Abstract
A novel approach to pancreatic cancer biomarker discovery has been developed, which employs a stable isotope labeled proteome (SILAP) standard coupled with extensive multidimensional separation coupled with tandem mass spectrometry (MS/MS). Secreted proteins from CAPAN-2 human pancreatic cancer derived cells were collected after conducting stable isotope labeling by amino acids in cell culture (SILAC). The resulting SILAP standard contained <0.5% of individual unlabeled proteins. Pooled sera from patients with early stage pancreatic cancer or controls were prepared, and an equal amount of the SILAP standard was added to each sample. Proteins were separated by isoelectric focusing (IEF) prior to two-dimensional liquid chromatography (2D-LC)–MS/MS analysis. A total of 1065 proteins were identified of which 121 proteins were present at 1.5-fold or greater concentrations in the sera of patients with pancreatic cancer. ELISA validation of these findings was successfully performed for two proteins, ICAM-1 and BCAM. Results of these studies have provided proof of principle that a SILAP standard derived from the CAPAN-2 secreted proteome can be used in combination with extensive multidimensional LC-MS/MS for the identification and relative quantitation of potential biomarkers of pancreatic cancer. This technique allows for the detection of low-abundance proteins, and focuses only on biologically relevant proteins derived from pancreatic cancer cells.
Keywords: SILAC, Pancreatic cancer, Serum biomarkers
Short abstract
CAPAN-2 human pancreatic ductal adenocarcinoma-derived cells were grown in [13C6, 15N1]leucine, [13C6,15N2]lysine. One-hundred twenty-one proteins were present at 1.5-fold or greater concentrations in the sera of patients with pancreatic cancer. Results of these studies have provided proof of principle that a SILAP standard derived from the CAPAN-2 secreted proteome can be used in combination with extensive multidimensional LC−MS/MS for the identification and relative quantification of potential biomarkers of pancreatic cancer.
Introduction
Pancreatic cancer is the fourth most common cause of cancer-related mortality in the United States.(1) The five-year survival rate is the lowest among all cancers, with estimates ranging from 0.4 to 4%. In 2007, an estimated 37 170 new cases of pancreatic were diagnosed, and an estimated 33 370 patients died as a result of their disease.(2) No effective screening biomarker exists for the early detection of pancreatic cancer. Carbohydrate antigen (CA) 19-9 is the most commonly used protein tumor marker for pancreatic cancer, however its use is largely limited to following the course of disease.3,4 This is primarily because CA 19-9 can be expressed in benign conditions such as cholangitis and chronic pancreatitis.5,6 Furthermore, CA 19-9 is not expressed at all by some pancreatic tumors, and in other tumors, it is often not detectable until pancreatic cancer is at a late and incurable stage.(7) Other potential biomarkers such as carcinoembryonic antigen (CEA), peanut agglutinin (PNA)-binding glycoproteins,(8)hTert (telomerase catalytic subunit),(9) and matrix metalloproteinase-2 (MMP-2)(10) also lack clinical efficacy. This situation has stimulated our search for biomarkers that can be used for the early detection of pancreatic cancer.(11)
Despite significant advances in proteomic methods and instrumentation, discovery of circulating disease biomarkers remains extremely challenging. We have developed a novel approach for identifying biomarkers of pancreatic cancer in human serum. The approach specifically addresses three of the major obstacles in biomarker discovery.
The first major obstacle is the accurate quantitation of large numbers of proteins. The difficulty of accurate protein quantitation is compounded by nonspecific losses suffered during extensive sample processing. For example, although immunoaffinity purification removes the major abundant serum proteins, significant losses of low abundant proteins bound to the high abundant proteins can occur.(12) Therefore, a stable isotope labeled proteome (SILAP) standard added to serum samples prior to immunopurification can act as a carrier for the low abundance proteins, controlling for and minimizing the possibility of such losses.13,14 The SILAP standard can also control for and help to prevent losses that can occur throughout the extensive workup procedure and LC-MS analysis, such as the nonspecific binding of peptides to glassware and surfaces.(15) Equally important, for any protein identified in the SILAP standard, the corresponding unlabeled serum protein can be quantified, if present. Absence of the unlabeled serum protein when the labeled protein is identified confers increased confidence that the protein is truly absent in the serum sample, as opposed to simply absent from the analysis due to sampling error, ion suppression or nonspecific loss.
The second major obstacle is the identification and characterization of biologically relevant proteins in serum. In unbiased shotgun analyses of serum samples, many of the proteins found to be differentially expressed are abundant serum proteins or nonspecific acute phase proteins, and not proteins related to the disease process. By using a SILAP standard derived from the secreted proteome of the CAPAN-2 pancreatic cancer cell line, low abundance and biologically relevant biomarker candidates can be identified and relatively quantitated. These proteins are generally present at much lower concentrations in serum than in the secreted CAPAN-2 proteome, making their quantitation and identification difficult without the use of a SILAP standard.
The third obstacle remains the extraordinary complexity of proteins present in human serum. In standard 2D-LC-MS/MS protocols, proteins are fractionated after trypsin digestion. Human serum contains proteins that are present over a wide dynamic range,16,17 so peptides from abundant proteins can become widely distributed, interfering with identification of lower abundance proteins in many of the fractions collected. One successful approach to improving the number of low abundance proteins identified has been to perform increasing numbers of orthogonal separation steps after tryptic digestion, either serially or in parallel. Some examples include 1D gel-electrophoresis,(18) IEF,(19) N-linked glycopeptide enrichment,(20) and cysteinyl peptide enrichment.(20) Immunoaffinity removal of abundant proteins(21) has also proved to be a robust and reproducible method for studying lower abundance proteins in serum.
Fewer methods have been developed to successfully integrate separation methods at the intact protein level with 2D-LC-MS/MS. IEF of intact proteins is one of the most common methods for separating complex protein mixtures. Historically, this has been performed as the first dimension of 2D gel electrophoresis. Few studies have been reported, however, demonstrating how this technique can be leveraged for sample separation prior to 2D-LC-MS/MS. One study using liquid phase IEF prior to trypsin digestion and reversed-phase 1D-LC-MS/MS suggested the feasibility of this approach.(22) A similar approach was taken to study plasma and amniotic fluid.(23) In this study, a proprietary IEF apparatus was used, prior to 1D-LC-MS/MS. However, only 73 and 69 proteins were identified in the respective samples. The power of IEF was suggested by a more comprehensive study, which included solution IEF followed by 1D-gel electrophoresis and LC-MS/MS, and was able to identify over 2000 serum proteins.(24) With the development of modern immobilized pH gradient (IPG) strips, the reproducibility and resolution of IEF separation has been significantly improved. IPG strip capacity provides another major advantage over other procedures, as up to 3 mg of protein can be readily loaded on an 18 cm strip.(25) In the current study, we have developed a method combining the use of a SILAP standard with immunoaffinity removal of abundant proteins and IEF-2D-LC-MS/MS analysis to identify a large number of pancreatic cancer associated biomarkers.
Experimental Procedures
General Workflow
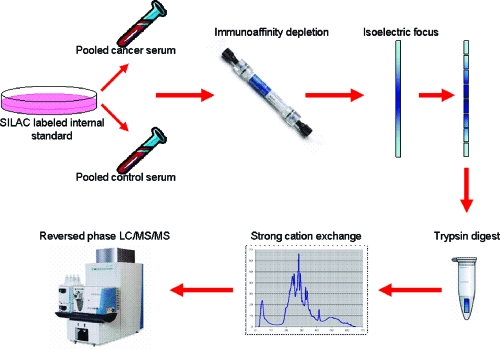
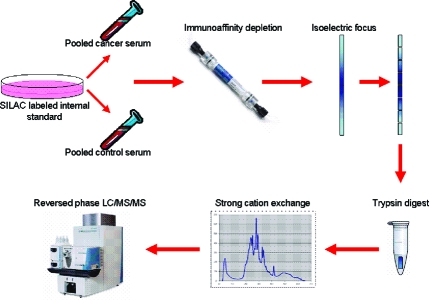
The workflow for sample processing is presented in Figure 1. Equal amounts of a SILAP standard were added to human serum samples, which were then subjected to immunopurification in order to remove high abundance proteins. Serum samples were then fractionated by IEF; the proteins were harvested, and digested with trypsin. Resulting tryptic peptides were then fractionated by strong cation exchange (SCX) chromatography and analyzed by reversed phase 2D-LC-MS/MS.
Figure 1.
General workflow for processing of serum samples. Pooled serum from pancreatic cancer patients or controls is mixed with SILAC labeled conditioned media from CAPAN-2 cells. Abundant proteins are removed by immunoaffinity depletion. Samples were separated by IEF. IPG strips were then cut into pieces and digested with trypsin. Tryptic peptides were separated by strong cation exchange, then analyzed by reversed-phase LC−MS/MS.
Preparation of a SILAP Standard
Human caucasian, pancreas, adenocarcinoma (CAPAN-2) cells were obtained from the Cell Culture Core of the University of Pennsylvania Center for Molecular Studies in Digestive and Liver Diseases. The cells were passaged at least five times using SILAC-based methodology.(26) SILAC was developed as a simple and accurate approach for MS-based quantitative proteomics. The method relies on the incorporation of amino acids with substituted stable isotopic nuclei, commonly 13C and 15N. In SILAC, cells are grown in culture media that contains a stable isotope form of a particular amino acid, commonly l-leucine or l-lysine. As these are essential amino acids, cells incorporate labeled amino acids into all newly synthesized proteins. Thus, with each cell doubling the cell population replaces at least half of the original form of the amino acid, eventually incorporating almost 100% of the labeled amino acid. The SILAP standard and unlabeled proteins from tissue or serum can then be combined and processed together. When tryptic peptides are analyzed by LC-MS, peptides from the labeled and unlabeled samples will differ by a predictable number of mass to charge (m/z) units. The ratio of peak intensities between the labeled and unlabeled peptides then accurately reflects relative protein levels. This differs significantly from the original use of SILAC, which used a single passage of isotopically labeled cells as the experimental model and the unlabeled cells as a control. Briefly, CAPAN-2 cells were grown to confluence in DME/F12 media (Sigma, St. Louis, MO) containing [13C6,15N1] leucine and [13C6,15N2] lysine (Cambridge Isotopes, Cambridge, MA) and 10% dialyzed FBS (Sigma, St. Louis, MO) to ensure complete labeling of cellular proteins as described previously.27,28 Cells were subsequently grown in serum-free media supplemented with insulin, transferrin and selenium (Sigma, St. Louis, MO). Conditioned media was collected every 2 days, pooled, filtered and aliquoted. The SILAP standard was isolated by concentrating 45 mL (2 mg total protein) of the SILAC conditioned media using 5000 MW cutoff spin-filters (Millipore, Billerica, MA).
Characterization of Proteins in the SILAP Standard from CAPAN-2 Cells
Proteins were incubated in boiling water for 5 min, reduced in 5 mM dithiothreitol (DTT) for 1 h at 60 °C, and alkylated with 15 mM iodoacetamide (IAA) for 30 min at room temperature in the dark. Tryptic peptides were obtained by digestion with sequencing grade trypsin (Promega, Madison, WI) (1:50 w/w) overnight at 37 °C. Proteins were characterized by standard 2D-LC-MS/MS analysis of the tryptic peptides as described below. There was <0.5% of individual identified unlabeled proteins in the SILAP standard.
Preparation of Serum Samples for IEF-2D-LC-MS/MS Analysis
Two pooled serum samples were created, one consisting of sera from 6 patients with known, early pancreatic cancer, the other from 6 patients with benign pancreatic disease (Table 1). All serum samples analyzed for this study were collected from patients with suspected, resectable pancreatic cancer or other pancreaticobiliary disease prior to surgical resection. Disease status was subsequently confirmed based on operative and surgical pathological reports. One mg of the SILAP standard was mixed with 300 μL of each pooled serum sample (∼20 mg of serum protein). Each sample was processed using an immunoaffinity column (Beckman-Coulter ProteomeLab IgY-12 LC2 column, Fullerton, CA), which depletes the 12 most abundant plasma proteins: albumin, IgG, transferrin, fibrinogen, IgA, α2-macroglobulin, IgM, α1-antitrypsin, haptoglobin, α1-acid glycoprotein, apolipoprotein A-I and apolipoprotein A-II. The immunoaffinity column was attached to an Hitachi EZChrome Elite HPLC. The immunoaffinity column used for these experiments removed approximately 95% of the protein from normal, human serum samples as determined by measuring serum protein concentration before processing, and protein concentration of the flow-through fraction. As expected, only a negligible amount of protein is lost from SILAP standard samples processed alone using this immunoaffinity column. After processing, we estimate each sample contained a mixture of approximately 1 mg of internal standard and 1 mg of depleted serum protein.
Table 1. Characteristics of Subjects Who Provide the 12 Serum Samples Used for the Pooled Samples and Initial Validation.
| age | gender | disease | |
|---|---|---|---|
| Cancer | 50 | M | Pancreatic cancer, resectable |
| 70 | M | Pancreatic cancer, resectable | |
| 66 | M | Pancreatic cancer, resectable | |
| 77 | M | Pancreatic cancer, resectable | |
| 68 | F | Pancreatic cancer, resectable | |
| 47 | F | Pancreatic cancer, resectable | |
| Control | 71 | F | Ampullary adenoma |
| 49 | F | Pancreatic pseudocyst | |
| 80 | M | Chronic pancreatitis | |
| 67 | F | Pancreatic serous adenoma | |
| 47 | F | Chronic pancreatitis | |
| 29 | F | Chronic pancreatitis |
Protein Separation by IEF
Protein concentrations were determined by Coomassie Protein Assay (Pierce Scientific, Milwaukee, WI). Two micrograms of each protein sample was precipitated using a standard methanol/chloroform protocol, then resuspended in 0.5% IPG buffer and DeStreak Rehydration Solution (GE/Amersham Biosciences, Piscataway, NJ), a commercially available, proprietary solution developed to prevent nonspecific oxidation and streaking, resulting in improved reproducibility. Isoelectric focusing of protein samples was performed using 18 cm 3−10 NL IPG strips (GE/Amersham Biosciences, Piscataway, NJ). The plastic backing was left in place during processing to facilitate handling. Each strip was cut into nine 2 cm pieces. Each piece was washed with 5%, then 1% trichloroacetic acid (TCA), dehydrated with 90% acetonitrile and allowed to air-dry. The approximate pI range of each of these IPG pieces was estimated as follows from information provided by the manufacturer: piece 1 (pI 3.0−4.0), piece 2 (pI 4.0−5.0), piece 3 (pI 5.0−5.4), piece 4 (pI 5.4−5.8), piece 5 (pI 5.8−6.0), piece 6 (pI 6.0−6.4), piece 7 (pI 6.4−7.6), piece 8 pI (7.6−9.0) and piece 9 (pI 9.0−10.0). Overnight in-strip digestion was performed for each of the 2 cm pieces using sequencing grade trypsin (Promega) diluted in 50 mM ammonium bicarbonate at 37 °C. The supernatant was removed and saved. Peptide extraction was performed by adding 1% trifluoroacetic acid/50% acetonitrile, and sonicating for 15 min. Extracted peptides were combined with the previously removed supernatant and concentrated by lyophilization.
Separation of Peptides by SCX Chromatography
The pH of each sample was lowered to 3 with 1% formic acid. An equal volume of SCX mobile phase A (10 mM ammonium formate, 25% acetonitrile, pH 3) was added to each sample. SCX chromatography was performed on a PolySulfethy A column (100 mm × 2.3 mm, 5 μm 300 Å, PolyLC) attached to an 1100 Series HPLC (Agilent, Santa Clara, CA). Samples were loaded for 5 min with mobile phase A, followed by a linear gradient for 30 min to 100% mobile phase B (500 mM ammonium formate, 25% acetonitrile, pH 6.8). Thirty-five 2 min fractions were collected, pooled into 10−13 fractions, lyophilized, and stored at − 80 °C awaiting further analysis. A total of 104 fractions from each of the pancreatic cancer serum and pancreatic control serum samples were generated for analysis by reversed phase LC-MS/MS. For characterization of the CAPAN-2 secreted proteome, 18 pooled fractions were prepared by SCX chromatography for analysis by reversed phase LC-MS/MS.
Reversed-Phase Capillary LC−MS/MS Analyses
Lyophilized peptides were reconstituted with 5% aqueous acetonitrile containing 0.1% formic acid for reversed phase separation. A nanoflow high pressure capillary LC system (Eksigent, Dublin, CA) coupled on line to a linear ion trap Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (LTQ-FT, Thermo Fisher, San Jose, CA) via an in-house-manufactured nanoelectrospray ionization interface was used to analyze peptide samples. The reversed-phase capillary column was prepared by slurry-packing Alltech Prosphere C18-AQ, 5 μm, 100 Å into an 18 cm long, 360 μm outer diameter (od) × 75 μm id fused silica capillary (New Objective, Woburn, MA) fritted with a polymerized solution containing potassium silicate and formamide. A trap column consisting of Alltech Prosphere C18, 10 μm, 300 Å slurry-packed into a 6 cm long, 360 μ od × 150 μm id fused silica capillary (New Objective, Woburn, MA) was also used. Mobile phases were 0.1% formic acid in water (A) and 0.1% formic acid in 100% acetonitrile (B). After loading 10 μL of peptides onto the column, the mobile phase was held at 95% A for 20 min. A linear gradient to 70% B was applied over 150 min. To identify the eluting peptides, the linear ion trap mass spectrometer was operated in a data-dependent MS/MS mode (m/z 300−2000) in which each full MS scan in the FT-ICR was followed by 7 MS/MS scans in the ion trap. The seven most intense precursor ions were dynamically selected in order of highest to lowest intensity and then subjected to collision-induced dissociation. The FT-ICR mass resolution was set at 50 000.
Protein Identification
Raw data were submitted to Bioworks Browser (Thermo Fisher, San Jose, CA) and batch searched through SEQUEST against the NCBI RefSeq database of human sequences (version updated 2/07). The database was indexed using the following criteria: strict trypsin cleavage rules with up to two internal cleavage sites; differential modifications of methionine oxidation, carboxyamidomethlyation on cysteine, [13C6,15N1]leucine and [13C6,15N2]lysine. All peptides shorter than six amino acids were removed from the data set. The remaining SEQUEST output files were further processed using the Trans-Proteomic Pipeline (Institute for Systems Biology, Seattle, WA) for analysis and validation of peptides and proteins using PeptideProphet and ProteinProphet, respectively. PeptideProphet peptide results where filtered using a minimum peptide probability of 0.3. ProteinProphet protein results were filtered using a minimum probability of 0.5. All proteins identified by a single unique peptide were eliminated. XPRESS software was originally developed for isotope coded affinity tag (ICAT) labeling experiments,(29) but which is equally applicable to other differential labeling approaches such as SILAC. Starting with an MS/MS spectra, either unlabeled or SILAC labeled, the program reconstructs reverse phase elution profiles for both the SILAC labeled and unlabeled precursor ions. Relative quantitation of light and heavy peptides was performed using XPRESS (also from the Trans-Proteomic Pipeline) with a parent mass tolerance of 0.2 mass units and mass differences of 7.027630 mass units on leucine and 7.93217 lysine, corresponding to [13C6,15N1]leucine and [13C6,15N2]lysine. The elution areas of the labeled and unlabeled precursor ions are determined and a ratio is generated. Manual validation by extracted ion monitoring was performed on differentially expressed peptides.
Bioinformatics
For identifying pathways to which proteins belong, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used as a pathway reference.(30) In this database, biological pathways have been curated manually and are constantly updated. GenBank Geninfo Identifier (GI) accession numbers from proteins identified were used as a query to the Protein Information Resource (PIR) database (http://pir.georgetown.edu). All human proteins corresponding to each GI accession number were extracted. A majority of the GI accession numbers produced multiple proteins. Individual proteins were then used as a query to the UniProt database to extract the corresponding KEGG identfications (IDs).(31) These conversions were performed to compensate for any errors during the assignment of KEGG IDs to GI accession numbers, thereby providing more accurate results. Individual KEGG IDs were searched against each pathway across the entire KEGG pathway database specific to the species Homo sapiens. As is typical of such analyses, not all GI accession numbers can be classified into functional pathways. Many GI accession numbers are implicated in multiple pathways. Some pathways such as cell communication and cell motility have considerable overlapping GI constituents. In a complementary analysis, Gene Ontology (GO) classifications were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Functional Annotation Tool (http://david.abcc.ncifcrf.gov) against GI accession numbers identified in the CAPAN-2 secreted proteome and in our human serum analyses.
Immunohistochemical Validation
Western blot analyses were performed to confirm presence of candidate proteins in CAPAN-2 secreted proteome, pooled cancer and control serum. Mouse monoclonal antibody to human matrix metalloproteinase-7 (MMP-7), goat polyclonal antibody to human 14-3-3 zeta (R&D Systems, Minneapolis, MN) and mouse monoclonal antibody to human cytokeratin 18 (CK18) (Abcam, Cambridge, MA) were purchased. Pooled serum proteins (15 μg)) from both the pancreatic cancer and control cohorts used during the discovery phase were diluted in LDS sample buffer (Invitrogen, Carlsbad, CA). Samples were incubated at 60 °C for 10 min, and separated on 4−12% Bis-Tris gels (NuPAGE Novex gels, Invitrogen, Carlsbad, CA). A SeeBlue Plus2 (Invitrogen, Carlsbad, CA) protein standard was used to estimate molecular weights. Samples were then transferred to nitrocellulose membrane (Invitrogen, Carlsbad, CA) and incubated with individual primary antibodies diluted 1:500 for 2 h at room temperature. Membranes were then incubated for 45 min with the appropriate secondary horseradish peroxidase-conjugated antibody (Sigma, St. Louis, MO) diluted 1:5000. Protein bands were then visualized by incubating membranes with ECL Plus detecting reagents (Amersham Biosciences, Piscataway, NJ). Serum and SILAP standard levels of MMP-7, intercellular adhesion molecule 1 (ICAM-1) and basal cell adhesion molecule (BCAM) were measured by enzyme-linked immunosorbent assay (ELISA). MMP-7 (R&D Systems, Minneapolis, MN), ICAM-1 (US Biological, Swampscott, MA) and BCAM (R&D Systems, Minneapolis, MN) ELISA kits were purchased and used according to manufacturer’s instructions.
Results
Characterization of Proteins Secreted by CAPAN-2 Cells
A total of 666 proteins in 442 protein groups were identified in the CAPAN-2 secreted proteome (Supplemental Table 1, Supporting Information). To assess biological relevance, we compared proteins secreted by the CAPAN-2 cell line to proteins found to be up-regulated in pancreatic tumors.(32) Twenty-seven of the 90 proteins previously identified as up-regulated in pancreatic tumors were also found in the CAPAN-2 secreted proteome. Gene Ontology (GO) functional annotation analysis could be performed on 368 of the 666 proteins from the CAPAN-2 secreted proteome. Forty-four of 367 proteins analyzed were classified as localizing to the extracellular region, and 36 of these proteins were also classified as secreted. Functional pathway analysis could be performed on 227 of the 666 proteins (Supplemental Table 2, Supporting Information). Proteins from key biological pathways such as cell-cell communication, proteasome, MAP kinase, Wnt signaling, focal adhesion, leukocyte transendothelial migration, insulin signaling and actin cytoskeleton were identified.
Proteins Characterized in Serum Samples
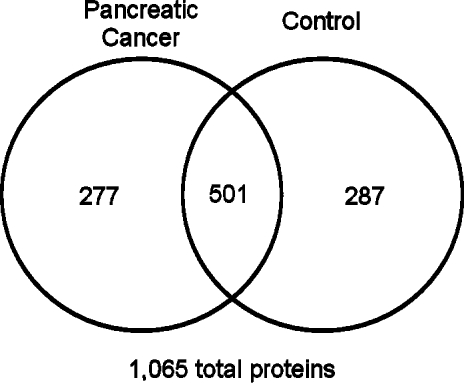
A total of 788 unique proteins from 568 functional protein groups were identified in control serum samples by stable isotope dilution IEF-2D-LC/MS/MS (Supplemental Table 3, Supporting Information). A total of 778 unique proteins from 529 functional groups were identified in the pancreatic cancer serum samples (Supplemental Table 4, Supporting Information). Together, a total of 1065 serum proteins were identified in both samples; 501 (47%) of these proteins were identified in both samples, with 277 proteins identified only in the pancreatic cancer serum sample, and 287 proteins identified only in the control sample (Figure 2).
Figure 2.
Venn diagram of proteins identified from cancer and control serum samples. Equal amounts of the SILAP standard were added to each serum sample.
Differential Secretion of Proteins into Pancreatic Cancer Serum
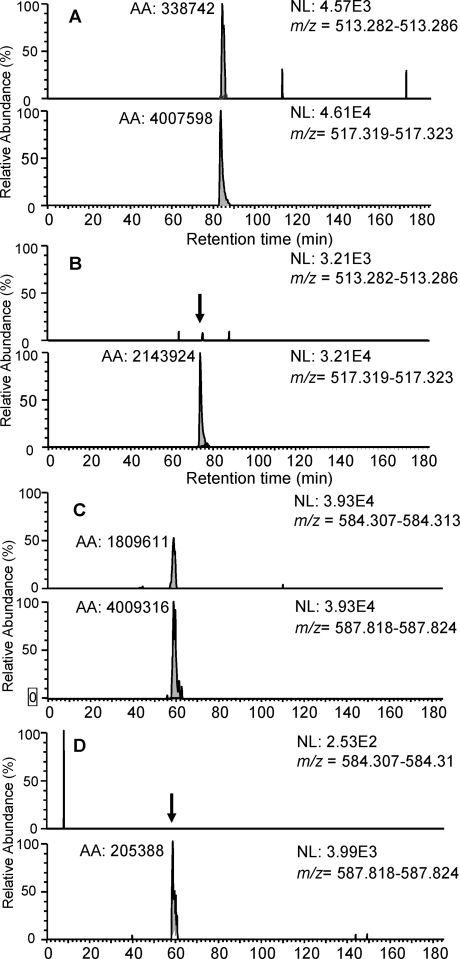
For each protein, an XPRESS ratio was generated representing the ratio of the unlabeled protein from the serum to the labeled protein from the SILAP standard. Proteins present in only the unlabeled form, and therefore absent in our SILAP standard, were excluded from further analysis. As the same amount of the SILAP standard was added to both pooled cancer and control serum samples, cancer and control XPRESS ratios could be compared for each protein. For 121 proteins, the relative amount in the cancer serum exceeded that in the control serum by 1.5-fold or more (Table 2). Two of these proteins, transgelin 2 and cathepsin D, were previously identified to be up-regulated in pancreatic tumors.(32) Manual review of extracted ion current spectra was performed to confirm differential XPRESS ratios. Examples of extracted ion current spectra are presented (Figure 3).
Table 2. Proteins with Increased Secretion into Pancreatic Cancer Serum Compared with Controls.
| entry no. | protein name | cancer to control ratio | entry no. | protein name | cancer to control ratio |
|---|---|---|---|---|---|
| 1 | RP26, ceramide kinase-like | 19980.00 | 62 | GANAB, alpha glucosidase II alpha subunit | 5.13 |
| 2 | ORP150 or HYOU1, oxygen regulated protein precursor | 1012.33 | 63 | PPIA, peptidylprolyl isomerase A | 5.00 |
| 3 | FLNA, filamin A, alpha | 623.55 | 64 | SHMT2, serine hydroxymethyltransferase 2 (mitochondrial) | 5.00 |
| 4 | UBE2L3, ubiquitin-conjugating enzyme E2L 3 isoform 1 | 332.22 | 65 | DDT, D-dopachrome tautomerase | 4.99 |
| 5 | LMO7, LIM domain only 7 | 268.20 | 66 | Eef1a1, eukaryotic translation elongation factor 1 alpha 1 | 4.49 |
| 6 | FILIP, filamin A interacting protein 1 | 221.67 | 67 | peroxiredoxin 6 | 4.47 |
| 7 | STXBP2 or Unc18−2, syntaxin binding protein 2 | 215.77 | 68 | catalase | 4.31 |
| 8 | CENPE, centromere protein E | 204.00 | 69 | S100P or MIG9, S100 calcium binding protein P | 4.16 |
| 9 | PTPRJ, protein tyrosine phosphatase, receptor type, J precursor | 136.00 | 70 | PREDICTED: similar to Phosphoglycerate mutase 1 (Phosphoglycerate mutase isozyme B) (PGAM-B) | 4.07 |
| 10 | ARHGEF15 or GEF 15, Rho guanine exchange factor 15 | 121.67 | 71 | hypothetical protein LOC345651 | 4.00 |
| 11 | PREDICTED: similar to Elongation factor 1-gamma (EF-1-gamma) (eEF-1B gamma), eukaryotic translation elongation factor 1 gamma | 120.94 | 72 | GTP-binding protein PTD004 isoform 1 | 3.83 |
| 12 | CLSTN1, calsyntenin 1 | 103.45 | 73 | SMAP1L, stromal membrane-associated protein 1-like | 3.65 |
| 13 | DAK, dihydroxyacetone kinase 2 | 95.57 | 74 | 14−3−3 theta, tyrosine 3\tryptophan 5 -monooxygenase activation protein, theta polypeptide | 3.65 |
| 14 | ARPC2, Actin related protein 2\3 complex subunit 2 | 88.20 | 75 | AXL, AXL receptor tyrosine kinase | 3.63 |
| 15 | MYH9, myosin, heavy polypeptide 9, nonmuscle | 63.26 | 76 | ACY1, aminoacylase 1 | 3.50 |
| 16 | PDXK, pyridoxal kinase | 62.54 | 77 | carbonic anhydrase I | 3.43 |
| 17 | ON or SPARC, secreted protein, acidic, cysteine-rich (osteonectin) | 56.72 | 78 | PLCL2, phospholipase C-like 2 | 3.29 |
| 18 | RKIP, prostatic binding protein | 55.82 | 79 | GPR126, G protein-coupled receptor 126 alpha 1, G protein-coupled receptor 126 beta 1, G protein-coupled receptor 126 alpha 2, G protein-coupled receptor 126 beta 2 | 3.20 |
| 19 | AST, aspartate aminotransferase 1 | 55.00 | 80 | 3‘(2’), 5′-bisphosphate nucleotidase 1 | 3.19 |
| 20 | cardiac muscle alpha Actin 1 proprotein | 36.22 | 81 | PSME1, proteasome activator subunit 1 | 3.15 |
| 21 | PKP2, plakophilin 2 | 26.33 | 82 | SERPINB1, serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 1 | 3.00 |
| 22 | CANT1, calcium activated nucleotidase 1 | 26.00 | 83 | GSTO1, glutathione-S-transferase omega 1 | 3.00 |
| 23 | S100A4, S100 calcium-binding protein A4 | 25.88 | 84 | moesin | 3.00 |
| 24 | HNRPD or AUF1, heterogeneous nuclear ribonucleoprotein D | 24.49 | 85 | cathepsin C isoform a preproprotein | 2.99 |
| 25 | RCN1, reticulocalbin 1 precursor | 23.62 | 86 | ADP-ribosylation factor 1 | 2.98 |
| 26 | ARHGDIA, Rho GDP dissociation inhibitor (GDI) alpha | 23.30 | 87 | TPI1, triosephosphate isomerase 1 | 2.96 |
| 27 | B-CAM, basal cell adhesion molecule precursor | 23.30 | 88 | ACTN1, actinin, alpha 1 | 2.80 |
| 28 | myosin regulatory light chain MRCL3, myosin regulatory light chain MRCL2 | 21.82 | 89 | NUDT5, nudix-type motif 5 | 2.79 |
| 29 | PLAP, placental alkaline phosphatase preproprotein | 20.33 | 90 | ERO1L, ERO1-like | 2.78 |
| 30 | LHPP, phospholysine phosphohistidine inorganic pyrophosphate phosphatase | 19.92 | 91 | uPA, urokinase plasminogen activator preproprotein | 2.64 |
| 31 | RRBP1, ribosome binding protein 1 | 19.67 | 92 | P4HB, prolyl 4-hydroxylase, beta subunit precursor | 2.63 |
| 32 | CAPZA1, F-Actin capping protein alpha-1 subunit | 19.42 | 93 | hypothetical protein LOC57653 | 2.59 |
| 33 | UBE2N, ubiquitin-conjugating enzyme E2N | 18.30 | 94 | MYEF2, myelin gene expression factor 2 | 2.56 |
| 34 | EFHD2, EF hand domain family, member D2 | 16.82 | 95 | RyR3, ryanodine receptor 3 | 2.56 |
| 35 | Chd7, chromodomain helicase DNA binding protein 7 | 16.75 | 96 | ras homologue gene family, member A | 2.51 |
| 36 | GGH, gamma-glutamyl hydrolase precursor | 16.03 | 97 | ENO1, enolase 1 | 2.50 |
| 37 | CAPZB, F-Actin capping protein beta subunit | 15.27 | 98 | DENN\MADD domain containing 2C | 2.50 |
| 38 | intercellular adhesion molecule 1 precursor | 14.00 | 99 | EPS8L1, epidermal growth factor receptor pathway substrate 8-like protein 1 isoform a | 2.50 |
| 39 | CLIC1, chloride intracellular channel 1 | 13.84 | 100 | proteasome alpha 5 subunit | 2.48 |
| 40 | DJ-1 protein | 13.19 | 101 | BAIAP2, BAI1-associated protein 2 | 2.45 |
| 41 | TPP1, tripeptidyl-peptidase I preproprotein | 12.51 | 102 | cathepsin D preproprotein | 2.44 |
| 42 | SDCBP2, syndecan binding protein 2 | 12.48 | 103 | Maspin (SERPINB5), serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 5 | 2.42 |
| 43 | major histocompatibility complex, class I, B, PREDICTED: similar to HLA class I histocompatibility antigen, B-18 alpha chain precursor (MHC class I antigen B*18) | 12.31 | 104 | PGLS, 6-phosphogluconolactonase | 2.35 |
| 44 | Sumo 2 or Sumo 3, PREDICTED: similar to SMT3 suppressor of mif two 3 homologue 2, small ubiquitin-like modifier protein 3 | 12.13 | 105 | 14−3−3zeta, tyrosine 3\tryptophan 5 -monooxygenase activation protein, zeta polypeptide, tyrosine 3\tryptophan 5 -monooxygenase activation protein, zeta polypeptide | 2.25 |
| 45 | BLVRB, biliverdin reductase B (flavin reductase (NADPH)) | 10.50 | 106 | Chaperonin | 2.21 |
| 46 | zinc finger protein 704 | 10.00 | 107 | heme binding protein 2 | 2.16 |
| 47 | S100A16, S100 calcium binding protein A16 | 9.10 | 108 | SH3BGRL3, SH3 domain binding glutamic acid-rich protein like 3 | 2.13 |
| 48 | SEMA7A, semaphorin 7A | 8.49 | 109 | keratin 8 | 2.02 |
| 49 | CHMP2B, chromatin modifying protein 2B | 8.49 | 110 | vinculin isoform VCL, vinculin isoform meta-VCL | 2.00 |
| 50 | NSD1, nuclear receptor binding SET domain protein 1 | 7.67 | 111 | collagen, type VI, alpha 1 precursor | 1.96 |
| 51 | myoglobin | 7.15 | 112 | podocalyxin-like precursor | 1.88 |
| 52 | STIP1, stress-induced-phosphoprotein 1 (Hsp70\Hsp90-organizing protein) | 7.00 | 113 | TCN1, transcobalamin I precursor | 1.85 |
| 53 | IFI30 or IP-30, interferon, gamma-inducible protein 30 preproprotein | 7.00 | 114 | BST2, bone marrow stromal cell antigen 2 | 1.81 |
| 54 | NME1-NME2 protein | 6.74 | 115 | keratin 18 | 1.76 |
| 55 | ephrin receptor EphA2 | 6.68 | 116 | HSPG2, heparan sulfate proteoglycan 2 | 1.73 |
| 56 | RNPEP, arginyl aminopeptidase (aminopeptidase B) | 6.50 | 117 | Ezrin, villin 2 | 1.69 |
| 57 | DENN\MADD domain containing 2A | 6.13 | 118 | cytosolic malate dehydrogenase | 1.65 |
| 58 | TAL1 or TALDO1, transaldolase 1 | 6.00 | 119 | MMP-7, matrix metalloproteinase 7 preproprotein | 1.61 |
| 59 | PVRL1 or CD111, poliovirus receptor-related 1 (herpesvirus entry mediator C; nectin) | 5.64 | 120 | TAGLN2, transgelin 2 | 1.57 |
| 60 | APLP2, amyloid beta (A4) precursor-like protein 2 | 5.50 | 121 | cystatin M precursor | 1.52 |
| 61 | ARMET, arginine-rich, mutated in early stage tumors | 5.18 |
Figure 3.
LC−MS ion current profiles of tryptic peptides obtained from pancreatic cancer and control serum proteins. Two pairs of chromatograms are presented for each tryptic peptide. The upper chromatogram is from the unlabeled peptide and the lower chromatogram is from the SILAP-derived standard peptide. (A) Syndecan binding protein 2 peptide R.RAEIKPGVR.E from pancreatic cancer serum. (B) Syndecan binding protein 2 peptide R.RAEIKPGVR.E from control serum. (C) Biliverdin reductase B peptide R.LQAVTDDHIR.M from pancreatic cancer serum. (D) Biliverdin reductase B peptide R.LQAVTDDHIR.M from control serum.
Western Blot Analyses
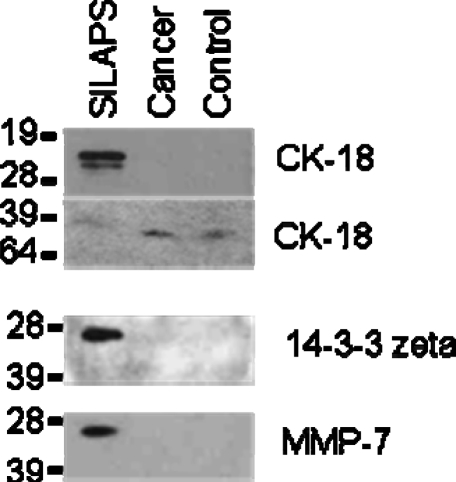
The protein ratios generated by XPRESS, indicated that levels of many candidate biomarkers were present at much higher levels in the SILAP standard than in either the cancer or control serum. For example, for MMP-7, the serum to SILAP standard ratio was 1:180.5 in the cancer sample and 1:290 in the control sample. For 14-3-3 zeta, the ratios were 1:95.0 and 1:213.4 (cancer and control, respectively). For CK18, the ratios were 1:127.0 and 1:223.1 (cancer and control, respectively). Western blot analysis for these three proteins confirmed this observation. MMP-7 and 14-3-3 zeta were clearly detected in the SILAP standard, but not detected in either the pooled cancer or control serum samples (Figure 4). The Western blot results for CK18 demonstrated two bands at approximately 21 kD and 26 kD in the SILAP standard, whereas the predicted molecular weight for CK18 approximates to 47 kD. During apoptosis, CK18 is known to be cleaved by caspase 3 and caspase 6 at Asp398, then Asp237, resulting in a 21 and 26 kD fragment.33,34 Neither cleavage product was observed in the cancer and control serum samples. A faint band was present in both serum samples at approximately 47 kD, which might have arisen from the intact CK18 protein. However, the intact protein was not detected in the SILAP standard.
Figure 4.
Western blot results for CK-18, 14−3−3 zeta and MMP-7. Bands representing 14−3−3 zeta and MMP-7 are clearly present in the SILAP standard, but not visible in either serum samples. Two bands at approximately 21 and 26 kD, likely representing specific caspase cleavage fragments of CK-18, are visible in the SILAP standard but not in the serum samples. A faint band at roughly 47 kD is present in both serum samples, and may represent the intact CK18.
Confirmation of Differential Protein Secretion into Serum by ELISA
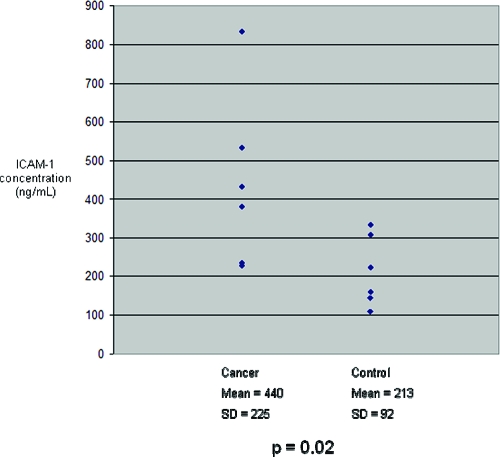
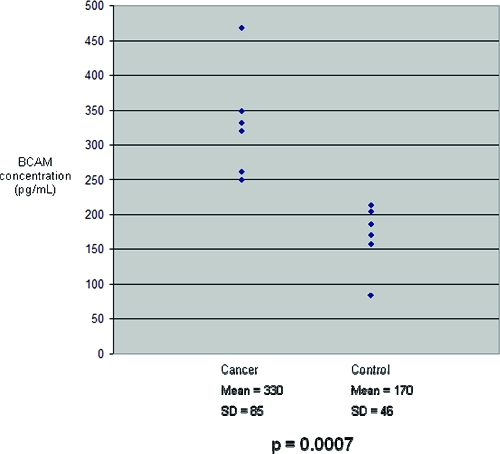
For improved sensitivity and to allow for quantitation, enzyme-linked immunosorbent assays (ELISA) were performed. ELISA kits were available for three candidate protein biomarkers: MMP-7, ICAM-1 and BCAM. Protein concentrations were measured in the SILAP standard and in the individual cancer and control serum samples. MMP-7 concentrations in the SILAP standard was 724.93 ng/mL. Because there was no significant difference in MMP-7 concentration between pooled cancer and control serum samples (data not shown), further validation was not pursued. Serum concentrations of MMP-7 averaged 19.962 ng/mL. Both ICAM-1 and BCAM concentrations were significantly elevated in the individual cancer serum samples compared with control samples (Figure 5 and Figure 6, respectively). These differences confirmed the findings of our SILAP standard discovery study. ICAM-1 and BCAM concentrations were measured in the SILAP standard, as well. BCAM concentration was 2904 pg/mL, while the ICAM-1 concentration was 17 ng/mL. It is noteworthy the concentrations of BCAM and MMP-7 were significantly greater in the SILAP standard than in human serum, whereas ICAM-1 was found at a much lower concentration in the SILAP standard.
Figure 5.
ICAM-1 concentrations in the six cancer and six control serum samples studied. Mean ICAM-1 concentration in cancer sera was significantly greater than in controls.
Figure 6.
BCAM concentrations in the six cancer and six control serum samples studied. Mean BCAM concentration in cancer sera was significantly greater than in controls.
Independent Validation of Biomarker Candidates
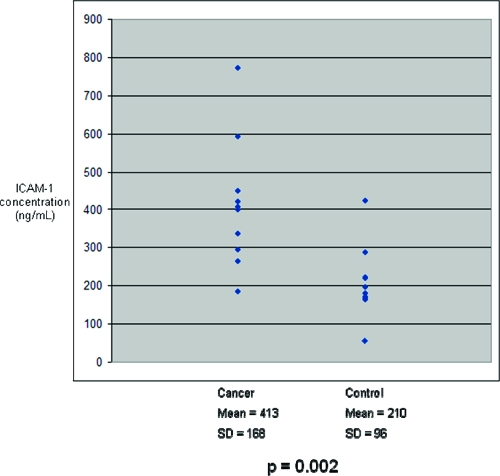
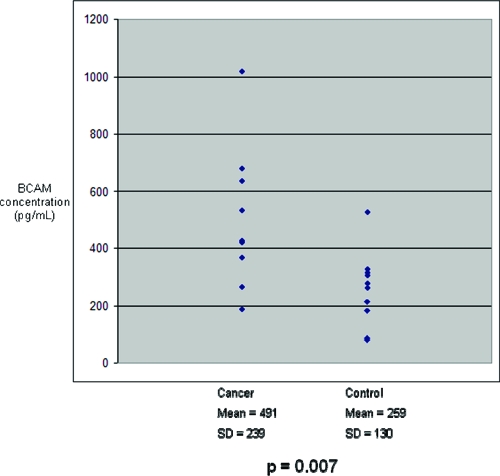
On the basis of the ELISA results, ICAM-1 and BCAM were chosen for further validation in an independent set of human serum samples. Ten patients with pancreatic cancer and 10 controls (Table 3) were analyzed. Similar to the patients whose sera were used in the discovery phase, these 20 serum samples were drawn from patients with suspected, resectable pancreatic cancer or other pancreaticobiliary disease prior to surgical resection. The nature of their disease was confirmed based on operative and surgical pathological reports. ICAM-1 and BCAM levels were significantly elevated in these cancer samples when compared with controls (Figure 7 and Figure 8, respectively). For both ICAM-1 and BCAM, regression analysis was performed to determine if patient age could account for differences in relative protein levels seen across all serum samples (Table 4). No significant association was found. Regression analysis was also performed to determine whether any correlation between BCAM and ICAM-1 levels (Table 4). No significant relationship was found when all samples were analyzed together, or when cancer and control samples were analyzed separately. Student’s t-tests were performed to determine if levels of ICAM-1 or BCAM were associated with patient gender or tumor resectability (Table 5). No association was found.
Table 3. Characteristics of Subjects Who Provided the 20 Serum Samples Used for Independent Validation of ICAM-1 and BCAM Levels.
| age | gender | disease | |
|---|---|---|---|
| Cancer | 69 | M | Pancreatic cancer, resectable |
| 58 | M | Pancreatic cancer, unresectable | |
| 75 | F | Pancreatic cancer, resectable | |
| 74 | F | Pancreatic cancer, unresectable | |
| 41 | F | Pancreatic cancer, unresectable | |
| 72 | F | Pancreatic cancer, resectable | |
| 19 | F | Pancreatic cancer, resectable | |
| 78 | M | Pancreatic cancer, resectable | |
| 59 | M | Pancreatic cancer, resectable | |
| 73 | M | Pancreatic cancer, unresectable | |
| Control | 76 | M | Duodenal polyp |
| 37 | M | Pancreatic pseudocyst | |
| 70 | M | Benign biliary stricture | |
| 37 | F | Mucinous cystic neoplasm, pancreas | |
| 66 | M | Macrocystic adenoma, pancreas | |
| 72 | M | Islet cell tumor | |
| 45 | M | Pancreatic pseudocyst | |
| 33 | F | Acinic cell cystadenoma, pancreas | |
| 56 | F | Mucinous cystic neoplasm, pancreas | |
| 64 | F | Mucinous cystic neoplasm, pancreas |
Figure 7.
ICAM-1 concentrations in ten cancer and ten control serum samples from an independent cohort. Mean ICAM-1 concentration in cancer sera was significantly greater than in controls.
Figure 8.
BCAM concentrations in ten cancer and ten control serum samples from an independent cohort. Mean BCAM concentration in cancer sera was significantly greater than in controls.
Table 4. Regression Analysis of Age versus BCAM Serum Levels, Age versus ICAM-1 Serum Levels, and ICAM-1 versus BCAM Serum Levels.
| R Square | F (Significance) | |
|---|---|---|
| Age v BCAM in Cancer | 0.003 | 0.847 |
| Age v BCAM in Control | 0.237 | 0.056 |
| Age v BCAM ALL | 0.000 | 0.946 |
| Age v ICAM-1 in Cancer | 0.018 | 0.625 |
| Age v ICAM-1 in Control | 0.175 | 0.106 |
| Age v ICAM-1 ALL | 0.003 | 0.759 |
| BCAM v ICAM-1 in Cancer | 0.004 | 0.806 |
| BCAM v ICAM-1 in Control | 0.061 | 0.376 |
| BCAM v ICAM-1 ALL | 0.064 | 0.163 |
No significant correlation was found between any of these variables, whether samples were analyzed by disease state (cancer or control) or altogether.
Table 5. Student’s t-Test to Assess the Effect of Patient Gender and Tumor Resectability on Serum BCAM and ICAM-1 Levelsa.
| P value | |
|---|---|
| Gender v BCAM in Cancer | 0.269 |
| Gender v BCAM in Control | 0.737 |
| Gender v BCAM in ALL | 0.746 |
| Gender v ICAM-1 in Cancer | 0.378 |
| Gender v ICAM-1 in Control | 0.816 |
| Gender v ICAM-1 in ALL | 0.880 |
| Resectability v BCAM in Cancer | 0.565 |
| Resectability v ICAM-1 in Cancer | 0.238 |
No significant effect of either variable was found.
Discussion
SILAC has become an important tool for proteomic studies in vitro; this study has demonstrated that SILAC can be used to prepare a SILAP standard, which can then be employed to quantify pancreatic cancer specific proteomic changes in human serum (Figure 1). Although we have targeted pancreatic cancer-specific proteins, the SILAP standard can be tailored to focus on any malignancy or disease for which a reasonable cell line model exists. Combining this SILAP standard approach with extensive, multidimensional LC-MS/MS, a large group of pancreatic cancer-specific proteins was identified (Figure 2) and shown to be differentially secreted into human serum (Table 2). This made it possible to identify differentially secreted proteins (Figure 4) that were below the limits of sensitivity of Western blot analyses (Figure 4).
The SILAP standard approach offers several advantages to the standard shotgun approach. As with any method integrating a labeled internal standard, the method allows for relative quantitation of the corresponding unlabeled serum proteins, while controlling for nonspecific losses during extensive sample processing. Limiting the analysis to serum proteins that were secreted from pancreatic cancer cells made it possible to filter out acute phase proteins and other abundant serum proteins, while simultaneously focusing on proteins with biological relevance to pancreatic cancer. Absolute quantitation by ELISA for ICAM-1 and BCAM confirmed the differences uncovered by the SILAP standard method, and demonstrates the sensitivity gain afforded by this method (Figures 5–8). For example, BCAM was present in control serum samples at the 100−200 pg/mL range (Figures 6 and 8).
The separation workflow integrates immunoaffinity removal of the twelve most abundant serum proteins and IEF at the intact protein level prior to standard 2D–LC (Figure 1). Implementation of IEF is relatively straightforward, as all equipment and consumables for IEF are commercially available. Indeed, IEF is widely used as the first dimension of 2D gel electrophoresis. As part of multidimensional LC-MS/MS, IEF is an attractive, orthogonal fractionation option. Along with SEC and anion exchange, it represents one of the few readily available options for sample fractionation at the intact protein level. As more dimensions of fractionation are integrated, several tradeoffs emerge. For example, extensive fractionation requires a greater amount of starting material. One advantage of IEF is the high sample capacity of IPG strips. Although, in this study, only 2D-LC was performed after IEF, it is certainly feasible to integrate more dimensions of fractionation in order to obtain a more comprehensive analysis. As more processing steps are added, the nonspecific loss of proteins increases. This is particularly important for low abundance proteins. Another, more difficult to address tradeoff is the exponential increase in MS data acquisition time that results from increased fractionation. In this study, the amount time required for data acquisition from a sample processed by IEF-2D-LC-MS/MS was approximately 18 days. Replicate analysis becomes impractical given these long analysis times. This problem only deepens as more layers of fractionation are added. With extensive fractionation, paradoxically, the redundant analysis of abundant proteins increases. Some of the most abundant peptides are sequenced hundreds of times. Development of algorithms and methods for the dynamic and static exclusion of these abundant peptides could improve the number of low abundance proteins identified.
The pancreatic cancer focused approach to biomarker discovery allows for the discovery of biologically relevant candidates. A number of proteins found in this study to be overexpressed in pancreatic cancer serum have been implicated in essential processes responsible for tumor growth and spread. For example, degradation of the extracellular matrix (ECM) is a key biological process that promotes the invasion of pancreatic tumors into their surrounding tissue. The cysteine protease cathepsin C and the aspartic protease cathepsin D have been implicated in ECM degradation, facilitating tumor growth, invasion and metastasis. Urokinase-type plasminogen activator (uPA), a serine protease, has similarly been implicated in the degradation of ECM. Cystatin M has been characterized as an inhibitor of lysosomal cysteine proteases, thereby acting as a tumor suppressor by inhibiting the growth and migration of tumor cells. While cystatin M expression in pancreatic cancer is not well studied, a recent study reports cystatin M is overexpressed in pancreatic tumors, and addition of functional cystatin M to culture media promotes cell proliferation.(35)
Another important pathway in carcinogenesis is apoptosis. Paradoxically, apoptosis is often increased in malignant tumors, usually accompanied by increased proliferation.(36) In breast cancer, increased apoptosis has also been associated with a worse prognosis.36−38 CK18 is a member of the intermediate filament system, and is expressed at high levels in epithelial cells and malignancies. During apoptosis, CK18 is cleaved by caspases and released into the extracellular compartment. Presence of caspase cleaved CK18 has been shown to be a reliable marker of apoptosis, while release of intact CK18 reflects necrosis.(39) Increased levels of caspase-cleaved CK18 have been measured in the serum of breast cancer patients when compared with controls.(40)
Angiogenesis is an important process for the growth and metastasis of pancreatic tumors. Heparan sulfate proteoglycan 2 (perlecan) is an ECM protein closely associated with vascular endothelia, and has been shown to play a key role in vasculogenesis. Modulation of cell-cell adherence also plays an important role in cancer invasion. A number of proteins important for the formation of adherens junctions, such as vinculin, alpha 1 actinin and poliovirus receptor-related 1 protein were found to be overexpressed in pancreatic cancer serum. Inflammation and leukocyte recruitment, through a number of proposed mechanisms, appear to play a key role in tumor progression and evasion of the immune response. ICAM-1 expressed on endothelial cells engages α1β2-integrin expressed by passing neutrophils. Transendothelial migration by leukocytes also is believed to occur via an ICAM-1 dependent manner. The proteins ezrin, radixin and moesin (ERM) have been hypothesized to act as linkers between ICAM-1 and cytoskeletal proteins (such as actin and vimentin) providing structural support as endothelial cells form channels to permit transendothelial migration.(41) While ICAM-1 expression in pancreatic tissue has been implicated in pancreatitis and pancreatic cancer, measurement of ICAM-1 as a serum marker of pancreatic cancer has not been previously described.
BCAM is a laminin receptor in the Ig superfamily. Little is known about the normal role and distribution of this protein, however, it appears to bind specifically to α5 chain containing laminin proteins. While BCAM expression in pancreatic cancer has not previously been described, overexpression in ovarian cancer(42) and some cutaneous malignancies have been reported.(43) Laminin α5 is widely expressed in basement membranes, and therefore some have hypothesized that BCAM may play a role during the process of tumor invasion.(44) Other investigators have noted the sequence similarity between BCAM and the melanoma antigen MUC18.(45) MUC18, an adhesive cell surface protein, has been hypothesized to play a role in cell-ECM interactions and may promote tumor invasion and metastasis.(46)
Conclusions
Sophisticated discovery strategies, like the one presented here, are now identifying many low abundance protein biomarker candidates. Such an approach, however, is impractical for replicate analysis, validation or for large scale clinical application. Standard immunological validation, however, remains challenging. High quality antibodies are often unavailable for proteins of interests, and development of ELISAs can be time-consuming and difficult. The SILAP standard strategy, however, provides a logical way forward. Candidate peptides for LC-multiple reaction monitoring (MRM)/MS analyses are identified and characterized during the discovery phase. The SILAP standard can then also be used for relative quantitation by LC-MRM/MS, a strategy that is currently under active validation. Absolute quantitation can also be performed, as candidate peptides identified during the discovery phase can then be synthesized as heavy isotope analogs and used as internal standards for standard stable isotope dilution LC-MRM/MS analyses. A recent study has shown that such an approach is capable of accurately measuring plasma proteins in the high pg range.(47) This approach will permit multiple biomarker candidates to be analyzed simultaneously. Simultaneous monitoring of even a fraction of the 121 candidate biomarkers discovered using our SILAP standard approach would be a major step forward toward a test that could detect pancreatic cancer at an early and curable stage.
Abbreviations:
SILAP, stable isotope labeled proteome; MS/MS, tandem mass spectrometry; SILAC, stable isotope labeling by amino acids in cell culture; IEF, isoelectric focusing; 2D-LC, two-dimensional liquid chromatography; IPG, immobilized pH gradient; SCX, strong cation exchange; CAPAN-2, Human Caucasian, pancreas, adenocarcinoma cells; KEGG, Kyoto Encyclopedia of Genes and Genomes; GI, GenBank Geninfo Identifier; GO, Gene Ontology; MMP-7, matrix metalloproteinase-7; CK18, cytokeratin 18; ICAM-1, intercellular adhesion molecule 1; BCAM, basal cell adhesion molecule; ELISA, enzyme-linked immunosorbent assay.
Acknowledgments
Supported by NIH grants P30ES013508, P30CA16520, RO195586, R25CA101871, and U54RR023567, an Institute for Translational Medicine and Therapeutics Research Fellowship, and the Foundation for Digestive Health and Nutrition Bernard L. Schwartz Designated Research Award in Pancreatic Cancer.
Supporting Information Available
Supplemental Tables 1−4. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Niederhuber J. E.; Brennan M. F.; Menck H. R. The National Cancer Data Base report on pancreatic cancer. Cancer 1995, 76, 1671–1677. [DOI] [PubMed] [Google Scholar]
- Jemal A.; Siegel R.; Ward E.; Hao Y.; Xu J.; Murray T.; Thun M. J. Cancer statistics, 2008. CA Cancer J. Clin. 2008, 58, 71–96. [DOI] [PubMed] [Google Scholar]
- Abrams R. A.; Grochow L. B.; Chakravarthy A.; Sohn T. A.; Zahurak M. L.; Haulk T. L.; Ord S.; Hruban R. H.; Lillemoe K. D.; Pitt H. A.; Cameron J. L.; Yeo C. J. Intensified adjuvant therapy for pancreatic and periampullary adenocarcinoma: survival results and observations regarding patterns of failure, radiotherapy dose and CA19−9 levels. Int. J. Radiat. Oncol. Biol. Phys. 1999, 44, 1039–1046. [DOI] [PubMed] [Google Scholar]
- Ritts R. E.; Pitt H. A. CA 19−9 in pancreatic cancer. Surg. Oncol. Clin. N. Am. 1998, 7, 93–101. [PubMed] [Google Scholar]
- Furuya N.; Kawa S.; Hasebe O.; Tokoo M.; Mukawa K.; Maejima S.; Oguchi H. Comparative study of CA242 and CA19−9 in chronic pancreatitis. Br. J. Cancer 1996, 73, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M. B.; Steinberg W. M.; Henry J. P. Elevated serum levels of tumor marker CA19−9 in acute cholangitis. Dig. Dis. Sci. 1988, 33, 1223–1225. [DOI] [PubMed] [Google Scholar]
- Rosty C.; Goggins M. Early detection of pancreatic carcinoma. Hematol. Oncol. Clin. North Am. 2002, 16, 37–52. [DOI] [PubMed] [Google Scholar]
- Ching C. K.; Rhodes J. M. Enzyme-linked PNA lectin binding assay compared with CA19−9 and CEA radioimmunoassay as a diagnostic blood test for pancreatic cancer. Br. J. Cancer 1989, 59, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara H.; Nakaizumi A.; Tatsuta M.; Baba M.; Takenaka A.; Uedo N.; Sakai N.; Yano H.; Iishi H.; Ohigashi H.; Ishikawa O.; Okada S.; Kakizoe T. Diagnosis of pancreatic cancer by detecting telomerase activity in pancreatic juice: comparison with K-ras mutations. Am. J. Gastroenterol. 1999, 94, 2513–2518. [DOI] [PubMed] [Google Scholar]
- Yokoyama M.; Ochi K.; Ichimura M.; Mizushima T.; Shinji T.; Koide N.; Tsurumi T.; Hasuoka H.; Harada M. Matrix metalloproteinase-2 in pancreatic juice for diagnosis of pancreatic cancer. Pancreas 2002, 24, 344–347. [DOI] [PubMed] [Google Scholar]
- Yu K. H.; Rustgi A. K.; Blair I. A. Characterization of proteins in human pancreatic cancer serum using differential gel electrophoresis and tandem mass spectrometry. J. Proteome Res. 2005, 4, 1742–1751. [DOI] [PubMed] [Google Scholar]
- Yocum A. K.; Yu K.; Oe T.; Blair I. A. Effect of immunoaffinity depletion of human serum during proteomic investigations. J. Proteome Res. 2005, 4, 1722–1731. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Weaver V. M.; Blair I. A. Analysis of protein expression during oxidative stress in breast epithelial cells using a stable isotope labeled proteome internal standard. J. Proteome Res. 2005, 4, 2007–2014. [DOI] [PubMed] [Google Scholar]
- Yocum A. K.; Busch C. M.; Felix C. A.; Blair I. A. Proteomics-based strategy to identify biomarkers and pharmacological targets in leukemias with t(4;11) translocations. J. Proteome Res. 2006, 5, 2743–2753. [DOI] [PubMed] [Google Scholar]
- Oe T.; Ackermann B. L.; Inoue K.; Berna M. J.; Garner C. O.; Gelfanova V.; Dean R. A.; Siemers E. R.; Holtzman D. M.; Farlow M. R.; Blair I. A. Quantitative analysis of amyloid beta peptides in cerebrospinal fluid of Alzheimer‘s disease patients by immunoaffinity purification and stable isotope dilution liquid chromatography/negative electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2006, 20, 3723–3735. [DOI] [PubMed] [Google Scholar]
- Yocum A. K.; Yu K.; Oe T.; Blair I. A. Effect of immunoaffinity depletion of human serum during proteomic investigations. J. Proteome Res. 2005, 4, 1722–1731. [DOI] [PubMed] [Google Scholar]
- Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J. Physiol. 2005, 563, 23–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everley P. A.; Bakalarski C. E.; Elias J. E.; Waghorne C. G.; Beausoleil S. A.; Gerber S. A.; Faherty B. K.; Zetter B. R.; Gygi S. P. Enhanced analysis of metastatic prostate cancer using stable isotopes and high mass accuracy instrumentation. J. Proteome Res. 2006, 5, 1224–1231. [DOI] [PubMed] [Google Scholar]
- Cargile B. J.; Sevinsky J. R.; Essader A. S.; Stephenson J. L. Jr.; Bundy J. L. Immobilized pH gradient isoelectric focusing as a first-dimension separation in shotgun proteomics. J. Biomol. Tech. 2005, 16, 181–189. [PMC free article] [PubMed] [Google Scholar]
- Liu T.; Qian W. J.; Gritsenko M. A.; Xiao W.; Moldawer L. L.; Kaushal A.; Monroe M. E.; Varnum S. M.; Moore R. J.; Purvine S. O.; Maier R. V.; Davis R. W.; Tompkins R. G.; Camp D. G.; Smith R. D. High dynamic range characterization of the trauma patient plasma proteome. Mol. Cell. Proteomics 2006, 5, 1899–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocum A. K.; Yu K.; Oe T.; Blair I. A. Effect of immunoaffinity depletion of human serum during proteomic investigations. J. Proteome Res. 2005, 4, 1722–1731. [DOI] [PubMed] [Google Scholar]
- Janini G. M.; Conrads T. P.; Veenstra T. D.; Issaq H. J. Development of a two-dimensional protein-peptide separation protocol for comprehensive proteome measurements. J. Chromatogr., B: Analyt. Technol. Biomed. Life Sci. 2003, 787, 43–51. [DOI] [PubMed] [Google Scholar]
- Michel P. E.; Crettaz D.; Morier P.; Heller M.; Gallot D.; Tissot J. D.; Reymond F.; Rossier J. S. Proteome analysis of human plasma and amniotic fluid by Off-Gel isoelectric focusing followed by nano-LC-MS/MS. Electrophoresis 2006, 27, 1169–1181. [DOI] [PubMed] [Google Scholar]
- Tang H. Y.; li-Khan N.; Echan L. A.; Levenkova N.; Rux J. J.; Speicher D. W. A novel four-dimensional strategy combining protein and peptide separation methods enables detection of low-abundance proteins in human plasma and serum proteomes. Proteomics 2005, 5, 3329–3342. [DOI] [PubMed] [Google Scholar]
- Sabounchi-Schutt F.; Astrom J.; Olsson I.; Eklund A.; Grunewald J.; Bjellqvist B. An immobiline DryStrip application method enabling high-capacity two-dimensional gel electrophoresis. Electrophoresis 2000, 21, 3649–3656. [DOI] [PubMed] [Google Scholar]
- Ong S. E.; Blagoev B.; Kratchmarova I.; Kristensen D. B.; Steen H.; Pandey A.; Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 2002, 1, 376–386. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Weaver V. M.; Blair I. A. Analysis of protein expression during oxidative stress in breast epithelial cells using a stable isotope labeled proteome internal standard. J. Proteome Res. 2005, 4, 2007–2014. [DOI] [PubMed] [Google Scholar]
- Yocum A. K.; Busch C. M.; Felix C. A.; Blair I. A. Proteomics-based strategy to identify biomarkers and pharmacological targets in leukemias with t(4;11) translocations. J. Proteome Res. 2006, 5, 2743–2753. [DOI] [PubMed] [Google Scholar]
- Han D. K.; Eng J.; Zhou H.; Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 2001, 19, 946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M.; Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R.; Bairoch A.; Wu C. H.; Barker W. C.; Boeckmann B.; Ferro S.; Gasteiger E.; Huang H.; Lopez R.; Magrane M.; Martin M. J.; Natale D. A.; O’Donovan C.; Redaschi N.; Yeh L. S. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, D115−D119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.; Yi E. C.; Donohoe S.; Pan S.; Eng J.; Cooke K.; Crispin D. A.; Lane Z.; Goodlett D. R.; Bronner M. P.; Aebersold R.; Brentnall T. A. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology 2005, 129, 1187–1197. [DOI] [PubMed] [Google Scholar]
- Caulin C.; Salvesen G. S.; Oshima R. G. Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis. J. Cell Biol. 1997, 138, 1379–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N. O.; Liao J.; Omary M. B. Apoptosis generates stable fragments of human type I keratins. J. Biol. Chem. 1997, 272, 33197–33203. [DOI] [PubMed] [Google Scholar]
- Hosokawa M.; Kashiwaya K.; Eguchi H.; Ohigashi H.; Ishikawa O.; Furihata M.; Shinomura Y.; Imai K.; Nakamura Y.; Nakagawa H. Over-expression of cysteine proteinase inhibitor cystatin 6 promotes pancreatic cancer growth. Cancer Sci. 2008, 99, 1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipponen P.; Aaltomaa S.; Kosma V. M.; Syrjanen K. Apoptosis in breast cancer as related to histopathological characteristics and prognosis. Eur. J. Cancer 1994, 30A, 2068–2073. [DOI] [PubMed] [Google Scholar]
- Berardo M. D.; Elledge R. M.; de Moor C.; Clark G. M.; Osborne C. K.; Allred D. C. bcl-2 and apoptosis in lymph node positive breast carcinoma. Cancer 1998, 82, 1296–1302. [PubMed] [Google Scholar]
- Zhang G. J.; Kimijima I.; Abe R.; Watanabe T.; Kanno M.; Hara K.; Tsuchiya A. Apoptotic index correlates to bcl-2 and p53 protein expression, histological grade and prognosis in invasive breast cancers. Anticancer Res. 1998, 18, 1989–1998. [PubMed] [Google Scholar]
- Kramer G.; Erdal H.; Mertens H. J.; Nap M.; Mauermann J.; Steiner G.; Marberger M.; Biven K.; Shoshan M. C.; Linder S. Differentiation between cell death modes using measurements of different soluble forms of extracellular cytokeratin 18. Cancer Res. 2004, 64, 1751–1756. [DOI] [PubMed] [Google Scholar]
- Ueno T.; Toi M.; Biven K.; Bando H.; Ogawa T.; Linder S. Measurement of an apoptotic product in the sera of breast cancer patients. Eur. J. Cancer 2003, 39, 769–774. [DOI] [PubMed] [Google Scholar]
- Ley K.; Laudanna C.; Cybulsky M. I.; Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007, 7, 678–689. [DOI] [PubMed] [Google Scholar]
- Maatta M.; Butzow R.; Luostarinen J.; Petajaniemi N.; Pihlajaniemi T.; Salo S.; Miyazaki K.; Autio-Harmainen H.; Virtanen I. Differential expression of laminin isoforms in ovarian epithelial carcinomas suggesting different origin and providing tools for differential diagnosis. J. Histochem. Cytochem. 2005, 53, 1293–1300. [DOI] [PubMed] [Google Scholar]
- Schon M.; Klein C. E.; Hogenkamp V.; Kaufmann R.; Wienrich B. G.; Schon M. P. Basal-cell adhesion molecule (B-CAM) is induced in epithelial skin tumors and inflammatory epidermis, and is expressed at cell-cell and cell-substrate contact sites. J. Invest Dermatol. 2000, 115, 1047–1053. [DOI] [PubMed] [Google Scholar]
- Kikkawa Y.; Miner J. H. Review: Lutheran/B-CAM: a laminin receptor on red blood cells and in various tissues. Connect. Tissue Res. 2005, 46, 193–199. [DOI] [PubMed] [Google Scholar]
- Campbell I. G.; Foulkes W. D.; Senger G.; Trowsdale J.; Garin-Chesa P.; Rettig W. J. Molecular cloning of the B-CAM cell surface glycoprotein of epithelial cancers: a novel member of the immunoglobulin superfamily. Cancer Res. 1994, 54, 5761–5765. [PubMed] [Google Scholar]
- Lehmann J. M.; Riethmuller G.; Johnson J. P. MUC18, a marker of tumor progression in human melanoma, shows sequence similarity to the neural cell adhesion molecules of the immunoglobulin superfamily. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 9891–9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshishian H.; Addona T.; Burgess M.; Kuhn E.; Carr S. A. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 2007, 6, 2212–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.