately inhibited scanning ribosomes as compared with a 5′UTR with no uORFs (Fig. 5). Secondly, human and mouse uORF1s are poorly translated and a significant proportion of ribosomes scan past uORF1 to initiate translation at a downstream AUG (Fig. 7). In contrast, the uORF1 of ATF4 is readily translated, which is a prerequisite for reinitiation at the uORF2 under basal conditions and at the main ATF4 ORF during eIF2α phosphorylation (17, 20). Instead, the presence of the GADD34 uORF2 was sufficient for translational control. An intact uORF2 reduces translation during unstressed conditions and is important for translational induction during stressed conditions, which incidentally are all properties that are observed with the endogenous GADD34 mRNA (Fig. 4, 5, 6, data not shown).
In addition, the features and arrangement of the GADD34 uORFs do not fit the model of reinitiation. The mouse GADD34 uORFs overlap by one nucleotide, whereas the human uORFs are separated by 30 nucleotides. In the case of the mouse GADD34 uORFs, if reinitiation plays a role in this regulation, ribosomes would have to reinitiate backwards (following translation of uORF1) to start translation at the AUG codon of uORF2. Although there have been reports of backward reinitiation, the extent of this was either very inefficient or requires a specialized RNA sequence that binds to ribosome-recruiting factors (i.e. eIF3) (42, 44). When extra codons were introduced to extend the overlap between the start and stop codons, the mutant mouse GADD34 5′UTR was still functional like the wild-type version, arguing against backward reinitiation (Fig. 6). In the case of the human GADD34, the uORFs are separated by a 30-nucleotide intercistronic spacer, which is considerably shorter than those separating uORFs in human ATF4 (87 nucleotides), ATF5 (110 nucleotides), and yeast GCN4 (198 nucleotides between uORF1 and uORF4) uORFs. Furthermore, whereas the upstream uORF of ATF4, ATF5, and GCN4 5′UTRs are short, uORF1 of the human GADD34 is relatively long (22 amino acids), which is predicted to greatly decrease the efficiency of reinitiation (32–34). These features argue that leaky scanning occurs on the GADD34 5′UTR to bypass uORF1 and initiate translation at uORF2.
The uORF2 of GADD34 is a strong barrier for scanning ribosomes, which efficiently represses GADD34 translation under basal conditions. A probable scenario is that, after translating uORF2, ribosomes dissociate, thus inhibiting translation of the GADD34 ORF. Alternatively, the uORF2 may mediate other effects. It is has been previously shown that the uORF in fungal mRNAs that encode a subunit of Arg-specific carbamoyl phosphate synthetase controls translation by stalling scanning ribosomes and mediating nonsense-mediated decay (45, 46). In this case, the translated coding sequence of the fungal uORF dictates translational control (47, 48). Interestingly, a comparison of mammalian GADD34 5′UTRs reveal that uORF2 is highly conserved. The human, chimp, and rat GADD34 5′UTRs contain non-overlapping uORFs, and the mouse and hamster versions contain overlapping uORFs. Although the uORF1s differ in length and are poorly conserved, the uORF2s are exactly 26 amino acids long and contain 18/26 identical amino acids. It remains to be determined whether the amino acid or nucleotide sequence composition within uORF2 plays a significant role in translational control during eIF2α phosphorylation. The high degree of conservation of uORF2 further strengthens the idea that translational control is mediated by this uORF specifically. However, it is unclear why a dispensable uORF1 has evolved within the GADD34 5′UTR. It is possible that uORF1 has an unexplored role under a different set of cellular stress conditions.
An unresolved question is how do scanning ribosomes bypass uORF2 and reach the GADD34 ORF when eIF2α is phosphorylated? Other than the reinitiation mechanism, additional mechanisms that can bypass eIF2α phosphorylation have been described. One such mechanism proposes that ribosomes are directly recruited to the 5′UTR downstream of uORF2, possibly through an IRES. The most unique example is via the cricket paralysis virus intergenic region IRES, which can bypass the requirement for all initiation factors to recruit the ribosome and can induce translation during eIF2α phosphorylation (31, 49). Another example is cat-1. In response to amino acid starvation, translation of cat-1 is induced, which is mediated in part by an IRES within its 5′UTR and requires translation of an uORF (50). However, our data indicate that an IRES-like mechanism does not direct GADD34 translation.3
Interestingly, an inhibitory uORF within the 5′UTR of ATF4 and GCN4 can also induce translation during eIF2α phosphorylation (17, 51). Moreover, the transcription factors C/EBPα and C/EBPβ, are also regulated by a single uORF in response to eIF2α phosphorylation (52, 53). These mechanisms are currently poorly understood and appear to be distinct from the reinitiation mechanism (17). Further studies are required to determine whether the mechanisms via these single uORFs and the GADD34 5′UTR share similar properties.
The finding that both transcriptional and translational mechanisms control GADD34 expression suggests that GADD34 is tightly regulated. Indeed, overexpression of GADD34 leads to apoptosis in some cell lines (54–56), and forced expression of the C-terminal region of GADD34 in mice causes dysfunction in glucose metabolism in the liver (53). Our results indicate that uORF2 regulation within the 5′UTR maintains low basal GADD34 expression during unstressed conditions and is important in the translational induction for optimal expression during cellular stress. Given that uORFs are predicted in ∼25% of all 5′UTRs, further characterization of GADD34 uORF translational control is warranted (57). The elucidation of the GADD34 5′UTR mechanism may shed light on how other stress-induced mRNAs are translated during eIF2α phosphorylation.
Supplementary Material
Acknowledgments
We thank Bruno Fonseca and Julianne Garrey for critical reading of the manuscript. Maria Hatzoglou kindly provided the mouse Hepa cells and Jim Johnson provided the CHOP antibody.
This work was funded by a Canadian Institutes for Health Research operating grant (to E. J.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; PERK, protein kinase R-like ER kinase; eIF2α, eukaryotic initiation factor 2α; ORF, open reading frame; uORF, upstream ORF; UTR, untranslated region; UPR, unfolded protein response; RACE, rapid amplification of cDNA ends; YFP, yellow fluorescent protein; DTT, dithiothreitol; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IRES, internal ribosome entry site; C/EBP, CAAT/enhancer-binding protein; GADD34, growth arrest DNA-inducible gene 34.
E. Jan, unpublished data.
References
- 1.Ron, D., and Harding, H. P. (2007) in Translational Control in Biology and Medicine (Mathews, M. B., Sonenberg, N., and Hershey, J., eds) pp. 345–368, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 2.Dever, T. E., Dar, A. C., and Sicheri, F. (2007) in Translational Control in Biology and Medicine (Mathews, M. B., Sonenberg, N., and Hershey, J., eds) pp. 319–344, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 3.Kostura, M., and Mathews, M. B. (1989) Mol. Cell. Biol. 9 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han, A. P., Yu, C., Lu, L., Fujiwara, Y., Browne, C., Chin, G., Fleming, M., Leboulch, P., Orkin, S. H., and Chen, J. J. (2001) EMBO J. 20 6909–6918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J. J., Throop, M. S., Gehrke, L., Kuo, I., Pal, J. K., Brodsky, M., and London, I. M. (1991) Proc. Natl. Acad. Sci. U. S. A 88 7729–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dever, T. E., Feng, L., Wek, R. C., Cigan, A. M., Donahue, T. F., and Hinnebusch, A. G. (1992) Cell 68 585–596 [DOI] [PubMed] [Google Scholar]
- 7.Harding, H. P., Zhang, Y., and Ron, D. (1999) Nature 397 271–274 [DOI] [PubMed] [Google Scholar]
- 8.Tardif, K. D., Mori, K., and Siddiqui, A. (2002) J. Virol. 76 7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding, H. P., Zeng, H., Zhang, Y., Jungries, R., Chung, P., Plesken, H., Sabatini, D. D., and Ron, D. (2001) Mol. Cell 7 1153–1163 [DOI] [PubMed] [Google Scholar]
- 10.Schneider, R., and Sonenberg, N. (2007) in Translational Control in Biology and Medicine (Mathews, M. B., Sonenberg, N., and Hershey, J., eds) pp. 401–432, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 11.Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S., and Kaufman, R. J. (2001) Mol. Cell 7 1165–1176 [DOI] [PubMed] [Google Scholar]
- 12.Alwine, J. C. (2008) Curr. Top. Microbiol. Immunol. 325 263–279 [DOI] [PubMed] [Google Scholar]
- 13.Pestova, T. V., Lorsch, J. R., and Hellen, C. U. (2007) in Translational Control in Biology and Medicine (Mathews, M. B., Sonenberg, N., and Hershey, J., eds) pp. 87–128, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 14.Proud, C. G. (2005) Semin. Cell Dev. Biol. 16 3–12 [DOI] [PubMed] [Google Scholar]
- 15.Rowlands, A. G., Panniers, R., and Henshaw, E. C. (1988) J. Biol. Chem. 263 5526–5533 [PubMed] [Google Scholar]
- 16.Harding, H. P., Novoa, I., Zhang, Y., Zeng, H., Wek, R., Schapira, M., and Ron, D. (2000) Mol. Cell 6 1099–1108 [DOI] [PubMed] [Google Scholar]
- 17.Lu, P. D., Harding, H. P., and Ron, D. (2004) J. Cell Biol. 167 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou, D., Palam, L. R., Jiang, L., Narasimhan, J., Staschke, K. A., and Wek, R. C. (2008) J. Biol. Chem. 283 7064–7073 [DOI] [PubMed] [Google Scholar]
- 19.Watatani, Y., Ichikawa, K., Nakanishi, N., Fujimoto, M., Takeda, H., Kimura, N., Hirose, H., Takahashi, S., and Takahashi, Y. (2008) J. Biol. Chem. 283 2543–2553 [DOI] [PubMed] [Google Scholar]
- 20.Vattem, K. M., and Wek, R. C. (2004) Proc. Natl. Acad. Sci. U. S. A 101 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, R. J., Kaminski, A., and Poyry, T. A. A. (2007) in Translational Control in Biology and Medicine (Mathews, M. B., Sonenberg, N., and Hershey, J., eds) pp. 197–224, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 22.Kenzelmann, M., Maertens, S., Hergenhahn, M., Kueffer, S., Hotz-Wagenblatt, A., Li, L., Wang, S., Ittrich, C., Lemberger, T., Arribas, R., Jonnakuty, S., Hollstein, M. C., Schmid, W., Gretz, N., Gröne, H. J., and Schütz, G. (2007) Proc. Natl. Acad. Sci. U. S. A 104 6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding, H. P., Zhang, Y., Zeng, H., Novoa, I., Lu, P. D., Calfon, M., Sadri, N., Yun, C., Popko, B., Paules, R., Stojdl, D. F., Bell, J. C., Hettmann, T., Leiden, J. M., and Ron, D. (2003) Mol. Cell 11 619–633 [DOI] [PubMed] [Google Scholar]
- 24.Novoa, I., Zeng, H., Harding, H., and Ron, D. (2001) J. Cell Biol. 153 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma, Y., and Hendershot, L. M. (2003) J. Biol. Chem. 278 34864–34873 [DOI] [PubMed] [Google Scholar]
- 26.Novoa, I., Zhang, Y., Zeng, H., Jungreis, R., Harding, H. P., and Ron, D. (2003) EMBO J. 22 1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marciniak, S., Yun, C., Oyadomari, S., Novoa, I., Zhang, Y., Jungreis, R., Nagata, K., Harding, H., and Ron, D. (2004) Genes Dev. 18 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johannes, G., Carter, M. S., Eisen, M. B., Brown, P. O., and Sarnow, P. (1999) Proc. Natl. Acad. Sci. U. S. A 96 13118–13123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jan, E., Kinzy, T. G., and Sarnow, P. (2003) Proc. Natl. Acad. Sci. U. S. A 100 15410–15415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jan, E., and Sarnow, P. (2002) J. Mol. Biol. 324 889–902 [DOI] [PubMed] [Google Scholar]
- 31.Fernandez, J., Yaman, I., Sarnow, P., Snider, M. D., and Hatzoglou, M. (2002) J. Biol. Chem. 277 19198–19205 [DOI] [PubMed] [Google Scholar]
- 32.Grant, C. M., Miller, P. F., and Hinnebusch, A. G. (1994) Mol. Cell. Biol. 14 2616–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak, M. (1987) Mol. Cell. Biol. 7 3438–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Child, S. J., Miller, M. K., and Geballe, A. P. (1999) J. Biol. Chem. 274 24335–24341 [DOI] [PubMed] [Google Scholar]
- 35.Kozak, M. (1991) J. Biol. Chem. 266 19867–19870 [PubMed] [Google Scholar]
- 36.Kozak, M. (1984) Nature 308 241–246 [DOI] [PubMed] [Google Scholar]
- 37.Kozak, M. (1986) Cell 44 283–292 [DOI] [PubMed] [Google Scholar]
- 38.Koritzinsky, M., Magagnin, M. G., van den Beucken, T., Seigneuric, R., Savelkouls, K., Dostie, J., Pyronnet, S., Kaufman, R. J., Weppler, S. A., Voncken, J. W., Lambin, P., Koumenis, C., Sonenberg, N., and Wouters, B. G. (2006) EMBO J. 25 1114–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brush, M. H., Weiser, D. C., and Shenolikar, S. (2003) Mol. Cell. Biol. 23 1292–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Connor, J. H., Weiser, D. C., Li, S., Hallenbeck, J. M., and Shenolikar, S. (2001) Mol. Cell. Biol. 21 6841–6850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sachs, M. S., and Geballe, A. P. (2006) Genes Dev. 20 915–921 [DOI] [PubMed] [Google Scholar]
- 42.Kozak, M. (2001) Nucleic Acids Res. 29 5226–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poyry, T. A., Kaminski, A., and Jackson, R. J. (2004) Genes Dev. 18 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poyry, T. A., Kaminski, A., Connell, E. J., Fraser, C. S., and Jackson, R. J. (2007) Genes Dev. 21 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaba, A., Jacobson, A., and Sachs, M. S. (2005) Mol. Cell 20 449–460 [DOI] [PubMed] [Google Scholar]
- 46.Gaba, A., Wang, Z., Krishnamoorthy, T., Hinnebusch, A. G., and Sachs, M. S. (2001) EMBO J. 20 6453–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang, P., Wang, Z., and Sachs, M. S. (2000) J. Biol. Chem. 275 26710–26719 [DOI] [PubMed] [Google Scholar]
- 48.Fang, P., Spevak, C. C., Wu, C., and Sachs, M. S. (2004) Proc. Natl. Acad. Sci. U. S. A 101 4059–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jan, E. (2006) Virus Res. 119 16–28 [DOI] [PubMed] [Google Scholar]
- 50.Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A. A., Koromilas, A. E., Zhou, L., Snider, M. D., Scheuner, D., Kaufman, R. J., and Hatzoglou, M. (2003) Cell 113 519–531 [DOI] [PubMed] [Google Scholar]
- 51.Mueller, P. P., and Hinnebusch, A. G. (1986) Cell 45 201–207 [DOI] [PubMed] [Google Scholar]
- 52.Calkhoven, C. F., Muller, C., and Leutz, A. (2000) Genes Dev. 14 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 53.Oyadomari, S., Harding, H. P., Zhang, Y., Oyadomari, M., and Ron, D. (2008) Cell Metab. 7 520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hollander, M. C., Poola-Kella, S., and Fornace, A. J., Jr. (2003) Oncogene 22 3827–3832 [DOI] [PubMed] [Google Scholar]
- 55.Adler, H. T., Chinery, R., Wu, D. Y., Kussick, S. J., Payne, J. M., Fornace, A. J., Jr., and Tkachuk, D. C. (1999) Mol. Cell. Biol. 19 7050–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hollander, M. C., Zhan, Q., Bae, I., and Fornace, A. J., Jr. (1997) J. Biol. Chem. 272 13731–13737 [DOI] [PubMed] [Google Scholar]
- 57.Iacono, M., Mignone, F., and Pesole, G. (2005) Gene (Amst.) 349 97–105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.