Summary
RNA enzymes have been made to undergo self-sustained replication in the absence of proteins, providing the basis for an artificial genetic system.
An RNA enzyme that catalyzes the RNA-templated joining of RNA was converted to a format whereby two enzymes catalyze each other’s synthesis from a total of four component substrates. These cross-replicating RNA enzymes were optimized so that they can undergo self-sustained exponential amplification at a constant temperature and in the absence of proteins or other biological materials. Amplification occurs with a doubling time of about one hour, and can be continued indefinitely. Populations of various cross-replicating enzymes were constructed and allowed to compete for a common pool of substrates. During a serial transfer experiment in which the population underwent overall amplification of >1025-fold, recombinant replicators arose and grew to dominate the population. RNA enzymes that undergo self-sustained replication can serve as an experimental model of a genetic system. Many such model systems could be constructed, allowing different selective outcomes to be related to the underlying properties of the genetic system.
The most fundamental process of biological systems is the replication of the genetic material, brought about by genetically-encoded enzymes. Genetic replication involves a plus-strand nucleic acid template that directs the synthesis of a complementary minus-strand, which in turn directs the synthesis of a new plus-strand. The number of both strands increases exponentially with repeated rounds of templated copying. A longstanding research goal has been to devise a non-biological system that undergoes replication in a self-sustained manner, that is, brought about by enzymatic machinery which is part of the system being replicated. One way to realize this goal, inspired by the notion of primitive RNA-based life, would be for an RNA enzyme to catalyze the replication of RNA molecules, including the RNA enzyme itself (1–4). This has now been achieved in a cross-catalytic system that involves two RNA enzymes that catalyze each other’s synthesis from a total of four component substrates. In this system, exponential growth continues indefinitely at constant temperature, with a doubling time of about 1 h. Furthermore, many such replicators can be constructed and allowed to compete for common resources, resulting in the emergence of new variants and survival of the fittest variants over time.
A well-studied class of RNA enzymes are the RNA ligases, which catalyze the RNA-templated joining of RNA molecules (5, 6). One such ligase is the “R3C” RNA enzyme, which was obtained using in vitro evolution (7). This enzyme binds two RNA substrates through Watson-Crick pairing and catalyzes nucleophilic attack of the 3′-hydroxyl of one substrate on the 5′-triphosphate of the other, forming a 3′,5′-phosphodiester and releasing inorganic pyrophosphate. The R3C ligase previously was configured so that it could self-replicate by joining two RNA molecules to produce another copy of itself (8). Self-replication was inefficient, however, because the substrates formed a non-productive complex that limited the extent of exponential growth. Even under the most favorable conditions, the doubling time was about 17 h and no more than two doublings could be achieved.
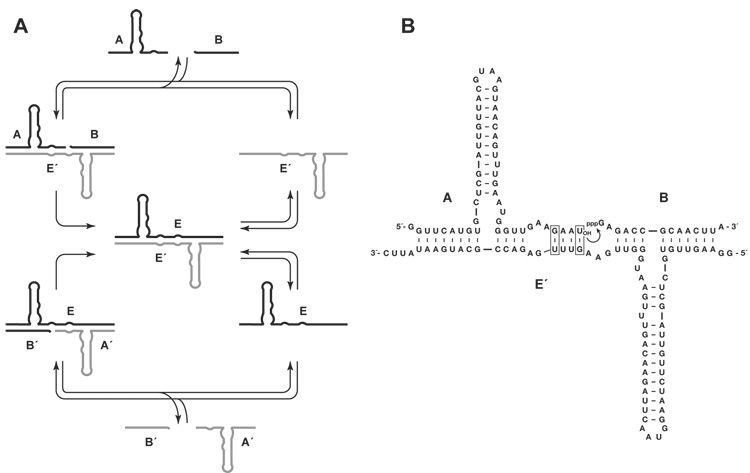
The R3C ligase then was converted to a cross-catalytic format (Fig. 1A), whereby a plus-strand RNA enzyme (E) catalyzed the joining of two substrates (A′ and B′) to form a minus-strand enzyme (E′), which in turn catalyzed the joining of two substrates (A and B) to form a new plus-strand enzyme (9). This too was inefficient because of the formation of non-productive complexes and the slow underlying rate of the two enzymes. Even with thermal cycling to disrupt the non-productive complexes and recycle the catalysts, it was not possible to achieve even a single doubling of the two enzymes (10). The enzyme E catalyzes the formation of E′ at a rate of 0.034 min−1 with a maximum extent of 20%, while E′ catalyzes the formation of E at a rate of 0.026 min−1 with a maximum extent of 11% (9). These reaction rates are about 10-fold slower than that of the parental R3C ligase (7), and when the two cross-catalytic reactions are carried out within a common mixture, the reaction rates are even slower (9).
Fig. 1.
Scheme for cross-catalytic replication of RNA enzymes. (A) The enzyme E′ (gray) catalyzes ligation of substrates A and B (black) to form the enzyme E, while E catalyzes ligation of A′ and B′ to form E′. The two enzymes dissociate to provide copies of both E and E′ that each can catalyze another reaction. (B) Sequence and secondary structure of the complex formed between the cross-replicating RNA enzyme and its two substrates (E′, A, and B are shown; E, A′, and B′ are the reciprocal). Curved arrow indicates the site of ligation. Boxed residues indicate the sites of critical wobble pairs that provide enhanced catalytic activity compared to the parental R3C ligase.
In order to achieve sustained exponential amplification, it thus became necessary to improve the catalytic properties of the cross-replicating RNA enzymes. This was done using in vitro evolution, optimizing the two component reactions in parallel and seeking solutions that would apply to both reactions when conducted in the cross-catalytic format (11). The 5′-triphosphate-bearing substrate was joined to the enzyme via a hairpin loop (B′ to E, and B to E′), and nucleotides within both the enzyme and the separate 3′-hydroxyl-bearing substrate (A′ and A) were randomized at a frequency of 12% per nucleotide position. The two resulting populations of molecules were subjected to six rounds of stringent in vitro selection, selecting for their ability to react in progressively shorter times, ranging from 2 h to 10 milliseconds. The shortest times were achieved using a quench-flow apparatus. Mutagenic PCR was performed after the third round to maintain diversity in the population. Following the sixth round, individuals were cloned from both populations and sequenced. There was substantial sequence variability among the clones, but all contained mutations just upstream from the ligation junction that resulted in a G•U wobble pair at this position.
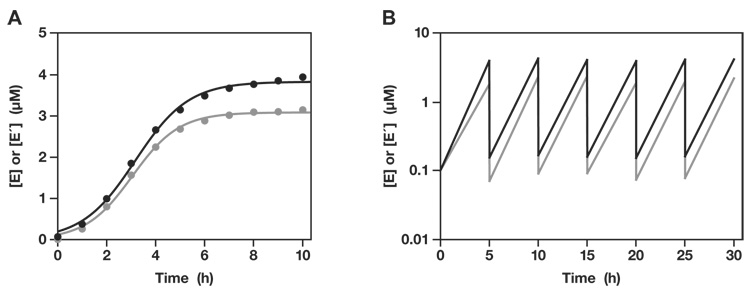
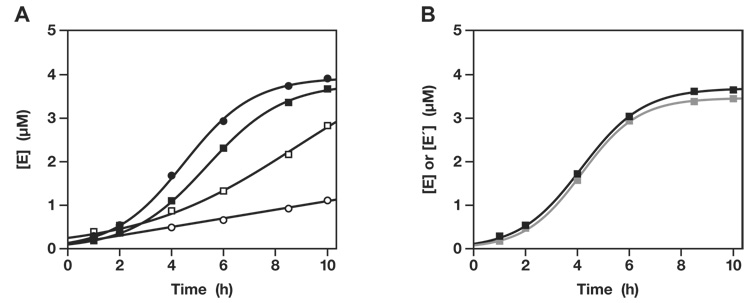
The G•U wobble pair was installed in both enzymes and both 3′-hydroxyl-bearing substrates (Fig. 1B). In the trimolecular reaction (with two separate substrates), the optimized enzymes, E and E′, exhibited a rate of 1.3 and 0.3 min−1 with a maximum extent of 92% and 88%, respectively. This was deemed sufficient to initiate exponential amplification. A reaction mixture was prepared containing 0.1 µM each of unlabeled E and E′, 5.0 µM each of [5′-32P]-labeled A and A′, 5.0 µM each of unlabeled B and B′, 25 mM MgCl2, and 50 mM EPPS buffer (pH 8.5), which was incubated at 42 °C for 10 h. Samples were taken from the mixture at various times, and the yield of newly-synthesized E and E′ was determined by separating the radiolabeled materials in a denaturing polyacrylamide gel. Both enzymes exhibited robust exponential growth, with more than 25-fold amplification after 5 h, followed by a leveling off as the supply of substrates became depleted (Fig. 2A). The data fit well to the logistic growth equation:
[E]t = a / (1 + be−ct), where [E]t is the concentration of E (or E′) at time t, a is the maximum extent of growth, b is the degree of sigmoidicity, and c is the exponential growth rate.
This equation is commonly used in population ecology to model the exponential growth of organisms subject to the carrying capacity of the local environment. For the enzymes E and E′, the exponential growth rate was 0.92 and 1.05 h−1, respectively.
Fig. 2.
Self-sustained amplification of cross-replicating RNA enzymes. (A) The yield of both E (black) and E′ (gray) increased exponentially before leveling off as the supply of substrates became exhausted. (B) Amplification was sustained by performing a serial transfer experiment, allowing ~25-fold amplification before transferring 1/25th of the mixture to a new reaction vessel that contained a fresh supply of substrates. The concentrations of E and E′ were measured at the end of each incubation.
Exponential growth can be continued indefinitely, so long as a supply of the four substrates is maintained. One way to achieve this is to carry out a serial transfer experiment in which a portion of a completed reaction mixture is transferred to a new reaction vessel that contains a fresh supply of substrates. Six successive reactions were carried out in this fashion, each 5 h in duration and transferring 1/25th of the material from one reaction mixture to the next. The first mixture contained 0.1 µM each of E and E′, but all subsequent mixtures contained only those enzymes that were carried over in the transfer. Exponential growth was maintained throughout 30 h total incubation, with an overall amplification of >108-fold for each of the two enzymes (Fig. 2B). This corresponds to 28 doublings in a process that was sustained by the enzymes themselves. No temperature cycling was required and the reaction mixtures did not contain any proteins or other biological materials.
A genetic system requires not only self-replication, but also the opportunity for many different genetic molecules to replicate, with their replication rate dependent on genetically-encoded functional properties. It is possible to construct many variants of the cross-replicating RNA enzymes that differ with respect to their “genotype” and associated “phenotype”. The genotype is defined as the regions of the enzyme that engage in Watson-Crick pairing with its cross-catalytic partner and that can vary in sequence without significantly affecting replication efficiency. These regions are located at the 5′ and 3′ ends of the enzyme (Fig. 1B). Other regions of Watson-Crick pairing between the two enzymes are tolerant of some sequence variation, albeit with some alteration of replication efficiency.
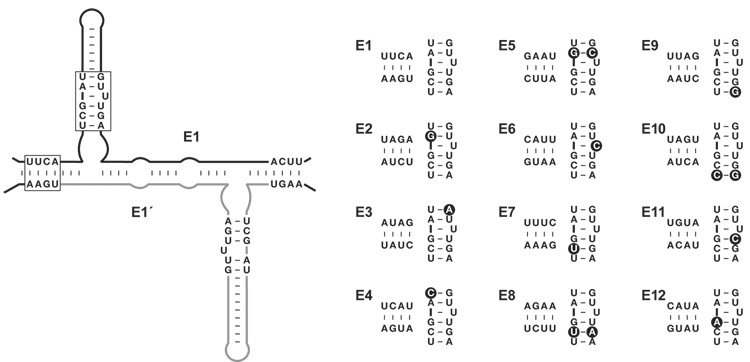
Four nucleotide positions at the 5′ end and four nucleotides at the 3′ end of the enzyme were chosen as the sites for genotypic variation (Fig. 3). A rule was adopted that each of these regions would contain one G•C and three A•U pairs so that there would be no substantial differences in base-pairing stability among the various genotypes. This allowed 32 possible pairs of complementary sequences for each region, of which 12 were chosen as a set of designated genotypes (Fig. 3). Each genotype was associated with a distinct phenotype, manifest as a particular sequence within the catalytic core of the enzyme. For simplicity, the same phenotype was associated with both members of a cross-replicating pair, although this need not be the case.
Fig. 3.
Twelve pairs of cross-replicating RNA enzymes were constructed. Four nucleotides at the 5′ and 3′ ends of the enzyme were chosen as the sites for genotypic variation, and 11 nucleotides within the catalytic core were chosen as the corresponding sites for phenotypic variation (boxed regions). The sequence of these regions for each of the 12 E enzymes is shown at the right. The corresponding E′ enzymes have a complementary sequence in the genotype region and the same sequence in the catalytic core. Alterations of the catalytic core relative to the E1 enzyme are highlighted by black circles.
Twelve pairs of cross-replicating enzymes were synthesized, as well as the 48 substrates (12 each of A, A′, B, and B′) necessary to support their exponential amplification. Each replicator was tested individually and demonstrated varying levels of catalytic activity and varying rates of exponential growth (fig. S1). Replication was somewhat faster in the presence of 25 compared to 15 mM MgCl2, but the lower concentration was chosen for subsequent studies because it is less likely to promote the use of mismatched substrates and renders the RNA less susceptible to hydrolysis. Of the 12 pairs of cross-replicating enzymes, the one shown in Fig. 1B (now designated E1 and E1′) had the fastest rate of exponential growth, achieving about 20-fold amplification after 5 h in the presence of 15 mM MgCl2. The various cross-replicating enzymes shown in Fig. 3 had the following rank order of replication efficiency: E1, E10, E5, E4, E6, E3, E12, E7, E9, E8, E2, E11. The top five replicators all achieved more than 10-fold amplification after 5 h, and all except E11 achieved at least 5-fold amplification after 5 h.
Two different serial transfer experiments were carried out involving mixtures of various cross-replicating enzymes and their corresponding substrates. The first was initiated with 0.1 µM each of E1–E4 and E1′–E4′, and 5.0 µM each of the 16 corresponding substrates. Sixteen successive reactions were carried out over a period of 70 h, transferring 1/20th of the material from one reaction mixture to the next (fig. S2A). Individuals were cloned from the population following the final reaction, and were sequenced to determine their genotype and to confirm the identity of their corresponding phenotype. Among 25 clones (sequencing E′ only), there was no dominant replicator (fig. S2B). E1′, E2′, E3′, and E4′ all were represented, as well as 17 clones that were the result of recombination between a particular A′ substrate and one of the three B′ substrates other than its original partner (or similarly for A and B).
Recombination occurs when an enzyme binds and ligates a mismatched substrate. In principle, any A could become joined to any B or B′, and any A′ could become joined to any B′ or B, resulting in 64 possible enzymes. The “genetic code” was designed so that cognate substrates have a binding advantage of several kcal/mol compared to non-cognate substrates (fig. S2C). However, once a mismatched substrate is bound and ligated, it forms a recombinant enzyme that can cross-replicate by drawing upon the corresponding set of four substrates. Recombinants can give rise to other recombinants, as well as revert back to non-recombinants. Mismatches are less likely to occur during the pairing of A and B′ regions compared to the pairing of A′ and B regions because the former enjoy the benefit of an additional base pair for matched substrates (Fig. 1B). Thus there are expected to be preferred pathways for mutation, primarily involving substitution among certain A′ and among certain B components (fig. S2D), although reflected in the identity of both members of a cross-replicating pair.
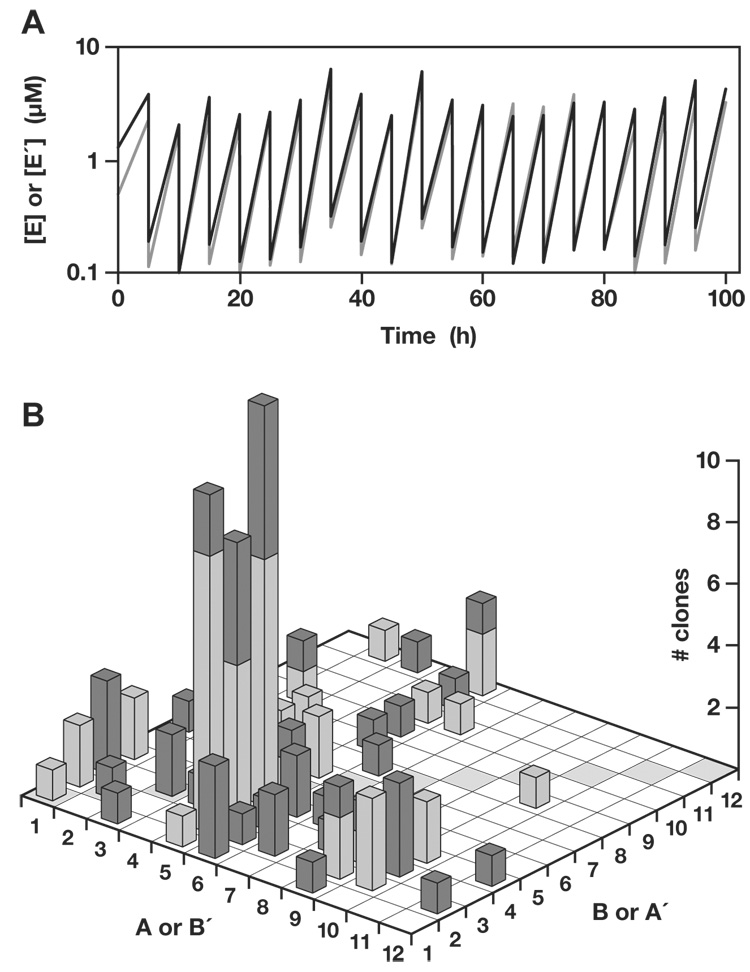
Another serial transfer experiment was initiated with 0.1 µM each of all 12 pairs of cross-replicating enzymes and 5.0 µM each of the 48 corresponding substrates. In this more complex mixture there was abundant opportunity for recombination, with 132 possible pairs of recombinant cross-replicating enzymes, as well as the 12 pairs of non-recombinant cross-replicators. Twenty successive reactions were carried out over a period of 100 h, transferring 1/20th of the material from one reaction mixture to the next, and achieving an overall amplification of >1025-fold (Fig. 4A). Again individuals were cloned from the final population and sequenced. Of 100 clones (sequencing 50 E and 50 E′), only 7 were non-recombinants (Fig. 4B). The distribution was highly non-uniform, with sparse representation of molecules containing components A6–A12 and B5–B12 (and reciprocal components B6′–B12′ and A5′–A12′). The most frequently represented components were A5 and B3 (and reciprocal components B5′ and A3′). The three most abundant recombinants were A5B2, A5B3, and A5B4 (and their cross-replication partners), which together accounted for one-third of all clones.
Fig. 4.
Self-sustained amplification of a population of cross-replicating RNA enzymes, resulting in selection of the fittest replicators. (A) Beginning with 12 pairs of cross-replicating RNA enzymes (Fig. 3), amplification was sustained by serial transfer for 20 successive rounds of ~20-fold amplification and 20-fold dilution. The concentrations of all E (black) and E′ (gray) molecules were measured at the end of each incubation. (B) Graphical representation of the observed genotypes among 50 E and 50 E′ clones (dark and light columns, respectively) that were sequenced following the last incubation. The A and B (or B′ and A′) components of the various enzymes are shown on the horizontal axes, with non-recombinant enzymes indicated by shaded boxes along the diagonal. The number of clones containing each combination of components is shown on the vertical axis.
The exponential growth rates of A5B2, A5B3, and A5B4 were compared to that of E1, the most efficient non-recombinant replicator. In the presence of their cognate substrates alone, E1 remained the most efficient replicator, but in the presence of all 48 substrates, the most efficient replicator was A5B3 (Fig. 5A). The exponential growth rate of E1 was 0.75 h−1 in the presence of its cognate substrates, but it exhibited only linear growth at a rate of 0.10 h−1 in the presence of all substrates. In contrast the exponential growth rate of A5B3 was 0.68 h−1 in the presence of its cognate substrates, and 0.33 h−1 in the presence of all substrates. When the A5B3 replicator was provided a mixture of substrates corresponding to the components of the three most abundant recombinants (A5, B2, B3, B4, B5′, A2′, A3′, and A4′), its exponential growth rate was 0.84 h−1, the highest measured for any replicator in the presence of 15 mM MgCl2 (Fig. 5B).
Fig. 5.
Exponential amplification of the starting cross-replicating enzymes (E1 and E1′) and of the most efficient cross-replicator (A5B3 and B5′A3′) that emerged during the serial transfer experiment involving all 48 substrates (Fig. 4). (A) Comparative growth of E1 (circles) and A5B3 (squares) in the presence of either their cognate substrates alone (filled symbols) or all substrates that were present during serial transfer (open symbols). (B) Growth of A5B3 (black) and B5′A3′ (gray) in the presence of the eight substrates (A5, B2, B3, B4, B5′, A2′, A3′, and A4′) that comprise the three most abundant cross-replicating enzymes.
The fitness of a pair of cross-replicating enzymes depends on several factors, including their intrinsic catalytic activity, exponential growth rate with cognate substrates, ability to withstand inhibition by other substrates in the mixture, and net rate of production through mutation among the various cross-replicators. The A5B3 recombinant and its cross-replication partner B5′A3′ have different catalytic cores (Fig. 3), and both exhibit robust activity. The A5B3 enzyme has a rate of 0.58 min−1 and maximum extent of 90%, which is comparable to E1 with a rate of 0.63 min−1 and maximum extent of 90% (measured in the presence of 15 mM MgCl2). The B5′A3′ enzyme has a rate of 0.66 min−1 and maximum extent of 90%, which is considerably more active than E1′ with a rate of 0.11 min−1 and maximum extent of 92%. The nearly equal rates of the A5B3 and B5′A3′ enzymes may account for their well-balanced rate of production throughout the course of exponential amplification (Fig. 5B). Other factors, however, such as substrate binding and product release, can influence the rate of exponential growth, which may explain why amplification of E1 with its cognate substrates outpaces that of A5B3. The selective advantage that A5B3 enjoys appears to derive from its relative resistance to inhibition by other substrates in the mixture (Fig. 5A) and its ability to capitalize on facile mutation among substrates B2, B3, and B4 and among substrates A2′, A3′, and A4′ (fig. S2D).
A population of cross-replicating RNA enzymes can serve as an experimental model of a genetic system. This model is greatly simplified compared to biological genetics because it involves only two genetic loci with, at present, only 12 alleles per locus. It is likely, however, that the number of alleles could be increased by exploiting more than four nucleotide positions at the 5′ and 3′ ends of the enzyme, and by relaxing the rule that these nucleotides form one G•C and three A•U pairs. One could construct many different genetic systems with alternative rule sets, resulting in alternative behaviors during the course of selective amplification. Different mixtures of enzymes and substrates and different reaction conditions are expected to lead to different outcomes, and these could then be related to the underlying properties of the genetic system.
In order to support greater complexity in a system of cross-replicating RNAs it will be necessary to constrain the set of substrates so that each enzyme can secure its own substrates without being overwhelmed by other substrates in the mixture. One way to do this is to choose a set of substrates that are more distinguishable than the ones used here. Another approach is to adjust the concentrations of the various substrates in proportion to their utilization by the population of enzymes. It is not clear how this would be done within the system, but it could be achieved using a deconstructive PCR procedure in which the population of newly-formed enzymes is used to generate a corresponding population of substrates (11). In this way both the successful enzymes and their component substrates are inherited from one generation to the next.
Another important challenge for an artificial genetic system is to support a broad range of encoded functions, well beyond replication itself. It is possible to insert a functional domain within the central stem-loop of the cross-replicating enzymes so that replication is dependent on execution of that encoded function (Lam & Joyce, unpublished results). It would be much more powerful, however, to have a system in which novel function emerges during the course of selective amplification. The self-sustained evolution of RNA with open-ended opportunities for discovering novel function likely has not occurred on Earth since the time of the RNA world, and continues to present an intriguing research opportunity.
Supplementary Material
References
- 1.Crick FHC. J. Mol. Biol. 1968;38:367. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 2.Szostak JW, Bartel DP, Luisi PL. Nature. 2001;409:387. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 3.Joyce GF. Nature. 2002;418:214. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- 4.Orgel LE. Crit. Rev. Biochem. Mol. Biol. 2004;39:99. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP, Szostak JW. Science. 1993;261:1411. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 6.Joyce GF. Annu. Rev. Biochem. 2004;73:791. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 7.Rogers J, Joyce GF. RNA. 2001;7:395. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul N, Joyce GF. Proc. Natl. Acad. Sci. USA. 2002;99:12733. doi: 10.1073/pnas.202471099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim D-E, Joyce GF. Chem. Biol. 2004;11:1505. doi: 10.1016/j.chembiol.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Kim K-S, Oh S, Yea SS, Yoon M-Y, Kim D-E. FEBS Lett. 2008;582:2745. doi: 10.1016/j.febslet.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supporting material on Science Online.
- 12.The authors thank Leslie Orgel for many stimulating discussions. This research was supported by grants from NASA (NNX07AJ23G) and NIH (R01GM065130), and by the Skaggs Institute for Chemical Biology at The Scripps Research Institute.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.