Abstract
Duplications and rearrangements of coding genes are major themes in the evolution of mitochondrial genomes, bearing important consequences in the function of mitochondria and the fitness of organisms. Yu et al. (BMC Genomics 2008, 9:477) reported the complete mt genome sequence of the oyster Crassostrea hongkongensis (16,475 bp) and found that a DNA segment containing four tRNA genes (trnK1, trnC, trnQ1 and trnN), a duplicated (rrnS) and a split rRNA gene (rrnL5') was absent compared with that of two other Crassostrea species. It was suggested that the absence was a novel case of "tandem duplication-random loss" with evolutionary significance. We independently sequenced the complete mt genome of three C. hongkongensis individuals, all of which were 18,622 bp and contained the segment that was missing in Yu et al.'s sequence. Further, we designed primers, verified sequences and demonstrated that the sequence loss in Yu et al.'s study was an artifact caused by placing primers in a duplicated region. The duplication and split of ribosomal RNA genes are unique for Crassostrea oysters and not lost in C. hongkongensis. Our study highlights the need for caution when amplifying and sequencing through duplicated regions of the genome.
Background
Because of its nature of maternal inheritance, mitochondrial (mt) genome has a fast rate of evolution and is particularly useful in phylogenetic analysis. The analysis of complete mt genome sequences provides not only information about nucleotide changes, but also insights into gene order and rearrangements that are indicative of major evolutionary changes.
We read with great interest an article appeared in a recent issue of BMC Genomics (9:477 2008) entitled 'Complete mitochondrial DNA sequence of oyster Crassostrea hongkongensis – a case of "Tandem duplication-random loss" for genome rearrangement in Crassostrea?' by Yu, Z.N., Wei, Z.P., Kong, X.Y., and Shi, W. [1]. Based on our data, we believe that an important part of Yu et al.'s paper is incorrect and would like to share our results with the readers of this Journal.
In their paper, Yu et al. (2008) reported that the complete mt genome of C. hongkongensis is 16,475 bp in length (GenBank accession number EU266073) and pointed out that 'A striking finding of this study is that a DNA segment containing four tRNA genes (trnK1, trnC, trnQ1 and trnN) and two duplicated or split rRNA genes (rrnL5' and rrnS) are absent from the genome, when compared with that of two other extant Crassostrea species, which is very likely a consequence of loss of a single genomic region present in ancestor of C. hongkongensis. It indicates this region seem to be a "hot spot" of genomic rearrangements over the Crassostrea mt-genomes' (p. 1, Abstract, line 14–19). We have independently sequenced the complete mt genomes of three C. hongkongensis individuals. All our three sequences contained the DNA segment that was reported missing in Yu et al.'s study. The discrepancy is not trivial as the loss of the duplicated region was central to Yu et al.'s hypothesis of a novel "tandem duplication and random loss" event during the evolution of C. hongkongensis. It was further suggested that this region was a "hot spot" for genomic rearrangement. Therefore, it is critical to determine if the loss of the duplicated region is real in view of the different sequences we obtained.
To determine which sequence is correct, we analyzed our sequences, compared them with Yu et al.'s sequence and sequences from other Crassostrea species, and designed primers to test the presence or absence of the genome region in question. Here we report that Yu et al.'s sequence is either incorrect or represent a rare mutation that is uncommon in C. hongkongensis.
Experimental design, results and discussion
The three complete mt genome sequences of C. hongkongensis that we independently obtained were submitted to GenBank: accession No. EU672834 for oyster HN from Hainan, FJ593172 for oyster BH45 from Guangxi, and FJ593173 for oyster H50 from Fujian. The length of the complete mt genome of C. hongkongensis reported by Yu et al. is 16,475 bp. The length of all three mt sequences that we obtained is 18,622 bp, which is 2,147 bp longer than that of Yu et al.'s. We aligned our sequences with that of Yu et al. and other Crassostrea species. We annotated our mt sequences according to that of C. gigas with minor revisions [2], and the results are presented in Table 1. Our sequence for C. hongkongensis has exactly the same gene order and arrangements as C. gigas, both containing the segment that is missing in Yu et al.'s sequence. The segment contains four tRNA genes, a duplicated rrnS and part of the split rrnL. The split rrnL is first discovered in C. virginica and appears to be unique for oysters [2].
Table 1.
Annotation of the mitochondrial genome of Crassostrea hongkongensis.
| Gene | Position | Size | Codon | Intergenic Nucleotides* | ||
| Nucleotide | Amino acids | Start | Stop | |||
| cox1 | 1–1617 | 1617 | 538 | ATA | TAA | 1 |
| rrnL 3'half | 1761–2472 | 712 | 143 | |||
| cox3 | 2575–3438 | 864 | 287 | ATA | TAA | 231 |
| trnI | 3439–3505 | 67 | 0 | |||
| trnT | 3506–3573 | 68 | 0 | |||
| trnE | 3595–3662 | 68 | 21 | |||
| cob | 3670–4875 | 1206 | 401 | ATA | TAA | 130 |
| trnD | 4984–5052 | 69 | 108 | |||
| cox2 | 5054–5755 | 702 | 233 | ATG | TAG | 1 |
| trnM1(ATG) | 5777–5842 | 66 | 21 | |||
| trnS1(AGN) | 5846–5915 | 70 | 3 | |||
| trnL2(UUR) | 5931–5997 | 67 | 15 | |||
| trnM2(ATG) | 6065–6129 | 65 | 67 | |||
| trnS2(UCN) | 6137–6204 | 68 | 7 | |||
| trnP | 6383–6451 | 69 | 178 | |||
| rrnS1 | 6452–7525 | 1074 | 0 | |||
| trnK1 | 7526–7594 | 69 | 0 | |||
| trnC | 7624–7691 | 68 | 29 | |||
| trnQ1(CAA) | 7709–7777 | 69 | 17 | |||
| rrnL 5'half | 7780–8384 | 605 | 1 | |||
| trnN | 8443–8508 | 66 | 58 | |||
| rrnS2 | 8509–9698 | 1190 | 0 | |||
| trnY | 9699–9764 | 66 | 0 | |||
| atp6 | 9770–10453 | 684 | 227 | ATG | TAG | 5 |
| trnG | 10966–11035 | 70 | 512 | |||
| trnV | 11644–11716 | 73 | 608 | |||
| nad2 | 11759–12757 | 999 | 332 | ATG | TAG | 42 |
| trnR | 12792–12858 | 67 | 34 | |||
| trnH | 12919–12983 | 65 | 60 | |||
| nad4 | 12986–14335 | 1350 | 449 | ATA | TAG | 2 |
| trnK2(AAA) | 14343–14417 | 75 | 7 | |||
| nad5 | 14419–16089 | 1671 | 556 | ATG | TAA | 1 |
| nad6 | 16101–16576 | 476 | 158 | ATT | TA- | 11 |
| trnQ2(CAA) | 16610–16678 | 69 | 33 | |||
| nad3 | 16684–17034 | 351 | 116 | ATG | TAG | 5 |
| trnL1(CUN) | 17070–17135 | 66 | 35 | |||
| trnF | 17171–17238 | 68 | 35 | |||
| trnA | 17259–17325 | 67 | 20 | |||
| nad1 | 17331–18266 | 936 | 311 | ATG | TAA | 5 |
| nad4L | 18270–18549 | 280 | 93 | ATG | T- | 3 |
| trnW | 18550–18618 | 69 | 22 | 0 | ||
"-" indicates termination codons completed via polyadenylation.
Underlined genes correspond to a segment missing in Yu et al.' sequence due to a sequencing artifact.
The three C. honghongensis oysters used in our study were from diverse populations (Hainan, Guangxi and Fujian) covering the entire geographic range of this species as we know (Guo et al., unpublished), and they were genetically identified using molecular markers prior to our study [3]. We compared one of our sequences with Yu et al.'s using BLAST http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi[4]. In the 16,475 shared nucleotides, there are 15 SNPs (single nucleotide polymorphisms) and the similarity between the two gnomes is 99.91%, suggesting that oysters used in our study and Yu et al.'s study are all C. hongkongensis. Sequence identity in major coding genes between our C. hongkongensis sequences and that of C. gigas is shown in Table 2. Considerable differentiation has occurred between the two sister-species at some genes (i.e., gene identity of 75.1% for nad2) despite the identical gene order. Analysis of all four C. hongkongensis mt sequences revealed 41 SNPs: 28 in coding and 13 in non-coding regions (Table 3). Of the 19 SNPs from protein coding genes, only one is non-synonymous, suggesting strong purifying selection. The non-synonymous mutation occurred at the atp6 gene in Yu's sequence only, and further studies are needed to determine whether it is a true SNP or sequencing error.
Table 2.
Sequence identity of major coding genes between Crassostrea hongkongensis and C. gigas.
| Gene | Number of nucleotides | Identity (%) | Number of amino acids | Identity (%) | ||
| C. hongkongensis | C. gigas | C. hongkongensis | C. gigas | |||
| cox1 | 1617 | 1617 | 87.4 | 538 | 538 | 98.0 |
| cox2 | 702 | 702 | 87.6 | 233 | 233 | 98.7 |
| cox3 | 864 | 876 | 82.7 | 287 | 291 | 89.7 |
| nad1 | 936 | 936 | 82.7 | 311 | 311 | 87.8 |
| nad2 | 999 | 999 | 75.1 | 332 | 332 | 72.6 |
| nad3 | 351 | 351 | 82.3 | 116 | 116 | 86.2 |
| nad4 | 1350 | 1353 | 78.3 | 449 | 450 | 81.8 |
| nd4L | 280 | 283 | 82.3 | 93 | 94 | 93.5 |
| nad5 | 1671 | 1671 | 77.1 | 556 | 556 | 80.2 |
| nad6 | 476 | 477 | 77.6 | 158 | 158 | 77.2 |
| cob | 1223 | 1248 | 79.5 | 401 | 412 | 88.0 |
| atp6 | 684 | 684 | 84.5 | 227 | 227 | 97.4 |
| rrnS1 | 1074 | 1037 | 93.2 | |||
| rrnS2 | 1190 | 1205 | 89.9 | |||
| rrnL 5' | 605 | 601 | 88.4 | |||
| rrnL 3' | 712 | 713 | 95.8 | |||
Table 3.
Single-nucleotide polymorphism (SNP) observed among four mitochondrial genome sequences of Crassostrea hongkongensis.
| Gene | Position | HN | BH45 | HC50 | Yu1 | Type |
| cox1 (1–1617) | 930 | C | C | A | C | Transversion, synonymous |
| 993 | C | C | C | T | Transition, synonymous | |
| 1395 | T | T | T | C | Transition, synonymous | |
| cox3 (2575–3438) | 2805 | G | A | G | G | Transition, synonymous |
| 3282 | G | T | G | G | Transversion, synonymous | |
| cob (3670–4875) | 4611 | T | C | T | T | Transition, synonymous |
| cob-trnD (4876–4983) | 4883 | C | C | C | T | Transition |
| cox2 (5054–5755) | 5533 | A | G | G | G | Transition, synonymous |
| cox2-trnS1(5756–5845) | 5845 | T | G | G | G | Transversion |
| rrnS1(6452–7525) | 7225 | C | T | C | - | Transition |
| 7325 | C | C | T | - | Transition | |
| rrnL-trnN (8385–8442) | 8407 | T | C | T | - | Transition |
| rrnS2 (8509–9698) | 8567 | C | T | C | - | Transition |
| 8832 | A | A | A | G | Transition | |
| 9036 | G | G | T | G | Transversion | |
| 9219 | T | C | T | T | Transition | |
| 9372 | C | T | C | C | Transition | |
| atp6 (9770–10453) | 10025 | T | T | T | C | Transition, nonsynonymous |
| atp6-trnG (10454–10965) | 10524 | G | T | G | G | Transversion |
| 10673 | G | A | G | G | Transition | |
| 10712 | C | C | T | C | Transition | |
| 10778 | A | G | G | G | Transition | |
| 10836 | C | T | C | C | Transition | |
| 10870 | T | C | T | T | Transition | |
| trnG-trnV (11036–11643) | 11502 | C | G | G | G | Transversion |
| 11509 | C | T | T | T | Transition | |
| 11638 | T | C | T | T | Transition | |
| trnV (11644–11716) | 11699 | T | C | T | C | Transition |
| 11701 | G | A | A | A | Transition | |
| nad2 (11759–12757) | 12070 | A | G | A | A | Transition, synonymous |
| nad4 (12986–14335) | 13232 | T | C | T | T | Transition, synonymous |
| 13678 | C | A | C | C | Transversion, synonymous | |
| nad5 (14419–16089) | 15159 | A | G | G | G | Transition, synonymous |
| 15216 | T | C | T | T | Transition, synonymous | |
| 15321 | G | G | T | G | Transversion, synonymous | |
| 15552 | A | A | A | G | Transition, synonymous | |
| 15645 | T | T | C | T | Transition, synonymous | |
| nad6-trnQ2 (16577–16609) | 16590 | C | C | T | C | Transition |
| nad3 (16684–17034) | 16986 | A | G | G | G | Transition, synonymous |
| nad1 (17331–18266) | 17876 | T | C | T | T | Transition, synonymous |
| 18134 | A | A | G | A | Transition, synonymous |
1Sequence for Yu is from Yu et al., 2008, and the other three sequences are from this study.
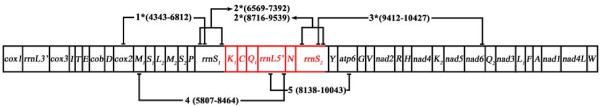
Yu et al. used ten pairs of primers to amplify the complete mt genome of C. hongkongensis (p. 11). We carefully studied the positions of each primer and located them in our mt genome sequences of C. hongkongensis (Figure 1). It occurred to us that Yu et al. might have failed to amplify the gene block of K1-C-Q1-rrnL5'-N-rrnS2 because some of their primers were placed in a duplicated region. As shown in Figure 1, primer pair 1* is located in gene cob and rrnS1 (or rrnS2), primer pair 2* is completely located within the duplicated gene rrnS (rrnS1 or/and rrnS2), and primer pair 3* is located in rrnS2 (or rrnS1) and atp6 (primer pairs 1*, 2* and 3* correspond to the third, the fourth and the fifth primer pairs in Yu et al.' paper, Table 4). Because these three primer pairs are either completely or partially (one of the two primers) located in the duplicated gene rrnS1 and rrnS2, they should theoretically amplify two fragments of different length, but in reality the smaller fragment may be preferentially amplified and sequenced. The length of shortest PCR products expected from the three primer pairs was 2,470 bp, 824 bp and 1,016 bp, respectively (Table 4). Primer pair 2* was completely located in the duplicated gene rrnS (rrnS1 or rrnS2); thus they may directly concatenate the sequence between the duplicated gene and artificially lose the gene block of K1-C-Q1-rrnL5'-N-rrnS2 (Figure 1). The block, 2,147 bp, may be too large to be amplified under competition with a smaller fragment.
Figure 1.
Position map of the primers used in amplifying fragments of C. hongkongensis mitochondrial genome. Above the gene map are the three pairs of primers used by Yu et al. (2008) and below are the two pairs of primers designed to confirm the existence of the gene block in red, which is reported missing by Yu et al. Gene segments are not drawn to scale.
Table 4.
Primers used to amplify fragments of Crassostrea hongkongensis mitochondrial genome.
| Order | Primer name | Sequence (5'-3') | Length | position | Product size (bp) |
| 1* | HK-4343F | TTAGAGTTCCGTTTCACCCG | 20 | 4343 | 2470, 4617 |
| HK-6812R | CTTTCGCTTCAATTTAGTTAGT | 22 | 6812, 8959 | ||
| 2* | HK-6569F | GGTTCTGGTATAATGTTAGCT | 21 | 6569, 8716 | 824, 2971 |
| HK-7392R | ATTACTCTCTTTTTACTCCC | 20 | 7392, 9539 | ||
| 3* | HK-9412F | CTAGGTCAGGTCGAAGTGCT | 20 | 7265, 9412 | 1016, 3163 |
| HK-10427R | AGAGCACAGGTGTTGGGTGA | 20 | 10427 | ||
| 4 | HK-5807F | GTCTCATAATCCGAAAGTGGTT | 22 | 5807 | 2658 |
| HK-8464R | CTTATACTTGGGCTACTTTCTT | 22 | 8464 | ||
| 5 | HK-8138F | GGTGCTCACTAAATCAGTATGT | 22 | 8138 | 1905 |
| HK-10043R | ATGAAGATAGTGACGGAAACCC | 22 | 10043 |
*Primer pairs from Yu et al. (2008). Because one or both of the primers are located in the duplicated rrnS gene, two fragments are expected but only the shorter one is actually amplified.
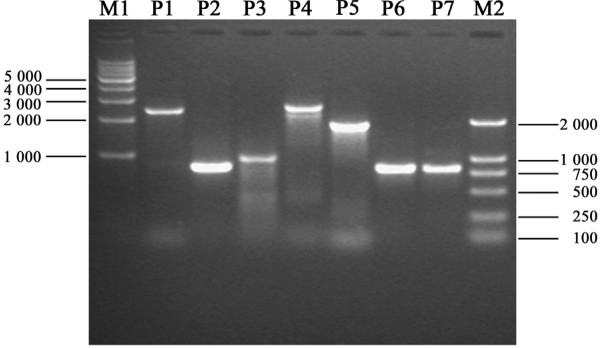
To test our hypothesis that the gene block between duplicated rrnS failed to amplify in Yu et al.'s study, we synthesized the three primer pairs used by Yu et al. (after removing mismatches based on our C. hongkongensis sequences to improve specificity). As expected, the three shorter products mentioned in Yu et al.' paper were successfully obtained (Table 4, Figure 2). We increased the elongation time for PCR trying to obtain the longer fragments, but failed probably because of distance between the duplicated genes (2,147 bp) is too long. We designed two new pairs of primers targeting the block between the duplicated rrnS genes, with one primer of each pair located in the rrnL gene that was supposed to be absent according to Yu et al. (Table 4, Fig, 1). The two new primer pairs designed by us successfully amplified and produced fragments of expected sizes, 2,658 and 1,905 bp (Table 4, Figure 2), proving that the gene block between the duplicated rrnS genes are actually there. To further confirm that the two products both contain the duplicated rrnS, each product was used as PCR template for amplification with the primers 2* that amplifies rrnS only; both PCR produced a fragment of the expected size (824 bp), the same as using genomic DNA as template (Figure 2). We also sequenced some of the fragments, and the sequences are the same as expected from the mt sequences we obtained. These results clearly demonstrate that the duplicated rrnS and the split rrnL exist in the mt genome of C. hongkongensis. There is no loss of the duplicated genes and the gene block between them. "Tandem duplication-random loss" is not a real feature of oyster mt genomes and has not occurred during the evolution of C. hongkongensis. The possibility of Yu et al. sequenced a rare mutant of C. hongkongensis is extremely low considering: 1) we sequenced three individuals from three diverse populations; 2) Yu and colleagues screened more than one individual; and 3) we duplicated their results with our samples. This is a clear case of PCR artifacts involving duplicated genes.
Figure 2.
PCR products amplified with different primers and separated on agarose gel electrophoresis. P1 - P3 are the products amplified using the primer pairs 1* – 3* and P4, P5 are the products amplified with the primers 4, 5 with genomic DNA template; while P6, P7 are the products amplified using the primers 2* with P4 and P5 as templates, respectively. M1: 1 Kb DNA Ladder marker 10000, 8000, 7000, 6000, 5000, 4000, 3000, 2000 and 1000 bp; M2: D2000 marker 2000, 1 000, 750, 500, 250 and 100 bp.
Conclusion
In conclusion, the complete mt genome of C. hongkongensis is 18,622 bp in length, and its gene order and arrangement are identical to that of C. gigas. The loss of a gene segment reported by Yu et al. (2008) was an artifact due to placing PCR primers in a duplicated gene, and the phenomenon of "tandem duplication-random loss" does not exist in the mt genome of C. hongkongensis. Our study highlights the need for caution when amplifying and sequencing through regions with tandem duplication. When tandem duplication is expected, it is important to design long PCR fragments and not place primers in duplicated regions. Cross-verification with different sets of primers should be considered.
Authors' contributions
JR did PCR, sequencing and initial analysis; XG and XL provided the samples; BL, XG, LX and GZ conceived the study; JR, BL and XG did most of the writing. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We acknowledge that Drs. Patrick Gaffney and Coren Milbury independently identified the potential problem in the published mt genome sequence and communicated with us during manuscript preparation. This work is supported by the MFG Fund of Chinese Academy of Sciences. Sample collection is supported by grants from US NOAA CBO (NA04NMF4570424) and NSFC (08201101B, 39825121 and 40406032).
Contributor Information
Jianfeng Ren, Email: rjf98yy@yahoo.com.cn.
Xiao Liu, Email: liuxiao@ms.qdio.ac.cn.
Guofan Zhang, Email: gfzhang@ms.qdio.ac.cn.
Bin Liu, Email: bliu@ms.qdio.ac.cn.
Ximing Guo, Email: xguo@hsrl.rutgers.edu.
References
- Yu ZN, Wei ZP, Kong XY, Shi W. Complete mitochondrial DNA sequence of oyster Crassostrea hongkongensis -a case of "Tandem duplication-random loss" for genome rearrangement in Crassostrea? BMC Genomics. 2008;9:477. doi: 10.1186/1471-2164-9-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbury CA, Gaffney PM. Complete mitochondrial DNA sequence of the eastern oyster Crassostrea virginica. Mar Biotechnol. 2005;7:697–712. doi: 10.1007/s10126-005-0004-0. [DOI] [PubMed] [Google Scholar]
- Wang H, Guo X. Identification of Crassostrea ariakensis and related oysters by multiplex species-specific PCR. J Shellfish Res. 2008;27:481–487. doi: 10.2983/0730-8000(2008)27[481:IOCAAR]2.0.CO;2. [DOI] [Google Scholar]
- Tatusova TA, Madden TL. Blast 2 sequences – a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]