Abstract
Stochastic synthesis of a ligand coupled to a nanoparticle results in a distribution of populations with different numbers of ligands per nanoparticle. This distribution was resolved and quantified using HPLC, and is in excellent agreement with the ligand/nanoparticle average measured by 1H NMR, GPC, and potentiometric titration, yet significantly more disperse than commonly held perceptions of monodispersity. Two statistical models were employed to confirm that the observed heterogeneity is consistent with theoretical expectations.
Stochastic ligand conjugation is a common strategy to produce practical quantities of functionalized nanoparticles. Analytical methods used to quantify the average nanoparticle to ligand ratio, such as NMR, UV/vis spectroscopy, and elemental analysis, provide no information about the distribution of ligands bound to each particle. In this study, we quantitatively analyze the HPLC trace of the conjugated nanoparticle, including “tailing” effects, and show that the heterogeneity implied by the entire trace is consistent with theoretical expectations regarding the dispersity of the sample. The width of the distribution exceeds typical community expectations regarding sample homogeneity and is not well represented by a conjugated nanoparticle showing the average number of conjugated ligands. Given the importance of nanoparticle conjugates for applications such as drug delivery (1–5), biomedical diagnostics (6, 7), and sensing (8, 9), it is paramount that a detailed understanding of the product distribution resulting from a stochastic synthesis process be gained.
A number of groups have endeavored to characterize the distribution of ligands on nanoparticles. A variety of methods have been employed including gel electrophesis (10), anion-exchange HPLC (11), ultra performance liquid chromatography (12), mass spectrometry (13), and fluorescence quenching (14). In addition, synthetic efforts have focused on making particles with controlled 1:1 ligand/particle ratios (15–18). If a stochastic synthesis process is used to generate a 1:1 ratio, what percentage of the sample will be functional? In this study, we focus on the distribution that exists for ensembles with approximately one ligand/particle. This study employs partially acetylated dendrimers because these materials have been shown to be most effective for targeted drug delivery applications (19, 20). The parent amine-terminated materials have been shown to non-selectively cause cell membrane permeability and are not suited for targeted drug delivery applications (21–23).
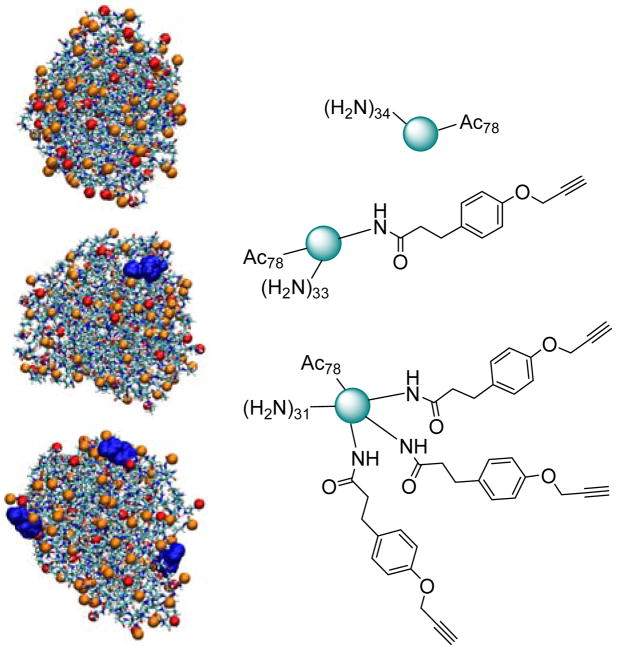
A series of reactions were conducted to conjugate varying amounts of 3-(4-(propynyloxy)phenyl)propanoic acid to the surface primary amines of a partially acetylated generation 5 poly(amidoamine) dendrimer (G5 PAMAM; G5(Ac)78(NH2)34) (Figure 1). Note that this ligand is suitable for “click” chemistry applications. These products were analyzed by 1H NMR spectroscopy and HPLC. The 1H NMR analysis, when combined with GPC and potentiometic titration data, gives information about the average number of ligands bound per particle. The HPLC data, when combined with a peak fitting analysis, provides both the distribution of ligands per dendrimer as well as the average number of ligands per dendrimer.
Figure 1.
Equilibrated molecular dynamics models and schematics of G5 PAMAM dendrimers with different numbers of ligands. Terminal amines (red), acetyl groups (orange), and ligands (blue) are depicted in the models. Corresponding schematic representations show the dendrimer in teal with terminal groups. PAMAM dendrimers are monodisperse, highly ordered, water soluble, polymeric nanoparticles (~4.5 nm diameter). Terminal amines can be used as coupling points to attach different ligands.
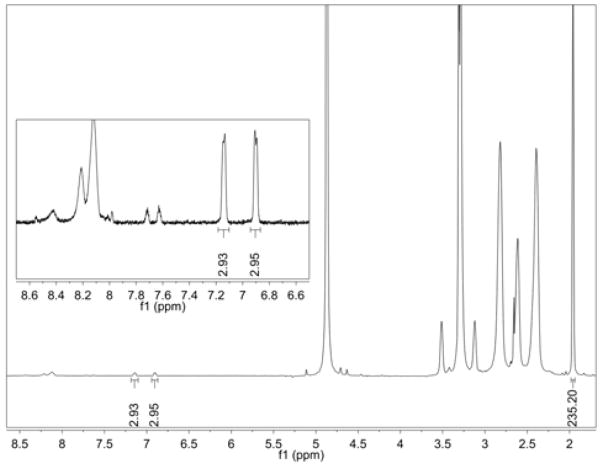
The ligand-dendrimer conjugates (samples A–D) were determined to have an average of 0.20, 0.60, 1.04, and 1.47 ligands per dendrimer by comparing the integration of the methyl protons in the terminal acetyl groups to the aromatic protons on the conjugated ligand (Figure 2). The number of acetyl groups per dendrimer was independently determined by first computing the total number of end groups from the number average molecular weight (GPC) and potentiometric titration data for G5-NH2(100%) as previously described (24). This value for the total number of end groups was applied to the ratio of primary amines to acetyl groups, obtained from the 1H NMR of the partially acetylated dendrimer, to compute the average number of acetyl groups per dendrimer. This determination is sensitive to both the total number of particle functional groups as well as the number that have been acetylated. The excellent dispersity characteristics of PAMAM dendrimers (PDI = 1.01) greatly facilitates this analysis.
Figure 2.
1H NMR spectrum for Sample D. The average number of ligands per dendrimer was determined by comparison of the aromatic protons on the ligand vs. the methyl protons in the acetyl group of the dendrimer. The number of acetyl groups was determined independently by GPC and potentiometric titration.
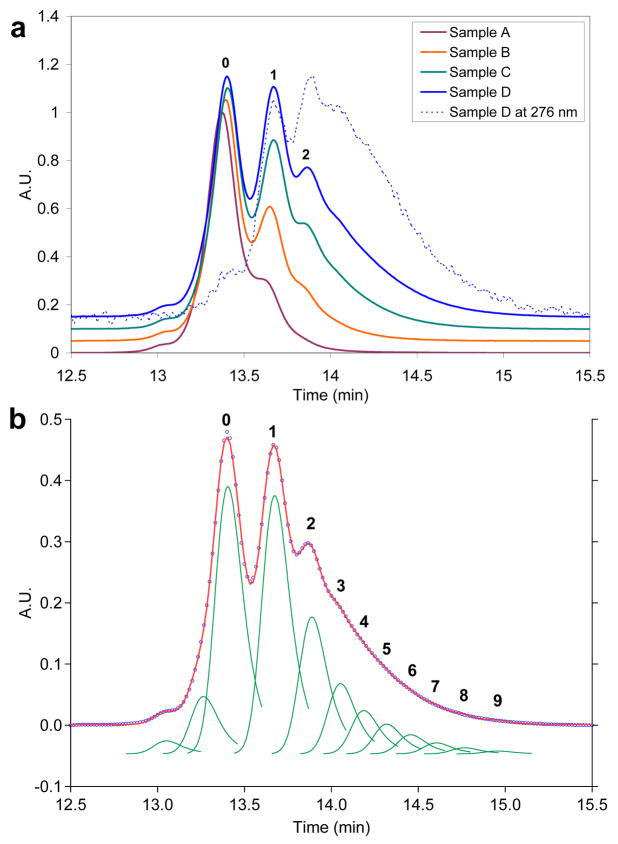
The HPLC elution profiles obtained at 210 nm for samples A–D are illustrated in Figure 3a, solid traces. The 210 nm wavelength was selected because it is convenient for monitoring the PAMAM dendrimers and is not significantly affected by varying amounts of conjugated ligands (25). The first large peak (0) appears at an elution time consistent with unmodified G5(Ac)78(NH2)34. The small peaks preceding peak 0 are also present in the original G5(Ac)78(NH2)34 sample and likely result from a small amount of lower generation dendrimer (26). The second large peak (1) was preliminarily assigned as G5(Ac)78(NH2)33(L)1 (L = NHCO(CH2)2C6H4OCH2C2H) based upon elution order (Figure 3a, solid traces A–D). Additional data supporting this assignment was obtained from the elution profile monitored at 276 nm (sample D, dashed trace). At 276 nm, an absorbance maximum for the ligand with minimal contribution from the dendrimer, peak 0 (G5(Ac)78(NH2)34) largely disappears as anticipated for dendrimer with no ligand conjugated. In this case, the first major feature observed (peak 1) was assigned as dendrimer containing one conjugated ligand (G5(Ac)78(NH2)33(L)1). Note that this is consistent with the assignment obtained based on monitoring dendrimer at 210 nm.
Figure 3.
HPLC elution data for dendrimer-ligand conjugates: (a) Dendrimer concentration monitored at 210 nm for Samples A–D are shown with solid lines. The elution profile of Sample D at 276 nm, which is the maximum absorbance for the ligand, is displayed with a dashed line. Absorbance is normalized to the peak maximum. (b) The elution profile at 210 nm for Sample D is shown in blue. Individual fitted peaks are presented in green and the summation of the fitted peaks are in red.
For the HPLC traces monitoring concentration of dendrimer at 210 nm, up to three distinct species are clearly resolved with a fourth apparent as an inflection point in trace D. Since the absorbance at 210 nm scales linearly with dendrimer concentration, we can use the area of each absorbance peak to obtain the relative concentration of each different dendrimer/ligand conjugate. Dilution studies of the conjugates demonstrate that Beer’s law is followed and that each of the fitted peaks has the same extinction coefficient (See Supporting Information). The data taken at 276 nm is useful in helping to confirm the HPLC peak assignments, but cannot be used quantitatively for concentration determinations because each dendrimer/ligand conjugate generates a dramatically different local concentration of ligand thus causing a deviation from Beer’s law.
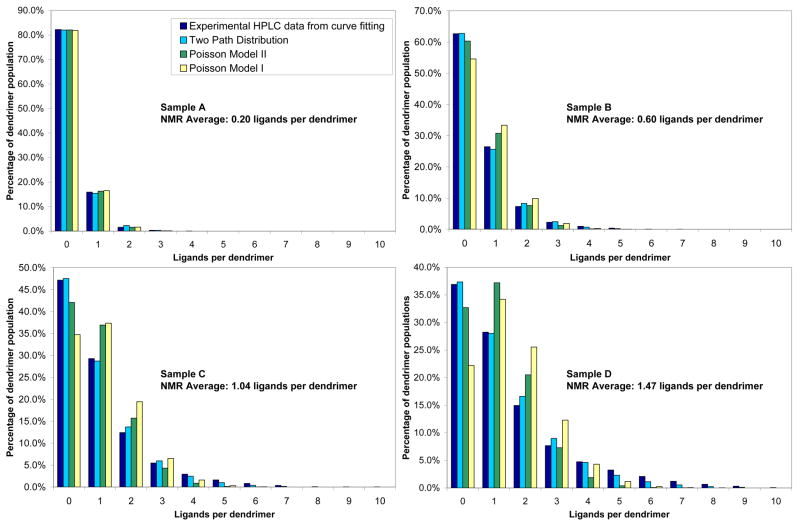
In order to quantitatively assess the relative concentration of each dendrimer/ligand conjugate present, it was necessary to apply a peak fitting procedure to the HPLC traces. The functional form of the dendrimer peaks was determined by fitting the elution profile of acylated dendrimer (G5(Ac)78(NH2)34) using Igor Pro 6.01. The peak shape, a Gaussian with an exponential decay tail to the right side of the elution peak, was then applied uniformly to all fitted peaks. The position and area of peaks 0–9 and the two lower generation impurity peaks at ~13 min were not constrained. The fit for sample D, which has a 1.47 ligand/dendrimer ratio measured by 1H NMR spectroscopy, is illustrated in Figure 3b. From these fits, the relative concentration of each dendrimer/ligand conjugate was determined (Figure 4) as well as the average ligand/dendrimer ratio (Table 1).
Figure 4.
Experimental and statistical distributions of ligand-dendrimer conjugates A–D. Experimental distribution is calculated from fitted peaks to the HPLC elution profiles.
Table 1.
Comparison of the average number of ligands per dendrimer independently computed by NMR and HPLC techniques with the three statistical models.
| NMR Average | HPLC Averagea | Poisson I (χ2 = 66)b | Poisson II (χ2 = 47) | Two Path (χ2 = 9) | |
|---|---|---|---|---|---|
| Sample A | 0.20 ± 0.02 | 0.20 ± 0.01 | 0.20 | 0.21 | 0.21 |
| Sample B | 0.60 ± 0.06 | 0.54 ± 0.02 | 0.60 | 0.53 | 0.54 |
| Sample C | 1.04 ± 0.10 | 0.98 ± 0.04 | 1.04 | 0.91 | 0.93 |
| Sample D | 1.47 ± 0.15 | 1.45 ± 0.06 | 1.47 | 1.17 | 1.32 |
Determination of HPLC error is discussed in the Experimental Methods section of the Supporting Information.
The Poisson I model uses the experimental NMR averages as input parameters.
The relative proportions of dendrimer species, resolved by HPLC and quantified through fitted peaks, were used to calculate a weighted average number of ligands per dendrimer. Table 1 displays this for each of the dendrimer-ligand conjugates. This weighted average is in excellent agreement with the average determined independently by the combined NMR/GPC/titration analysis. Indeed, it is this comparison that gives confidence in the physical meaning of the peak fitting procedure. Significantly, the HPLC data produces additional distribution information that could not be extracted from the combined NMR/GPC/titration analysis.
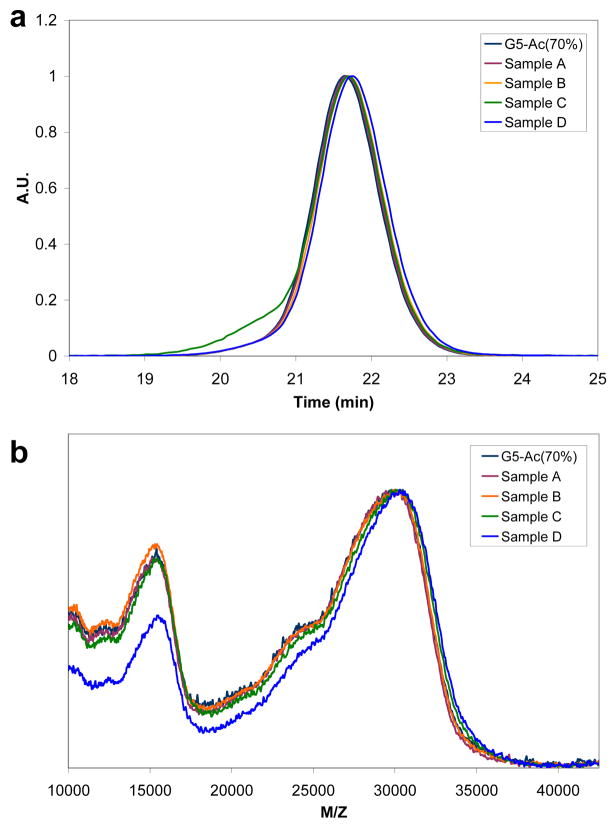
The resolution of the various conjugated species by HPLC stands in stark contrast to the results obtained by NMR (Figure 2) in which no resolution of the various numbers of ligands conjugated is obtained. Gel permeation chromatography (GPC) exhibits a trend towards longer retention time but does not resolve the components (Figure 5a). Note that the longer retention time indicates the conjugates are effectively smaller, as measured by GPC, although the light scattering and refractive index detectors verify that the mass has increased upon conjugation. MALDI-TOF also exhibits a trend to higher mass, but once again the individual components are not resolved (Figure 5b).
Figure 5.
Standard analytical techniques that are commonly utilized to characterize nanoparticle conjugates fail to detect the different dendrimer-ligand populations: (a) The light scattering data of four dendrimer-ligand samples and partially acetylated dendrimer starting material, as separated by Gel Permeation Chromatography (GPC), clearly demonstrates the challenges in detecting the different populations based on differences in size. The single peak resolution achieved by GPC is in stark contrast to the multiple peak resolution achieved by HPLC (See Supporting Information for GPC conditions). (b) Similarly, molecular weight analysis of the same material by Matrix-Assisted, Laser-Desorption Time-of-Flight Mass Spectroscopy (MALDI-TOF) cannot detect significant differences between the five samples. (See Supporting Information for MALDI-TOF conditions)
Three statistical models were employed for comparison with the experimentally determined distributions (Figure 4, Table 1). Poisson model I assumed that ligand conjugation with the nanoparticle proceeds in a stochastic fashion. The total number of available attachment points on the dendrimer surface (34) and the average ligand/dendrimer ratio determined by NMR were used as input. This fit gave a χ2 per degree of freedom of 66. In Poisson model II, the ligand/dendrimer ratios were allowed to vary as fitting parameters in a simultaneous χ2 minimization using all four data sets. This fit gave a χ2 per degree of freedom of 47. Both Poisson models match the 0.20 and 0.60 ligand/dendrimer distributions quite well but begin to deviate from the experimental data as the ligand/dendrimer ratio increases.
In order to better reproduce the experimental ligand distributions, a two path kinetic model was used (See Supporting Information). It allowed for deviations from the Poisson distribution by varying the activation energy of the reaction as a function of n ligands on the nanoparticle (eq 1). This two path model was also motivated by previous publications which indicate product amide autocatalysis should be expected for this reaction (27–29).
| (1) |
The two paths correspond to attaching a ligand far from other ligands or near enough to previously attached ligands so that the barrier for attachment is reduced. These rates are fed into a master equation for the concentrations, cn, of dendrimers with n ligands attached: ċn = Rn−1cn−1 − Rncn. A1,2 were adjusted to account for the number of available sites for the two paths. The solutions of the master equations were fit to all four data sets simultaneously using five independent parameters (Ea1-Ea2, and four independent values for the A’s), resulting in a χ2 per degree of freedom of 9. The best fit reproduced the experimental changes in concentration which is a strong confirmation that the approach had physical meaning for this system. The difference in activation barrier between the two paths could be extracted from the fit and was determined to be Ea1 – Ea2 = 4.0 ± 1.5 kJ/mol or 0.041 ± 0.016 eV.
Typically, analyses of functionalized nanoparticles only determine the average number of ligands per nanoparticle. In most cases, even HPLC does not resolve the distribution. Indeed, the “tailing” observed in the plots (Figure 3) is frequently ascribed to non-ideal interaction between the analyte and the chromatography support. In this instance, we provide qualitative and quantitative data to demonstrate that tailing results from the distribution of ligands bound to dendrimers. This claim is supported by the following pieces of information: 1) no tailing is observed for the partially acetylated sample (see Figure S3) 2) the lower conversion samples (A,B) exhibit well-defined peaks 3) dendrimers with a large number of conjugated ligands appear in “tail” region (see Figure S3) 4) the HPLC fits quantitatively agree with the NMR spectra in terms of the assessment of the average number of conjugated ligands.
The resolution of the distribution provides insight into the meaning of average ligand/dendrimer ratios. For the average of 0.20 ligands/dendrimer, the distribution is comprised of over 81% unmodified dendrimer, about 16% dendrimer with one ligand attached, and less than 3% dendrimer with two ligands in fairly good accord with common expectations. However, the dendrimer-ligand conjugate with an average of 1.47 contains 0 to 9 ligands per dendrimer with the largest population of dendrimers actually containing no ligands. Note that both the random distribution and the two path kinetic model accurately predict the breadth of the population. These experimentally measured breadths modeled herein challenge the commonly held perceptions that such a sample would be comprised of a narrower distribution predominantly containing dendrimers with 1 or 2 ligands attached. Furthermore, the average number misrepresents the functionally active portion of the dendrimer sample and does not make it apparent that the most common species contains no ligands and would therefore be inactive.
The deviation at larger ligand/dendrimers ratios of the experimental distribution from a Poisson analysis is quite interesting. The conjugation probability would be expected to decrease due to site blocking effects. Thus, the observation of an increased probability is particularly intriguing. A straightforward explanation of the activation energy barrier difference of ~4 kJ/mol is product amide autocatalysis. This has the interesting implication of creating non-random spatial distribution of ligands on the polymer surface. These observations are of general interest since amide/ester couplings are broadly used for conjugation reactions by the nanoparticle community.
In this paper, we have quantitatively analyzed the ligand/dendrimer distributions arising from conjugation of approximately one ligand on average to a spherical nanoparticle with excellent agreement between experimental measurements and theoretical analysis. Accurate understanding of these distributions for functional nanomaterials can lead to more informed applications and predictions of nanoparticle structure, function, and activity. It is expected that the details of the conjugation chemistry, site saturation effects, and steric blocking caused by the conjugated ligand can all affect the distributions obtained. Studies more closely examining these parameters will be the subject of future reports.
Supplementary Material
Experimental methods, Beer’s Law analysis of dendrimer absorbance at 210 nm, GPC and MALDI-TOF analysis, additional HPLC profiles for the partially acetylated dendrimer starting material and a dendrimer-ligand conjugate with an average of 3.1 ligands, and detailed description of the Two Path Kinetic Model. This information is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under award 1 R01 CA119409, and with Federal funds from the National Science Foundation under award DMS 0554587.
References
- 1.Tong R, Cheng JJ. Anticancer polymeric nanomedicines. Polymer Reviews. 2007;47:345–381. [Google Scholar]
- 2.Gu FX, Karnik R, Wang AZ, Alexis F, Levy-Nissenbaum E, Hong S, Langer RS, Farokhzad OC. Targeted nanoparticles for cancer therapy. Nano Today. 2007;2:14–21. [Google Scholar]
- 3.Rawat M, Singh D, Saraf S, Saraf S. Nanocarriers: Promising vehicle for bioactive drugs. Biological & Pharmaceutical Bulletin. 2006;29:1790–1798. doi: 10.1248/bpb.29.1790. [DOI] [PubMed] [Google Scholar]
- 4.Patri AK, Majoros IJ, Baker JR. Dendritic polymer macromolecular carriers for drug delivery. Current Opinion in Chemical Biology. 2002;6:466–471. doi: 10.1016/s1367-5931(02)00347-2. [DOI] [PubMed] [Google Scholar]
- 5.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotechnology. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 6.Nie SM, Xing Y, Kim GJ, Simons JW. Nanotechnology applications in cancer. Annual Review of Biomedical Engineering. 2007;9:257–288. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 7.Jain KK. Nanotechnology in clinical laboratory diagnostics. Clinica Chimica Acta. 2005;358:37–54. doi: 10.1016/j.cccn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Rossi LM, Quach AD, Rosenzweig Z. Glucose oxidase-magnetite nanoparticle bioconjugate for glucose sensing. Analytical and Bioanalytical Chemistry. 2004;380:606–613. doi: 10.1007/s00216-004-2770-3. [DOI] [PubMed] [Google Scholar]
- 9.Park SJ, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 10.Sperling RA, Pellegrino T, Li JK, Chang WH, Parak WJ. Electrophoretic separation of nanoparticles with a discrete number of functional groups. Advanced Functional Materials. 2006;16:943–948. [Google Scholar]
- 11.Claridge SA, Liang HYW, Basu SR, Frechet JMJ, Alivisatos AP. Isolation of discrete nanoparticle - DNA conjugates for plasmonic applications. Nano Letters. 2008;8:1202–1206. doi: 10.1021/nl0802032. [DOI] [PubMed] [Google Scholar]
- 12.Cason CA, Oehrle SA, Fabre TA, Girten CD, Walters KA, Tomalia DA, Haik KL, Bullen HA. Improved methodology for monitoring poly(amidoamine) dendrimers surface transformations and product quality by ultra performance liquid chromatography. Journal of Nanomaterials 2008 [Google Scholar]
- 13.Tracy JB, Kalyuzhny G, Crowe MC, Balasubramanian R, Choi JP, Murray RW. Poly(ethylene glycol) ligands for high-resolution nanoparticle mass spectrometry. Journal of the American Chemical Society. 2007;129:6706–6707. doi: 10.1021/ja071042r. [DOI] [PubMed] [Google Scholar]
- 14.Casanova D, Giaume D, Moreau M, Martin JL, Gacoin T, Boilot JP, Alexandrou A. Counting the number of proteins coupled to single nanoparticles. Journal of the American Chemical Society. 2007;129:12592–12593. doi: 10.1021/ja0731975. [DOI] [PubMed] [Google Scholar]
- 15.Huo Q, Worden JG. Monofunctional gold nanoparticles: synthesis and applications. Journal of Nanoparticle Research. 2007;9:1013–1025. [Google Scholar]
- 16.Fu AH, Micheel CM, Cha J, Chang H, Yang H, Alivisatos AP. Discrete nanostructures of quantum dots/Au with DNA. Journal of the American Chemical Society. 2004;126:10832–10833. doi: 10.1021/ja046747x. [DOI] [PubMed] [Google Scholar]
- 17.Derfus AM, Chen AA, Min DH, Ruoslahti E, Bhatia SN. Targeted quantum dot conjugates for siRNA delivery. Bioconjugate Chemistry. 2007;18:1391–1396. doi: 10.1021/bc060367e. [DOI] [PubMed] [Google Scholar]
- 18.Ackerson CJ, Jadzinsky PD, Jensen GJ, Kornberg RD. Rigid, specific, and discrete gold nanoparticle/antibody conjugates. Journal of the American Chemical Society. 2006;128:2635–2640. doi: 10.1021/ja0555668. [DOI] [PubMed] [Google Scholar]
- 19.Kukowska-Latallo JF, Candido KA, Cao ZY, Nigavekar SS, Majoros IJ, Thomas TP, Balogh LP, Khan MK, Baker JR. Nanoparticle targeting of anticancer drug improves therapeutic response in animal model of human epithelial cancer. Cancer Research. 2005;65:5317–5324. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]
- 20.Thomas TP, Majoros IJ, Kotlyar A, Kukowska-Latallo JF, Bielinska A, Myc A, Baker JR. Targeting and inhibition of cell growth by an engineered dendritic nanodevice. Journal of Medicinal Chemistry. 2005;48:3729–3735. doi: 10.1021/jm040187v. [DOI] [PubMed] [Google Scholar]
- 21.Hong SP, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi XY, Balogh L, Orr BG, Baker JR, Holl MMB. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: Hole formation and the relation to transport. Bioconjugate Chemistry. 2004;15:774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 22.Hong SP, Leroueil PR, Janus EK, Peters JL, Kober MM, Islam MT, Orr BG, Baker JR, Holl MMB. Interaction of polycationic polymers with supported lipid bilayers and cells: Nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chemistry. 2006;17:728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 23.Leroueil PR, Hong SY, Mecke A, Baker JR, Orr BG, Holl MMB. Nanoparticle interaction with biological membranes: Does nanotechnology present a janus face? Accounts of Chemical Research. 2007;40:335–342. doi: 10.1021/ar600012y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majoros IJ, Thomas TP, Mehta CB, Baker JR. Poly(amidoamine) dendrimer-based multifunctional engineered nanodevice for cancer therapy. Journal of Medicinal Chemistry. 2005;48:5892–5899. doi: 10.1021/jm0401863. [DOI] [PubMed] [Google Scholar]
- 25.Islam MT, Majoros IJ, Baker JR. HPLC analysis of PAMAM dendrimer based multifunctional devices. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2005;822:21–26. doi: 10.1016/j.jchromb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Shi XY, Bi XD, Ganser TR, Hong SP, Myc LA, Desai A, Holl MMB, Baker JR. HPLC analysis of functionalized poly(amidoamine) dendrimers and the interaction between a folate-dendrimer conjugate and folate binding protein. Analyst. 2006;131:842–848. doi: 10.1039/b602546c. [DOI] [PubMed] [Google Scholar]
- 27.Menger FM, Eliseev AV, Khanjin NA. A self-replicating system - new experimental data and a new mechanistic interpretation. Journal of the American Chemical Society. 1994;116:3613–3614. [Google Scholar]
- 28.Menger FM, Eliseev AV, Khanjin NA, Sherrod MJ. Evidence for an alternative mechanism to a previously proposed self-replicating system. Journal of Organic Chemistry. 1995;60:2870–2878. [Google Scholar]
- 29.Titskii GD, Litvinen LM. Nucleophilic catalysis of carboxamides and related compounds in acylation. Zhurnal Obshchei Khimii. 1970;40:2680–2688. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental methods, Beer’s Law analysis of dendrimer absorbance at 210 nm, GPC and MALDI-TOF analysis, additional HPLC profiles for the partially acetylated dendrimer starting material and a dendrimer-ligand conjugate with an average of 3.1 ligands, and detailed description of the Two Path Kinetic Model. This information is available free of charge via the Internet at http://pubs.acs.org.