Abstract
The muscle specific ubiquitin E3 ligase MuRF1 has been implicated as a key regulator of muscle atrophy under a variety of conditions, such as during synthetic glucocorticoid treatment. FOXO class transcription factors have been proposed as important regulators of MuRF1 expression, but its regulation by glucocorticoids is not well understood. The MuRF1 promoter contains a near-perfect palindromic glucocorticoid response element (GRE) 200 base pairs upstream of the transcription start site. The GRE is highly conserved in the mouse, rat, and human genes along with a directly adjacent FOXO binding element (FBE). Transient transfection assays in HepG2 cells and C2C12 myotubes demonstrate that the MuRF1 promoter is responsive to both the dexamethasone (DEX)-activated glucocorticoid receptor (GR) and FOXO1, whereas coexpression of GR and FOXO1 leads to a dramatic synergistic increase in reporter gene activity. Mutation of either the GRE or the FBE significantly impairs activation of the MuRF1 promoter. Consistent with these findings, DEX-induced upregulation of MuRF1 is significantly attenuated in mice expressing a homodimerization-deficient GR despite no effect on the degree of muscle loss in these mice vs. their wild-type counterparts. Finally, chromatin immunoprecipitation analysis reveals that both GR and FOXO1 bind to the endogenous MuRF1 promoter in C2C12 myotubes, and IGF-I inhibition of DEX-induced MuRF1 expression correlates with the loss of FOXO1 binding. These findings present new insights into the role of the GR and FOXO family of transcription factors in the transcriptional regulation of the MuRF1 gene, a direct target of the GR in skeletal muscle.
Keywords: forkhead transcription factor class O, muscle RING finger 1, glucocorticoid receptor
skeletal muscle is a dynamic tissue that has the capacity to continuously regulate its size in response to a variety of external cues, including mechanical load, neural activity, hormones/growth factors, stress, and nutritional status. In addition, skeletal muscle serves as the most significant repository for protein in the body, a source that is tapped to provide a pool of amino acids for tissue repair and gluconeogenesis under conditions of starvation and other metabolic stresses. Muscle loss or “atrophy” occurs as the result of a number of disparate conditions, including aging, immobilization, metabolic diseases, cancer, and neurodegenerative diseases, and as a serious side effect of therapeutic corticosteroid hormone treatment (15, 27, 32). The recently identified E3 ubiquitin ligase, muscle RING finger 1 (MuRF1), is proposed to be a key regulator of the atrophy process given that 1) it is expressed predominantly in skeletal muscle (3), 2) it is upregulated under a variety of atrophy conditions (3, 12, 25), and 3) deletion of the gene in mice results in significant muscle sparing following denervation (3). Although the full physiological functions of MuRF1 are not yet known, it is often assumed that it functions in some manner to regulate protein degradation since it is expressed early in the atrophy process and its peak expression usually occurs during maximum muscle loss. For example, MuRF1 has been shown to play a direct role in myosin heavy chain ubiquitination and degradation during synthetic glucocorticoid treatment (5). However, MuRF1 may have additional important functions in skeletal muscle, such as inhibition of protein synthesis during starvation conditions (22) as well as regulating carbohydrate metabolism (16). Despite considerable interest in MuRF1 as a regulator of skeletal muscle mass and metabolism, there are limited data on the transcriptional regulation of the MuRF1 gene.
Both natural and synthetic glucocorticoid hormones are potent inducers of skeletal muscle atrophy (14). Glucocorticoids exert their physiological actions primarily via a nuclear pathway to directly affect target gene transcription. Natural glucocorticoids such as cortisol and corticosterone, as well as synthetic glucocorticoids such as dexamethasone (DEX) and prednisolone, exert their biological effects predominantly via the glucocorticoid receptor (GR) (56). The GR is a member of the nuclear receptor superfamily and acts as a ligand-dependent transcription factor. Skeletal muscle expresses significant levels of GR, as do cultured myotubes. In the absence of ligand, the GR is found largely in the cytoplasm in a large complex that includes chaperones such as heat shock protein 90. Upon ligand binding, the GR becomes localized in the cell nucleus and binds to DNA sequences called glucocorticoid response elements (GREs). The consensus GRE sequence is AGAACANNNTGTTCT, where two GRs bind to each six-nucleotide half-site of the palindome as a homodimeric complex (2). The three-base pair (bp) spacer sequence (NNN) can consist of any nucleotide combination, although that particular sequence length is critical. However, the perfect consensus sequence is rarely found in native glucocorticoid-responsive promoters, with two or more nucleotide differences often found in either or both half-sites (45). In addition, GR may positively or negatively influence transcription of target genes in a GRE- and homodimer-independent manner via interaction with promoter-bound STAT5, AP-1, or NF-κB transcription factors (reviewed in Ref. 50). Development of a mouse strain expressing a dimerization-deficient GR [GR homodimerization mutuant (GRdim)] has allowed the identification of physiological processes and specific gene expression that are dependent on the classical GRE binding activity of the GR vs. an indirect action through other transcription factors (38, 39).
Although the synthetic glucocorticoid DEX induces MuRF1 mRNA accumulation in vivo as well as in cultured myotubes (3, 48), it is currently unknown whether the gene is directly regulated by ligand-bound GR in vivo. DEX induction could be mediated by the increased expression of other transcription factors and associated coactivators that in turn bind to and activate the MuRF1 promoter. For example, the class O type forkhead transcription factors (FOXO), including FOXO1, FOXO3a, and FOXO4, have recently been implicated as key regulators of gene expression during skeletal muscle atrophy (42, 48). FOXO1, FOXO3a, and FOXO4 are all expressed in skeletal muscle (17), and FOXO1 and FOXO3a mRNA in particular are upregulated during fasting and DEX treatment (9, 25). Constitutively active FOXO proteins can activate the endogenous MuRF1 gene (42, 48); however, there is no information on the direct effect of FOXO proteins on the MuRF1 promoter.
Recently, it has been demonstrated that upregulation of MuRF1 expression following DEX treatment or starvation of C2C12 cells can be suppressed by IGF-I (41, 42, 48). The mechanism by which IGF-I is able to suppress MuRF1 transcription is believed to be at least in part via the phosphatidylinositol 3-kinase/Akt pathway. Akt phosphorylates members of the FOXO class of forkhead transcription factors (40, 60), and phosphorylated FOXOs are sequestered in the cytoplasm by 14-3-3 proteins, thereby inhibiting transcription of FOXO target genes. In addition, FOXO proteins lacking Akt phosphorylation sites prevent IGF-I inhibition of DEX induction of MuRF1 (48). Nevertheless, significant questions remain as to whether nuclear FOXO transcription factors alone are sufficient to activate transcription of the MuRF1 gene. In the present study, we provide a detailed analysis of the regulatory elements governing MuRF1 induction following glucocorticoid treatment, with particular attention to the role of the GR and specific FOXO transcription factors.
MATERIALS AND METHODS
Cell culture.
HepG2 and C2C12 cells were cultured in DMEM supplemented with 10% FBS, nonessential amino acids, and antibiotics and grown at 37°C in 5% CO2. C2C12 cells were cultured in DMEM, supplemented with 10% FBS, nonessential amino acids, and antibiotics, and grown at 37°C in 5% CO2. C2C12 myoblasts were differentiated to myotubes by switching confluent cells (usually 24–48 h postsplit) to DMEM supplemented with 2% charcoal dextran-treated FBS, nonessential amino acids, and antibiotics and grown for an additional 48–96 h at 37°C in 5% CO2. All cell culture reagents except charcoal dextran-treated FBS (Hyclone) were from Invitrogen.
Plasmids.
The pSG5-GR construct was a kind gift from Dr. Stoney Simons, the pcDNA3-FoxO1 construct was provided by Dr. Masahiko Negishi, pCMV5-cMyc-FoxO3a was provided by Dr. Dominic Accili, and pcDNA3-FoxO4 was provided by Dr. Karen Arden. The pcDNA3.1-GRwt construct contains the full-length mouse GR cDNA, and the pcDNA3.1-GRdim construct was derived from pcDNA3.1-GRwt by overlap PCR introducing the A458T point mutation and a novel BsrGI restriction site, similar to the strategy used to create the same point mutation in the GRdim mouse targeting vector described previously (38). The mouse MuRF1 proximal promoter region was obtained by PCR from bacteria artificial chromosome (BAC) clones RP23-40E12 (Children's Hospital Oakland Research Institute). BAC DNA was isolated from bacterial cultures using the BACMAX DNA purification kit from Epicentre according to the manufacturer's protocol. Primers were designed to amplify approximately −5,000, −2,000, −1,000, and −500 bp from near the transcription start site. The primer sequences used to generate the MuRF1 promoter fragments are MuRF1-Pro5000-F 5′-GGA CAG TGC ATC ATG ACC CAG-3′, MuRF1-Pro2000-F 5′-CCA GAA CTA CAC CAG AAA CTC-3′, MuRF1-Pro1000-F 5′-GGA GCT GGG AAT ATA GAC TTG-3′, MuRF1-Pro500-F 5′-CCT TAG AGC TGT TCA GAA TCC AG-3′, and MuRF-Pro-R 5′-CAC TCG GAT CCT CTT TGT CTT C-3′. PCR was performed using TaqPlus Long (Strategene). The resulting PCR products were then subcloned into the pGEMT-EZ vector (Promega) and sequenced to confirm that the correct amplicon had been obtained. The promoter fragments were then digested out of the pGEMT-EZ vector with EcoRI, blunted, and cloned into the SmaI site of pGL3-Basic vector (Promega), resulting in fusion with the firefly luciferase reporter gene. The recombinant plasmids were then subjected to restriction digest analysis and sequenced to confirm correct orientation. MuRF1 promoter fragments, fused to the secreted alkaline phosphatase (SEAP) reporter gene, were constructed by digesting the pGEMT-EZ recombinant plasmids with EcoRI and subcloning the MuRF1 fragments into the EcoRI site of the pSEAP-Basic vector. These constructs were then subjected to restriction digestion analysis and sequenced to confirm correct orientation. Oligonucleotides corresponding to the predicted FOXO binding site [FOXO binding element (FBE)] and the GRE in the MuRF1 promoter, as well as a string of six diaminofluorescein (Daf-16) binding elements (6X-DBE), were designed and ordered from Invitrogen. The oligo sequences are MuRF1-FBE-F 5′-CTA GTT CTT GTT TAC GAC C-3′, MuRF1-FBE-R 5′-CTA GGG TCG TAA ACA AGA A-3′, MuRF1-GRE-F 5′-CTA GGC TCT GAA CAG TCT GTT CTT GTT-3′, MuRF1-GRE-R 5′-CTA GAA CAA GAA CAG ACT GTT CAG AGC-3′, 6X-DBE-F 5′-CTA GAA GTA AAC AAC TAT GTA AAC AAC TAT AAG TAA ACA ACT ATG TAA ACA ACT ATA AGT AAA CAA CTA TGT AAA CAA GAT C-3′, and 6X-DBE-R 5′-CTA GGA TCT TGT TTA CAT AGT TGT TTA CTT ATA GTT GTT TAC ATA GTT GTT TAC TTA TAG TTG TTT ACA TAG TTG TTT ACT T-3′. The complementary oligo sequences were annealed by mixing and heating to 95°C and slowly cooling to 25°C over a 35-min period using a thermocycler. The annealed oligos were then ligated into the SpeI site of the thymidine kinase (TK)-Luc vector (kindly provided by Dr. Ronald Evans). The resulting recombinant plasmids were then restriction digested to confirm the presence of an insert and sequenced to determine the number and orientation of the concatemerized oligos. Site-directed mutagenesis of the FBE and GRE in the pGL3- and pSEAP-MuRF1-Pro500 constructs was performed essentially as described in the site-directed mutagenesis kit protocol from Strategene. The primers used to mutate the FBE and GRE are: MuRF1Pro-GRE-Mut-F 5′-CCT GGC TCT GGT CAG TCT GAC CTT GTT TAC G-3′, MuRF1Pro-GRE-Mut-R 5′-CGT AAA CAA GGT CAG ACT GAC CAG AGC CAG G-3′, MuRF1Pro-FBE-Mut-F 5′-CTG TTC TTG GTG ACG ACC CCC-3′, and MuRF1Pro-FBE-Mut-R 5′-GGG GGT CGT CAC CAA GAA CAG-3′. The resulting clones were sequenced to confirm that the correct mutation had been obtained. The mutated nucleotides are underlined.
Cell culture reporter gene and Northern blot assays.
One hundred twenty-five to 150 × 103 HepG2 cells/well were plated into 12-well plates and cultured for 24 h or until an approximate confluency of 30–40% was reached. Using FuGene 6 (Roche), 1 μg of total DNA/well was transiently transfected [including 0.250 μg/well of the indicated reporter construct, 0.070 μg/well of SV40-Renilla luciferase (Promega), 0.250 μg/well GR expression vector (i.e., pSG5-GR, pcDNA3.1-GRwt, or pcDNA3.1- GRdim), 0.125 μg/well FOXO expression vector, i.e., pcDNA3-FoxO1, pCMV5-cMyc-FoxO3a, or pcDNA3-FOXO4, and pBluescript as filler DNA] for 12–16 h. Cells were then treated for 18–24 h with either vehicle, 1 μM DEX (from a 1-mM stock in ethanol; Sigma), 20 ng/ml IGF-I (Sigma), or both as indicated. Dual-luciferase reporter assays were performed in HepG2 cells essentially as described previously (7). Briefly, transfected cells were lysed with 1X passive lysis buffer (Promega), scraped, transferred to microfuge tubes, and centrifuged at high speed for 5 min to clear cellular debris, and then 25 μl of supernatant from each tube (corresponding to each well) was transferred to a 96-well plate. Both working firefly buffer (25 mM glycylglycine, pH 8.0, 5 mM K2HPO4, 4 mM EGTA, 15 mM MgSO4, 4 mM ATP, 1.25 mM DTT, 0.1 mM CoA, 80 μM d-luciferin) and working Renilla buffer (1.1 M NaCl, 2.2 mM Na2EDTA, 0.22 M K2HPO4, pH 5.1, 0.5 mg/ml BSA, 1.5 mM NaN3, and 1.5 μM coelenterazine) were prepared fresh prior to each assay. d-luciferin was purchased from ICN. Five milligrams of luciferin was dissolved in 18 ml of 25 mM glycylglycine (pH 8.0), aliquoted, and stored at −80°C. Coelenterazine (Promega) was dissolved in EtOH to a final concentration of 3 mM. All other chemicals were purchased from Sigma. Following treatments, cells were lysed and supernatants analyzed for luciferase activity using a Beckman Coulter LD 400 luminometer programmed to dispense 100 μl of working firefly buffer with a 1-s delay and a 10-s integration, followed by injection of 100 μl of working Renilla buffer with a 2-s delay and a 10-s integration. Firefly luciferase activities were normalized to Renilla luciferase activity to correct for variations in transfection efficiency.
For C2C12 transfection experiments, 125–150 × 103 cells/well were plated into 12-well plates and cultured as described above for 24 h. Using FuGene 6 (Roche), 1 μg of total DNA/well was transiently transfected (including 0.250 μg/well of the indicated reporter construct, 0.125 μg/well of a TK-β-galactosidase construct, and pBluescript as filler DNA) for 12–16 h. Cells were then switched to DMEM containing 2% charcoal/dextran-treated FBS. Twenty-four hours following the switch, the cells were treated with vehicle and either 1 μM DEX (from a 1-mM stock in ethanol; Sigma), 20 ng/ml IGF-I (Sigma), or both as indicated. Medium from each well of the transfected differentiated C2C12 myotubes was sampled immediately prior to ligand treatment and then every 24 h following ligand treatment for 3 days. Fresh ligand was added to the differentiated C2C12 myotubes every 24 h. Ten to 15 microliters of each collected medium was then processed and analyzed using the Great EscAPe SEAP Detection Kit (BD Biosciences), following the instructions of the manufacturer. At the end of the experiment, the C2C12 myotubes were lysed and β-galactosidase activities determined and used to correct for variations in SEAP activities resulting from variations in transfection efficiencies.
For C2C12 Northern blot experiments, cells were differentiated and treated as above, and total RNA was isolated using using RNeasy columns (Qiagen) according to the manufacturer's instructions. Northern analysis was conducted using random hexamer-labeled MuRF1 and rpL8 cDNAs essentially as described previously (8).
Animal studies.
The generation of GRdim mice was described previously (38). The mice were backcrossed to Balb/c mice for more than 20 generations. Homozygous mutants and wild-type controls were obtained by intercrossing heterozygous GRdim mice. Animals were housed individually in ventilated cages under specific pathogen-free conditions with ad libitum access to food and water and a normal dark-light cycle. For the 6- and 24-h DEX treatments, 10 mg/kg water-soluble DEX (Sigma) was given by intraperitoneal injection, and 50 mg/l DEX was subsequently provided in the drinking water. For the 3- and 8-day DEX treatments, 50 mg/l water-soluble DEX was provided in the drinking water and replenished every 2nd day. At each time point, animals were weighed and euthanized, and spleens as well as tibialis anterior and gastrocnemius muscles were dissected and weighed. Muscles were frozen in liquid nitrogen until further processing. Splenocytes were isolated by passing the freshly isolated spleen through a 40-μm nylon mesh, washed in PBS, and subsequently treated with OptiLyse (Beckman Coulter) to remove the erythrocytes. Total splenocyte numbers were determined by counting the cells under the microscope with a Neubauer chamber. Total RNA from treated wild-type and GRdim gastrocnemius muscle was isolated and analyzed by Northern hybridization using MuRF1, FOXO1 and FOXO3a, and rpl32 cDNA probes as described above. All animal experimentation was conducted in accordance with standards of human animal care and approved by the Lower Saxony state authorities (Niedersächisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Braunschweig, Germany).
Chromatin immunoprecipitation.
Chromatin immunoprecipitations (ChIP) were performed as modified by Metivier et al. (30) and Nelson et al. (35). Chemicals were obtained from Sigma unless stated otherwise. C2C12 cells were grown to confluence in 100-mm tissue culture plates in DMEM supplemented with 10% FBS. Cells were then shifted to 2% FBS and allowed to differentiate into myotubes for 4 days. One plate per time point for the 1-μM DEX time course treatment or four sets of plates in quadruplicate were treated with 1 μM DEX, 20 ng/ml R3-IGF-I, or a combination of 1 μM DEX and 20 ng/ml R3-IGF-I for 15 or 30 min. Untreated cells were used as a control. At the indicated times, medium was removed and cells were incubated with 1% formaldehyde in PBS for 10 min at 37°C. Formaldehyde cross-linking was quenched by adding glycine to a final concentration of 125 mM and incubating for an additional 10 min at room temperature. Cells were washed twice in PBS, scraped in 10 mM Tris·HCl, pH 8.0, 150 mM NaCl, and 1 mM EDTA, and collected by centrifugation at 3,000 g for 5 min. Cell pellets were resuspended in 400 μl of 1% SDS, 10 mM EDTA, 50 mM Tris·HCl, pH 8.0, 10 mM β-glycerophosphate, 1 mM sodium orthovanadate, and protease inhibitor cocktail (Roche). Cells were sonicated twice for 30 s each on setting “3” and “pulsed” using a Branson microtip sonicator. The resulting lysates were then centrifuged at full speed in an Eppendorf microfuge for 10 min. Supernatants were diluted to 10 ml with ChIP dilution solution (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris·HCl, pH 8.0, 167 mM NaCl, protease inhibitor cocktail, 10 mM β-glycerophosphate, and 1 mM sodium vanadate). The indicated antibody [anti-HA tag IgG HA probe (Y-11), anti-FOXO1 IgG (H-128), and anti-glucocorticoid receptor (M-20) were obtained from Santa Cruz Biotechnology] was added to the diluted chromatin at a concentration of 1 μg/ml, and samples were incubated overnight at 4°C with tumbling. Fifty microliters of protein A (50% slurry) preabsorbed with sheared salmon sperm DNA was added and incubated with tumbling at 4°C for 2 h. Immunoprecipitations were transferred to Bio-Rad minicolumns (prerinsed with 500 μl of ChIP dilution solution), with an additional 500-μl dilution solution to rinse the tubes and ensure complete transfer of beads to the minicolumns. Columns were then washed twice each with 1 ml TSEI (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris·HCl, pH 8.0, 150 mM NaCl), TSEII (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris·HCl, pH 8.0, 500 mM NaCl), and TSEIII (1 mM EDTA, 10 mM Tris·HCl, pH 8.0, 1% NP-40, 1% sodium deoxycholate, 0.25 M LiCl) followed by three washes with Wash Buffer IV [10 mM Tris (pH 8.0), 1 mM EDTA]. Washed beads were transferred to new tubes with 400 μl of Wash Buffer IV, adding an additional 500 μl Wash Buffer IV to the columns to ensure that all beads were transferred. The beads were then pelleted by centrifugation at 3,000 g for 2 min. After the supernatant was removed by careful aspiration, 100 μl of 10% Chelex-100 (Bio-Rad) was added to the beads, followed by vortexing and incubation at 95°C for 10 min to reverse the cross-linking. Two microliters of 20 U/ml proteinase K solution (Invitrogen) was added, followed by vortexing and incubation at 55°C for 30 min and heat inactivation at 95°C for 10 min. After centrifugation at full speed in an Eppendorf microfuge for 2 min, the supernatant was transferred to a new tube and combined with a second extraction of the Chelex beads with 100 μl of water. Input (nonimmunoprecipitated chromatin) was prepared by adding 100 μl of 10% Chelex directly to 100 μl of the diluted chromatin extract and processed as above. Quantitative PCR of input and immunoprecipitated chromatin samples was performed using 2 μl of DNA as template and PerkinElmer 2X SYBR Green master mix on an ABI 7700. Cycling conditions were one cycle at 95°C for 10 min, followed by 40 cycles at 95°C for 10 s and 60°C for 60 s. Primers flanking the putative mouse MuRF1 GRE/FBE region were forward 5′-TATCTGGCTCTCCCCTGAAC-3′ and reverse 5′-CCTCAAAGATTTGGCCCTCT-3′. Values for each time point and hormone treatment were normalized to input values. For agarose gel analysis of PCR products, reactions were stopped at 30 cycles, and samples from each treatment group were pooled and run on a 2% agarose gel containing ethidium bromide.
RESULTS
The MuRF1 promoter contains a conserved near-perfect palindromic glucocorticoid response element.
Since MuRF1 is known to be induced by synthetic glucocorticoids, as well as in several catabolic conditions associated with elevated endogenous glucocorticoids, we sought to identify the key glucocorticoid-responsive elements in the gene. We began by isolating BAC clones encompassing the entire mouse MuRF1 transcription unit and amplifying 5,000 bp upstream of the transcription start site of the gene. Human, rat, and mouse promoter sequences were aligned to detect conserved sequences that may be functionally relevant for MuRF1 expression. The MuRF1 (Fig. 1A) promoter shows a high degree of homology across species for about 400 bp upstream of the transcription start site, and there are additional sporadic pockets of homology far upstream of the proximal promoter, 3′ of the transcription unit, and within some introns (not shown). We searched for potential transcription factor binding sites by scanning the promoters for published and well-established consensus sequences. Previous reports identified and characterized consensus Class O forkhead (FOXO) binding sites in the promoter of the muscle atrophy-related muscle atrophy F-box gene (42), and we found multiple potential AT-rich FBEs in the MuRF1 promoter as well, including one overlapping the consensus TATA box (Fig. 1A). Importantly, the MuRF1 promoter has a near-perfect consensus GRE ∼200 bp upstream of the transcription start site, supporting our hypothesis that this gene could be a direct target of the ligand-activated GR. The potential GREs from the human, mouse, and rat MuRF1 genes differ only by a single nucleotide in the 3-bp spacer region, whose exact sequence is not critical for GR binding (Fig. 1B). Interestingly, a consensus FBE was found directly adjacent to the putative MuRF1 GRE, raising the possibility that FOXO and the GR function together to regulate the MuRF1 promoter. In addition to FOXO and GR binding sites, we detected a cluster of potential NF-κB response elements in the MuRF1 promoter, consistent with the upregulation of this promoter by proinflammatory cytokines and upon activation of NF-κB (4). Several C/EBP sites were also detected in the MuRF1 promoter, which is of interest because C/EBPβ and -δ are upregulated by DEX in skeletal muscle cells (57, 58). Finally, numerous conserved consensus muscle-specific E-box sites were found in the promoter, as expected given its highly selective cardiac and skeletal muscle expression pattern.
Fig. 1.
Schematic of the muscle RING finger 1 (MuRF1) promoter and sequence alignment of the proximal regulatory regions. A: promoter sequences from mouse, rat, and human MuRF1 [5,000 base pairs upstream of the transcription start site (+1) through the first exon] were downloaded from the Ensembl database (www.ensembl.org) and aligned using the ClustalW algorithm. Approximate positions of potential transcription factor binding sites are indicated in the schematics of the MuRF1 promoter at top and/or boxed in the alignment at bottom. The forkhead transcription factor class O (FOXO) forkhead binding site (G/A)TAAA(T/C)AA (black ovals), glucocorticoid response element (GRE) (A/T)GAACANNNTGTTC(A/T) (hatched rectangle), CCAAT/enhancer-binding protein (C/EBP) TT(G/T)NGNAA (◊), NF-κB consensus binding sequence GGG(G/A)N(C/T)(C/T)(C/T)CC (gray hexagons), and muscle-specific E-box CANGTG (MyoD, etc.); N represents any nucleotide. The sequence alignments for ∼400 base pairs upstream and 26 base pairs downstream of the rat, mouse, and human MuRF1 promoters are shown below the corresponding promoter schematics. Identical sequences for the indicated regions are highlighted in black. Arrow indicates transcription start site. B: comparison of a consensus GRE and perfect palindrome GRE and putative GREs from the mouse (mMuRF1), rat (rMuRF1), and human MuRF1 (hMuRF1) promoters. Arrows indicate half-sites.
The MuRF1 promoter is a direct target of activated GRs.
Since the mere presence of consensus transcription factor binding sites does not guarantee their functional importance, MuRF1 promoter fragments, or the predicted GRE and FBE in a minimal heterologous promoter, were placed in front of the firefly luciferase reporter and transiently transfected into HepG2 cells along with expression vectors for the murine GR and/or FOXO family members. For these initial studies, HepG2 cells were chosen for their ease of transfection and their previous use in transfection studies of gluconeogenic gene promoters that are also regulated by both GR and FOXO family members (23, 24, 52). Consistent with the presence of the consensus GRE within 500 bp of the transcription start site, DEX was able to induce all MuRF1 promoter constructs containing between 500 and 5,000 bp (Fig. 2A). Indeed, the isolated MuRF1 GRE is sufficient to support potent DEX-induced transcription either alone (14-fold) or as a multimer (19- to 45-fold) (Fig. 2B).
Fig. 2.
Dexamethasone (DEX)-activated glucocorticoid receptor (GR) induces the MuRF1 proximal promoter. A: HepG2 cells were transfected with luciferase reporter constructs (Luc) containing varying lengths of the MuRF1 (pGL3-MuRF1) promoter, an SV40-Renilla luciferase reporter construct, and a mouse GR expression vector (pSG5-GR). Cells were treated with or without 1 μM DEX for 24 h. Luciferase activities in cell extracts were normalized to Renilla luciferase activity to control for transfection efficiency. Numbers on the y-axis indicate distance from the transcription start site fused to the reporter pGL3-Basic, a promoterless LUC vector. B: the isolated MuRF1 GRE supports DEX-mediated transcriptional activation of a heterologous promoter. The DEX regulation of the thymidine kinase (TK) promoter was tested with 0 (control), 1 (1XGRE), 3 (3XGRE), and 4 (4XGRE) inserted GREs. The arrows below the indicated GRE constructs labeled on the y-axis depict the orientation of each individual GRE oligonucleotide. In A and B, fold induction was obtained by dividing values from DEX-treated samples by the mean of values from matched untreated samples. Each condition was done in triplicate, and error bars reflect SD.
FOXO transcription factors differentially activate the MuRF1 promoter.
Next, we tested whether the native MuRF1 promoter would respond to cotransfected FOXO1, FOXO3a, or FOXO4 expression vectors, since these proteins are candidates for intermediate DEX-inducible regulatory factors. As shown in Fig. 3A, FOXO factors only marginally induce the proximal MuRF1 promoter (2- to 3-fold). As with the GRE, we tested whether the isolated FOXO binding site (FBE) would respond to cotransfected FOXO1, FOXO3a, or FOXO4 expression vectors; in this context, only FOXO3a and FOXO4 induced transcription of the reporter containing a single FBE (Fig. 3B). However, FOXO1 is able to activate multimerized DBE or the MuRF1 FBE, suggesting that this FOXO family member is most active as a multimeric complex rather than a monomer (Fig. 3C).
Fig. 3.
FOXO transcription factor induction of the MuRF1 promoter. A: HepG2 cells were transfected with 500-base pair MuRF1-promoter-LUC constructs, SV40-Renilla luciferase reporter construct, and expression vectors for murine FoxO1, FoxO3A, or FoxO4 (pcDNA3-FoxO1, pcDNA3-FoxO3A, or pcDNA3-FoxO4) or pcDNA3 alone (dash). Luciferase activities in cell extracts were normalized to Renilla luciferase activity to control for variations in transfection efficiency. Each point was done in triplicate; error bars reflect SD. B: the isolated MuRF1 FOXO binding element (FBE) supports FOXO3a- and FOXO4- but not FOXO1-mediated transcriptional activation of a heterologous promoter. FOXO regulation of the TK promoter was tested by inserting a single FBE and cotransfecting HepG2 cells with the control pcDNA3 vector alone (dash) or each FOXO expression vector as in A. C: concatamerized FOXO binding elements from the MuRF1 promoter (FBE-4X) and Daf-16 binding elements (DBE-6X) support transcriptional activation of the TK promoter by FOXO1. Open bars, normalized luciferase values from the cells transfected with the TK-Luc-4XFBE and TK-Luc-6XDBE in the absence of exogenous FOXO1 expression; black bars, normalized luciferase values from the TK-Luc-4XFBE- and TK-Luc-6XDBE-transfected cells in the presence of exogenous FOXO1 expression. Data were processed as in A and B.
The MuRF1 promoter is synergistically activated by FOXO1 and GR.
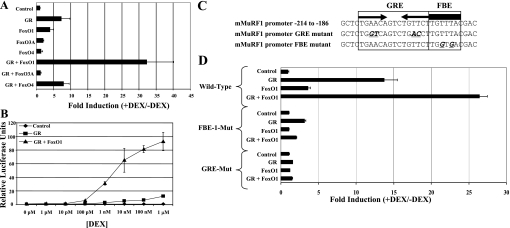
Remarkably, however, FOXO1 showed strong synergistic activation with the GR on the 500-bp MuRF1 promoter (Fig. 4A). This potent synergy was consistently observed (20- to 40-fold) over multiple experiments and on all promoter fragments tested ≤5,000 bp from the transcription start site. FOXO3a and FOXO4 did not synergize with the GR; indeed, expression of FOXO3a somewhat inhibits GR activation of the MuRF1 promoter. The GR-FOXO1 synergy is also reflected in dose-response experiments, where this combination supports strong activation of the MuRF1 promoter with as little as 1 nM DEX in the culture medium (Fig. 4B). Both the GRE and the adjacent FBE play an important role in the observed GR-FOXO synergy. DEX induction of the 500-bp MuRF1 promoter is nearly abolished when the GRE is mutated and is completely abolished when the GRE is intact but the adjacent FOXO site is mutated (Fig. 4, C and D).
Fig. 4.
DEX-activated GR synergizes with FOXO1 to potently induce the MuRF1 promoter. A: HepG2 cells were transfected with the pGL3-MuRF1 promoter (−500) reporter construct, SV40-Renilla luciferase reporter construct, pSG5-GR, and/or pcDNA3-FOXO1, pcDNA3-FoxO3A, or pcDNA3-FOXO4. Cells were treated with or without 1 μM DEX for 24 h. Firefly luciferase activities in cell extracts were normalized to Renilla luciferase activity to control for variations in transfection efficiency. Fold induction was obtained by dividing values from DEX-treated samples by the mean of values from matched untreated samples. Each point was done in triplicate, and errors reflect SD. B: HepG2 cells were transfected with pGL3-MuRF1 promoter (−500), SV40-Renilla, and pBluescript as filler DNA (control) and either GR (GR) or GR and FoxO1 (GR + FoxO1) expression vectors. Twenty-four hours posttransfection, cells were treated with the indicated DEX concentration and incubated overnight. Each point was done in triplicate. C and D: mutation of either the GRE or the FBE is sufficient to abolish DEX-induced MuRF1 promoter activity. HepG2 cells were transfected with either the wild-type MuRF1 promoter construct (−500) or MuRF1 promoter constructs that have either the GRE mutated (GRE-Mut) or the FBE mutated (FBE-Mut) as shown in C in combination with expression vectors for FoxO1 and/or GR as indicated. The cells where then treated and assayed for luciferase activity as in A.
DEX induction of MuRF1 expression is inhibited in GRdim mice that express a dimerization-deficient GR.
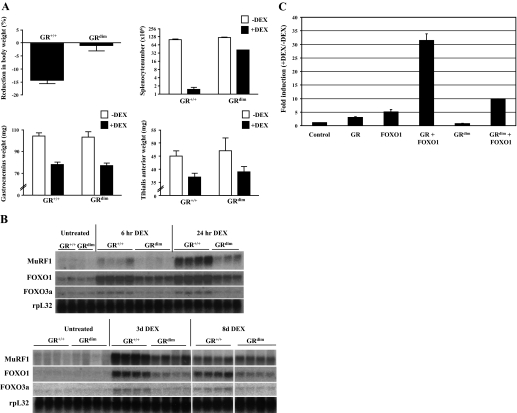
The functional significance of the near-perfect GRE in the MuRF1 promoter was tested in vivo through the use of the GRdim mouse strain that expresses a GR with a point mutation in the DNA binding domain that prevents binding of the GR to classical palindromic GREs (38). This mutant GR still retains the ability to be tethered to target genes via protein-protein interactions such as the NF-κB or AP-1 transcription factor complexes. Both wild-type and GRdim mice were treated with water-soluble DEX either by injection (6- and 24-h time points) or in the drinking water (24-h, 3-day, and 8-day time points). The experiment was terminated at 8 days of treatment due to the severe weight loss observed in the wild-type mice, which was not observed in the GRdim mice (P < 0.001). As shown in Fig. 5A, despite the inhibited overall body weight decline in GRdim vs. wild-type mice in response to DEX, the degree of atrophy induced in the tibilialis anterior and gastrocnemius muscles was not statistically different in the two strains. By contrast, DEX-induced splenocyte apoptosis was strongly inhibited in GRdim mice (P < 0.001).
Fig. 5.
DEX-induced MuRF1 expression is inhibited in mutant GR homodimerization mutant (GRdim) mice vs. wild-type GR+/+ mice. A: GR+/+ and GRdim mice were treated with DEX in their drinking water for 8 days, weighed (top left), and killed for determination of splenocyte number (top right) as well as gastrocnemius (bottom left) and tiblialis anterior (bottom right) weights. Open bars, vehicle treated; black bars, DEX treated. GR+/+, n = 8; GRdim, n = 7. B: Northern analysis of MuRF1, FOXO1, and FOXO3a expression in gasctocnemius muscle from DEX-treated GR+/+ and GRdim mice. B, top: control, 6-h-, and 24-h-treated mice. B, bottom: control, 3-day-, and 8-day-treated mice. Only results from 4 of the 8-day-treated GR+/+ and GRdim mice are presented. rpL32 expression is shown as a loading control below each set of Northerns. Bars represent means ± SE. C: effect of wild-type GR and the homodimerization mutant GR on DEX induction of the MuRF1 promoter in a transient transfection assay. HepG2 cells were transfected with the pGL3-MuRF1 promoter (−500) reporter construct, SV40-Renilla luciferase reporter construct (control), pcDNA3.1-GRwt (GR) or pcDNA3.1-GRdim (GRdim), and/or pcDNA3-FoxO1 (FOXO1). Cells were treated with or without 1 μM DEX for 24 h. Firefly luciferase activities in cell extracts were normalized to Renilla luciferase activity to control for variations in transfection efficiency. Fold induction was obtained by dividing values from DEX-treated samples by the mean of values from matched untreated samples. Each point was done in triplicate, and errors reflect SD.
We next examined MuRF1 expression as well as FOXO1 and FOXO3a expression in GRdim vs. GR+/+ mice (Fig. 5B). MuRF1 expression is clearly upregulated by 6 h after DEX injection and continues to increase ≤3 days with continuous DEX treatment in wild-type mice. MuRF1 induction is strongly inhibited in GRdim mice, which is particularly apparent at the earliest time points. After 8 days of DEX treatment, however, MuRF1 expression declines in wild-type mice until it is essentially equal to its induced expression in GRdim animals. In addition, FOXO1 and FOXO3a are also rapidly induced by DEX in wild-type mice, in agreement with a previous study (9), and expression is reduced in GRdim mice at all time points tested. We then tested the ability of the homodimerization mutant GR expressed in GRdim mice to upregulate the proximal MuRF1 promoter, again using transient transfection assays (Fig. 5C). The homodimerization mutant GR lost the ability to induce the MuRF1 promoter on its own, as expected, and showed a strongly reduced, but not completely abolished, ability to augment FOXO1 induction of the promoter. These transfection results may at least partially explain the residual DEX-induced expression of MuRF1 in GRdim mice.
The GR and FOXO1 bind to the endogenous MuRF1 promoter in C2C12 myotubes.
Given the strong dependence on GR homodimerization in vivo for full MuRF1 gene induction, we next sought to determine the role of the GRE and FOXO1 sites in cultured skeletal myotubes. As we observed in HepG2 cells, the 500-bp MuRF1 promoter is also DEX inducible in differentiated C2C12 mouse skeletal myotubes (Fig. 6A). DEX induction of the promoter is nearly abolished when the GRE is mutated, and the adjacent FOXO site is intact and completely abolished when the GRE is intact but the adjacent FOXO site is mutated (Fig. 6B). Furthermore, addition of 20 ng/ml IGF-I strongly inhibits DEX induction of the endogenous MuRF1 gene, as has been reported previously (Fig. 6C) (41, 48), and IGF-I likewise inhibits DEX induction of the MuRF1 promoter (Fig. 6D). Therefore, the 500-bp proximal promoter of MuRF1 contains the critical elements necessary to respond positively to glucocorticoids (via synergy with a forkhead site) and negatively to IGF-I in a manner identical to the endogenous gene in differentiated myotubes.
Fig. 6.
DEX induces the MuRF1 promoter in differentiated C2C12 myotubes. A: C2C12 myoblasts were transfected with a reporter construct containing 500 base pairs of the MuRF1 promoter fused to the secreted alkaline phosphatase (SEAP) gene. The myoblasts were then differentiated by switching to low-serum medium followed by treatment with 1 μM DEX over a period of 3 days. The medium was sampled every 24 h to measure for SEAP activity. Conditions were done in triplicate and SEAP numbers normalized with β-galactosidase to correct for variations in transfection efficiency. Each point was done in triplicate, and errors reflect SD. B: mutation of either the GRE or the FBE is sufficient to abolish DEX-induced MuRF1 promoter activity in C2C12 cells. C2C12 myoblasts were transfected with either the wild-type MuRF1 promoter construct (−500) or MuRF1 promoter constructs that have either GRE-Mut or FBE-Mut SEAP constructs, differentiated, and treated with 1 μM DEX over a period of 3 days. The medium was sampled every 24 h to measure for SEAP activity as in A. Each time point was done in triplicate and normalized with β-galactosidase to correct for variations in transfection efficiency. C: Northern blot analysis of C2C12 cells differentiated for 48 h and then treated with DEX (10 μM), IGF-I (20 ng/ml), or DEX (10 μM) + IGF-I (20 ng/ml) for 24, 48, and 72 h. Ligand was refreshed every 24 h. D: IGF-I potently inhibits DEX-induced activation of the MuRF1 promoter. C2C12 myotubes were treated for 24 h with either 1 μM DEX, 20 ng/ml IGF-I, or 1 μM DEX and 20 ng/ml IGF-I followed by sampling of the medium for SEAP activity, as described in A. Fold induction was obtained by dividing values from DEX-treated samples by the mean of values from matched untreated samples. Each point was done in triplicate, and errors reflect SD.
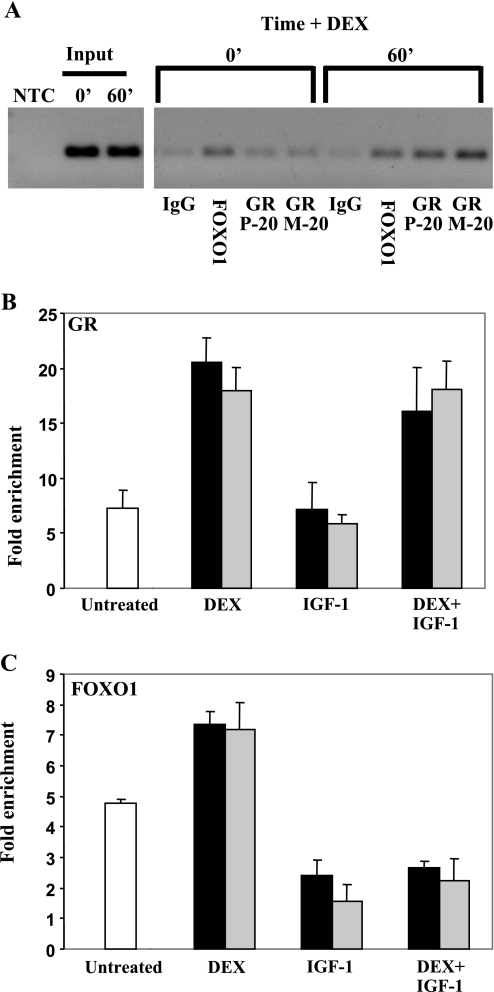
To determine whether FOXO1 and GR can directly occupy the MuRF1 promoter in vivo, we established a modified ChIP protocol suitable for use with differentiated C2C12 myotubes. C2C12 myoblasts were plated on 100-mm dishes and cultured until confluent and then differentiated for 4 days in reduced serum medium. Myotubes were then treated with vehicle, 1 μM DEX, 20 ng/ml IGF-I, or 1 μM DEX plus 20 ng/ml IGF-I for the indicated times. Sonicated chromatin from fixed cells was analyzed with control IgG, an anti-FOXO1 antibody, or two anti-GR antibodies. Both GR and FOXO1 are present on the endogenous MuRF1 promoter in untreated C2C12 myotubes (Fig. 7A), and binding of each factor is enhanced by DEX treatment as soon as 15 min after ligand addition (Fig. 7, B and C), especially the GR. GR binding to the MuRF1 promoter is unaffected by IGF-I (Fig. 7B), although the DEX induction of the endogenous gene or the transfected MuRF1 promoter is strongly inhibited. However, IGF-I addition rapidly induces the loss of FOXO1 from the MuRF1 promoter in the presence or absence of DEX (Fig. 7C). We also attempted to detect FOXO3a binding to the MuRF1 promoter by ChIP as well; only one of two commercially available antibodies we tested (Upstate vs. Santa Cruz) gave a weak positive signal, and the binding pattern was highly similar to FOXO1 (not shown).
Fig. 7.
GR and FOXO1 associate directly with the MuRF1 promoter in C2C12 myotubes. A: C2C12 myotubes were treated with 1 μM DEX for 0 or 60 min (1 10-cm plate/time point), and cross-linked chromatin was immunoprecipitated with normal rabbit IgG, an anti-FOXO1 antibody, or one of two anti-GR antibodies (P20 or M20). After reversal of cross-links, immunoprecipitated MuRF1 promoter fragments were detected by PCR using primers flanking the predicted GRE and FOXO sites in the mouse MuRF1 promoter, followed by agarose gel electrophoresis. PCRs of a no-template control (NTC) and input DNAs are shown at left. B and C: C2C12 myotubes were treated with vehicle (open bars), 1 μM DEX, 20 ng/ml R3-IGF-I, or 1 μM DEX + 20 ng/ml R3-IGF-I for 15 min (black bars) or 30 min (gray bars) before harvest. Four plates for each treatment were processed in parallel as in A. Values (fold enrichment) are expressed as the mean MuRF1 promoter copies immunoprecipitated with either the anti-GR antibody (B) or anti-FOXO1 antibody (C) divided by that immunoprecipitated by a nonspecific antibody (anti-HA), as determined by quantitative PCR using the same primer pairs as in A. Error bars reflect SE.
DISCUSSION
Although it is known that MuRF1 expression increases under numerous atrophy-inducing conditions (11), the mechanism of transcriptional regulation of this gene is poorly understood. The present study provides new insights into how the GR and the FOXO family of transcription factors regulate the transcription of the MuRF1 gene. Specifically, we demonstrate here that 1) the MuRF1 proximal promoter is directly activated by glucocorticoids via a conserved GRE that depends strongly on GR homodimerization for full activation, at least in the earliest time points after DEX treatment, 2) FOXO1 and GR synergistically and specifically induce the MuRF1 proximal promoter, and 3) IGF-I inhibition of DEX-induced MuRF1 expression correlates with the loss of FOXO1 binding to the endogenous MuRF1 promoter. Importantly, these data also reveal that not all FOXO family members equally activate the FOXO binding motif in the MuRF1 promoter.
The presence of an essentially perfect palindromic GRE in the MuRF1 proximal promoter likely explains the strong induction of the gene by exogenous synthetic glucocorticoids (3, 48) and how the gene is induced under a variety of catabolic conditions associated with increased endogenous corticosteroid levels (25, 53). Each half-site of the MuRF1 GRE is perfectly conserved between the human, rat, and mouse genes, with the only difference found in the spacer region that, nevertheless, still retains the three-base pair length necessary for GR homodimer binding (43). It is surprisingly rare to find such a perfect GRE in an endogenous glucocorticoid target gene promoter, although the specific sequences of the divergent half-sites found in most direct GR target genes are strongly conserved across species (45, 46). Most GREs, such as the pair found in the well-studied phosphoenolpyruvate carboxykinase (PEPCK) gene in the liver (18) or the first-described GREs in the mouse mammary tumor virus long-terminal repeat (37), have multiple base pair changes compared with the perfect palindrome consensus GRE that is present in the MuRF1 promoter. Previous studies on the estrogen-regulated family of vitellogenin genes in the frog liver demonstrate that a single perfect estrogen response element supports essentially the same fold induction by estradiol as a pair of imperfect palindromic elements (29). The physiological relevance of having such a well-conserved consensus GRE in the MuRF1 promoter is an interesting and important question.
Analysis of MuRF1 induction by DEX in GRdim mice revealed a critical role for GR homodimerization, which is most evident at the earliest time points examined in these experiments. Interestingly, despite the clear overall reduction in MuRF1 expression, as well as FOXO1 and FOXO3a, the degree of muscle atrophy in GRdim mice after 8 days of DEX treatment was exactly the same in the mutant as in wild-type mice. Consequently, the residual level of MuRF1 expression in GRdim mice may well be enough to support full DEX-induced atrophy. The rapid rise and gradual decline of MuRF1 expression we observed in DEX-treated Balb/c mice is in agreement with experiments in rats dosed with the synthetic glucocorticoid prednisolone (1). Concerning the mechanism of residual MuRF1 induction in GRdim mice, it is conspicuous that cotransfection of FOXO1 with the homodimerization mutant GR supports at least modest induction of the MuRF1 promoter in transient transfection assays, whereas the induction without FOXO1 is completely lost (Fig. 5). This is in line with the previous observation that the GRdim receptor on its own fails to bind to a palindromic GRE. Nevertheless, it is conceivable that FOXO1, when present together, still tethers the homodimerization mutant GR to the composite GRE-FBE motif, since protein-protein interactions with other transcription factors are not compromised by the GRdim mutation. Regardless, it will be important to identify DEX-responsive genes that are less affected than MuRF1 and FOXOs in GRdim mice that may ultimately be more important for the loss of muscle mass in response to DEX. For example, activation of the N-end rule ubiquitin pathway was recently implicated in MyoD degradation in response to DEX in C2C12 cells (49). In addition, transgenic overexpression of MuRF1 was not sufficient to induce atrophy but led to altered levels of enzymes involved in carbohydrate metabolism (16). Thus, consideration of these and other studies, and our present results with GRdim mice, may force a reevaluation of the role of MuRF1 (and possibly even FOXO1 and FOXO3a) in the reduction of muscle mass in catabolic states, although the protein certainly may still play a significant role in structural protein degradation in denervation-induced atrophy. Further analysis of the GRdim model may allow for a genetic dissection of the induction of atrophy vs. metabolic effects of glucocorticoids in this important GR target tissue.
Several recent papers have implicated FOXO family transcription factors as being important for the upregulation of gene expression during skeletal muscle atrophy, including after DEX treatment or nutrient starvation in C2C12 cells. For example, Stitt et al. (48) reported that activated FOXO1 is necessary but not sufficient to activate the MuRF1 gene in cultured myotubes; however, the effects of FOXO3a or FOXO4 were not reported. Interestingly, selective transgenic overexpression of FOXO1 in skeletal muscle produced mice with significantly smaller muscles than wild-type litter mates without apparent upregulation of MuRF1 (19). The lack of MuRF1 induction in these instances is not surprising given our present findings. Assuming that FOXO1 was the only transcription factor overexpressed in these transgenic mice, and that circulating corticosteroid levels are normal, then MuRF1 would not necessarily be upregulated. The reduction in muscle mass observed in these mice may be due to the activation of other atrophy-inducing pathways. For example, overexpression of FOXO1 leads to an upregulation of 4E-BP1 and inhibition of mTOR signaling, resulting in a decrease in protein synthesis (47).
There is important precedence for the collaborative role of FOXO1 on GR target genes from studies of inversely regulated insulin and glucocorticoid-responsive genes in the liver. The IGF-binding protein-1 (10) and glucose-6-phosphatase promoters contain GREs with directly adjacent and even overlapping FOXO binding sites, and the PEPCK (13) and pyruvate dehydrogenase kinase-4 (24) genes have FOXO sites in relatively close proximity to their GREs. In each case, integrity of the FOXO site is important for maximal glucocorticoid responsiveness.
In addition, like MuRF1, the ability of FOXO1 and FOXO3a to have differential effects on a single promoter has recently been demonstrated by Onuma et al. (36) for the glucose-6-phosphatase promoter. It is intriguing that key genes involved in gluconeogenesis have clustered functional GREs and FOXO binding sites in critical glucocorticoid and insulin-sensitive enhancers reminiscent of our findings with the muscle-specific MuRF1 gene promoter. It is tempting to speculate that if indeed one role of MuRF1, as an E3 ubiquitin ligase, is to target specific skeletal muscle proteins for degradation, then a gene involved in providing a source of amino acid substrates for the liver to use to make glucose de novo might logically be under similar regulatory control as important gluconeogenic enzymes. The common regulatory strategy of MuRF1 expression with liver gluconeogenic enzymes also supports the notion of a potential prominent role for MuRF1 in skeletal muscle glucose metabolism.
The role of FOXO1 as a key component of a composite glucocorticoid-responsive unit in MuRF1 gene regulation is further emphasized by our ChIP data. IGF-I not only virtually eliminates DEX upregulation of the 500-bp MuRF1 promoter and the endogenous MuRF1 gene (see Fig. 7) but also rapidly clears the MuRF1 promoter of FOXO1 binding without affecting GR binding. On the PEPCK enhancer in rat liver H4IIE cells, insulin treatment causes a rapid loss of FOXO binding in control and DEX-treated cells, with significant reduction already apparent by as little as 3 min after insulin addition (13). In this case, GR binding was reduced to ∼40% compared with the DEX only-induced level after 30 min of treatment, which we did not observe upon IGF-I addition to C2C12 cells. Nevertheless, the most impressive change in promoter occupancy examined in both our experiments and the PEPCK gene experiments was decreased FOXO1 binding. In addition to inhibition of FOXO1 (and to a lesser extent GR) binding to the PEPCK promoter in response to insulin treatment, dramatic changes in histone posttranslational modifications that paralleled decreased transcription were observed (13). Whether these changes occur on the MuRF1 promoter in response to DEX and/or IGF-I treatment remains an interesting question, as is whether removal of FOXO1 is the key event triggering the reversal of transcriptional activation and associated chromatin modifications induced by DEX.
The exact nature of the synergy between the GR and FOXO1 is clearly an important area for further investigation because it is also not well understood for liver gluconeogenic genes. FOXO1 has been demonstrated to physically interact in solution with several steroid hormone receptors, such as the estrogen, progesterone, and androgen receptors (21, 26, 44, 59), but not, to date, with the glucocorticoid receptor. The functional significance of this interaction (either repressing or enhancing receptor activity) varies greatly depending on the cell type and target gene promoter context. For example, FOXO1 synergistically activates the IGF-binding protein-1 promoter with ligand-activated progesterone receptors in endometrial adenocarcinoma cells but not in endometrial fibroblasts (20). FOXO1, FOXO3a, and FOXO4 all contain an LXXLL motif that can interact with a ligand-induced pocket on the hormone binding domain of steroid receptors (59). Since FOXO1 binding to the MuRF1 promoter is (at least modestly) enhanced by DEX treatment of C2C12 cells along with the strong recruitment of GR (Fig. 7), a situation also observed on the PEPCK and glucose-6-phosphatase genes (13, 51), these proteins may be mutually bound and stabilized on the DEX-activated MuRF1 promoter. Synergy may also be achieved via corecruitment of transcriptional coactivators, such as histone acetyltransferases like CBP/p300, that have been shown to be independently recruited by the liganded GR or FOXO1 in other contexts (28, 34). Irrespective of the precise nature of the GR-FOXO1 synergy, our results reveal that each factor is required to create an optimal glucocorticoid-inducible MuRF1 promoter.
There are several important implications of our findings on MuRF1 gene regulation. First, the activated GR may be an important player in the control of muscle atrophy, but the presence of nuclear FOXO1 is critical to achieve full induction of the MuRF1 promoter and, quite possibly, multiple glucocorticoid-regulated genes in skeletal muscle. Furthermore, the glucocorticoid concentration needed to induce MuRF1 is significantly reduced in the presence of FOXO1. A logical prediction on the basis of these results is that conditions with both elevated corticosteroids and active FOXO1 would lead to the greatest degree of muscle wasting and altered metabolism. In fact, diabetes mellitus presents such a condition (53). In several models of diabetes, Akt is less active, potentially leading to decreased inhibition of FOXO activity and/or increased nuclear localization (55). Elevated nuclear FOXO1 would be available to interact with the GR on the MuRF1 promoter to further induce the gene above the level induced by corticosteroids alone. Prolonged glucocorticoid exposure also leads to insulin resistance (54), which might in turn lead to higher MuRF1 induction, accelerated muscle atrophy, and metabolic changes as part of a vicious cycle. In contrast, the need for both nuclear FOXO1 and activated GR to fully activate MuRF1 may be protective and prevent unnecessary muscle breakdown. For example, following intense exhaustive exercise, circulating cortisol levels increase (6, 31), yet elevated cortisol is generally not associated with an increase in the breakdown of contractile proteins, nor is loss of muscle mass a consequence of endurance training (33). Thus, the need for additional factors to fully activate MuRF1 gene expression could protect the muscle from breakdown under certain physiological conditions.
In summary, our results demonstrate a potent synergy between the GR and FOXO1 in transcriptional control of an important gene linked to skeletal muscle atrophy and metabolic control. Further understanding the molecular details of the opposing effects of corticosteroids and insulin/IGF-I on gene expression in skeletal muscle, the breadth of the GR-FOXO1 cooperation on muscle gene expression in general, and the very nature of this robust synergy should have important ramifications for preventing inappropriate induction of genes involved in muscle atrophy and altered metabolism in catabolic disease states.
GRANTS
This work was supported by a Kirchstein National Research Service Award to D. S. Waddell and National Institute of Diabetes and Digestive and Kidney Diseases Grant RO1-DK-075801 to J. D. Furlow and S. C. Bodine.
Acknowledgments
We acknowledge Dr. Frank Gannon and members of the Gannon Laboratory at the European Molecular Biology Laboratory for generous support and advice on the ChIP experiments.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Almon RR, DuBois DC, Yao Z, Hoffman EP, Ghimbovschi S, Jusko WJ. Microarray analysis of the temporal response of skeletal muscle to methylprednisolone: comparative analysis of two dosing regimens. Physiol Genomics 30: 282–299, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beato M, Chalepakis G, Schauer M, Slater EP. DNA regulatory elements for steroid hormones. J Steroid Biochem 32: 737–747, 1989. [DOI] [PubMed] [Google Scholar]
- 3.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294: 1704–1708, 2001. [DOI] [PubMed] [Google Scholar]
- 4.Cai D, Frantz JD, Tawa NE Jr, Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell 119: 285–298, 2004. [DOI] [PubMed] [Google Scholar]
- 5.Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab 6: 376–385, 2007. [DOI] [PubMed] [Google Scholar]
- 6.Duclos M, Corcuff JB, Arsac L, Moreau-Gaudry F, Rashedi M, Roger P, Tabarin A, Manier G. Corticotroph axis sensitivity after exercise in endurance-trained athletes. Clin Endocrinol (Oxf) 48: 493–501, 1998. [DOI] [PubMed] [Google Scholar]
- 7.Dyer BW, Ferrer FA, Klinedinst DK, Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal Biochem 282: 158–161, 2000. [DOI] [PubMed] [Google Scholar]
- 8.Furlow JD, Berry DL, Wang Z, Brown DD. A set of novel tadpole specific genes expressed only in the epidermis are down-regulated by thyroid hormone during Xenopus laevis metamorphosis. Dev Biol 182: 284–298, 1997. [DOI] [PubMed] [Google Scholar]
- 9.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J 375: 365–371, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan L, Pan H, Unterman TG. Insulin response sequence-dependent and -independent mechanisms mediate effects of insulin on glucocorticoid-stimulated insulin-like growth factor binding protein-1 promoter activity. Endocrinology 146: 4274–4280, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Glass DJ Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol 37: 1974–1984, 2005. [DOI] [PubMed] [Google Scholar]
- 12.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA 98: 14440–14445, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall RK, Wang XL, George L, Koch SR, Granner DK. Insulin represses phosphoenolpyruvate carboxykinase gene transcription by causing the rapid disruption of an active transcription complex: a potential epigenetic effect. Mol Endocrinol 21: 550–563, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Hasselgren PO Glucocorticoids and muscle catabolism. Curr Opin Clin Nutr Metab Care 2: 201–205, 1999. [DOI] [PubMed] [Google Scholar]
- 15.Hasselgren PO, Fischer JE. Muscle cachexia: current concepts of intracellular mechanisms and molecular regulation. Ann Surg 233: 9–17, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirner S, Krohne C, Schuster A, Hoffmann S, Witt S, Erber R, Sticht C, Gasch A, Labeit S, Labeit D. MuRF1-dependent regulation of systemic carbohydrate metabolism as revealed from transgenic mouse studies. J Mol Biol 379: 666–677, 2008. [DOI] [PubMed] [Google Scholar]
- 17.Hribal ML, Nakae J, Kitamura T, Shutter JR, Accili D. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J Cell Biol 162: 535–541, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai E, Stromstedt PE, Quinn PG, Carlstedt-Duke J, Gustafsson JA, Granner DK. Characterization of a complex glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol 10: 4712–4719, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem 279: 41114–41123, 2004. [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Jia L, Stallcup MR, Coetzee GA. The role of protein kinase A pathway and cAMP responsive element-binding protein in androgen receptor-mediated transcription at the prostate-specific antigen locus. J Mol Endocrinol 34: 107–118, 2005. [DOI] [PubMed] [Google Scholar]
- 21.Kim JJ, Buzzio OL, Li S, Lu Z. Role of FOXO1A in the regulation of insulin-like growth factor-binding protein-1 in human endometrial cells: interaction with progesterone receptor. Biol Reprod 73: 833–839, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koyama S, Hata S, Witt CC, Ono Y, Lerche S, Ojima K, Chiba T, Doi N, Kitamura F, Tanaka K, Abe K, Witt SH, Rybin V, Gasch A, Franz T, Labeit S, Sorimachi H. Muscle RING-finger protein-1 (MuRF1) as a connector of muscle energy metabolism and protein synthesis. J Mol Biol 376: 1224–1236, 2008. [DOI] [PubMed] [Google Scholar]
- 23.Krones-Herzig A, Mesaros A, Metzger D, Ziegler A, Lemke U, Bruning JC, Herzig S. Signal-dependent control of gluconeogenic key enzyme genes through coactivator-associated arginine methyltransferase 1. J Biol Chem 281: 3025–3029, 2006. [DOI] [PubMed] [Google Scholar]
- 24.Kwon HS, Huang B, Unterman TG, Harris RA. Protein kinase B-alpha inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes 53: 899–910, 2004. [DOI] [PubMed] [Google Scholar]
- 25.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 18: 39–51, 2004. [DOI] [PubMed] [Google Scholar]
- 26.Li P, Lee H, Guo S, Unterman TG, Jenster G, Bai W. AKT-independent protection of prostate cancer cells from apoptosis mediated through complex formation between the androgen receptor and FKHR. Mol Cell Biol 23: 104–118, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch GS, Schertzer JD, Ryall JG. Therapeutic approaches for muscle wasting disorders. Pharmacol Ther 113: 461–487, 2007. [DOI] [PubMed] [Google Scholar]
- 28.Ma H, Hong H, Huang SM, Irvine RA, Webb P, Kushner PJ, Coetzee GA, Stallcup MR. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol 19: 6164–6173, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez E, Wahli W. Cooperative binding of estrogen receptor to imperfect estrogen-responsive DNA elements correlates with their synergistic hormone-dependent enhancer activity. EMBO J 8: 3781–3791, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115: 751–763, 2003. [DOI] [PubMed] [Google Scholar]
- 31.Minetto MA, Lanfranco F, Baldi M, Termine A, Kuipers H, Ghigo E, Rainoldi A. Corticotroph axis sensitivity after exercise: comparison between elite athletes and sedentary subjects. J Endocrinol Invest 30: 215–223, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Mitch WE, Goldberg AL. Mechanisms of muscle wasting. The role of the ubiquitin-proteasome pathway. N Engl J Med 335: 1897–1905, 1996. [DOI] [PubMed] [Google Scholar]
- 33.Nader GA Concurrent strength and endurance training: from molecules to man. Med Sci Sports Exerc 38: 1965–1970, 2006. [DOI] [PubMed] [Google Scholar]
- 34.Nasrin N, Ogg S, Cahill CM, Biggs W, Nui S, Dore J, Calvo D, Shi Y, Ruvkun G, Alexander-Bridges MC. DAF-16 recruits the CREB-binding protein coactivator complex to the insulin-like growth factor binding protein 1 promoter in HepG2 cells. Proc Natl Acad Sci USA 97: 10412–10417, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson JD, Denisenko O, Sova P, Bomsztyk K. Fast chromatin immunoprecipitation assay. Nucleic Acids Res 34: e2, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onuma H, Vander Kooi BT, Boustead JN, Oeser JK, O'Brien RM. Correlation between FOXO1a (FKHR) and FOXO3a (FKHRL1) binding and the inhibition of basal glucose-6-phosphatase catalytic subunit gene transcription by insulin. Mol Endocrinol 20: 2831–2847, 2006. [DOI] [PubMed] [Google Scholar]
- 37.Payvar F, DeFranco D, Firestone GL, Edgar B, Wrange O, Okret S, Gustafsson JA, Yamamoto KR. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell 35: 381–392, 1983. [DOI] [PubMed] [Google Scholar]
- 38.Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93: 531–541, 1998. [DOI] [PubMed] [Google Scholar]
- 39.Reichardt HM, Tuckermann JP, Gottlicher M, Vujic M, Weih F, Angel P, Herrlich P, Schutz G. Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J 20: 7168–7173, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rena G, Guo S, Cichy SC, Unterman TG, Cohen P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J Biol Chem 274: 17179–17183, 1999. [DOI] [PubMed] [Google Scholar]
- 41.Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am J Physiol Endocrinol Metab 287: E591–E601, 2004. [DOI] [PubMed] [Google Scholar]
- 42.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117: 399–412, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoneveld OJ, Gaemers IC, Lamers WH. Mechanisms of glucocorticoid signalling. Biochim Biophys Acta 1680: 114–128, 2004. [DOI] [PubMed] [Google Scholar]
- 44.Schuur ER, Loktev AV, Sharma M, Sun Z, Roth RA, Weigel RJ. Ligand-dependent interaction of estrogen receptor-alpha with members of the forkhead transcription factor family. J Biol Chem 276: 33554–33560, 2001. [DOI] [PubMed] [Google Scholar]
- 45.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet 3: e94, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci USA 105: 5745–5749, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Southgate RJ, Neill B, Prelovsek O, El-Osta A, Kamei Y, Miura S, Ezaki O, McLoughlin TJ, Zhang W, Unterman TG, Febbraio MA. FOXO1 regulates the expression of 4E-BP1 and inhibits mTOR signaling in mammalian skeletal muscle. J Biol Chem 282: 21176–21186, 2007. [DOI] [PubMed] [Google Scholar]
- 48.Stitt TN, Drujan D, Clarke BA, Panaro FJ, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin liganses by inhibiting FOXO transcription factors. Mol Cell 14: 1–14, 2004. [DOI] [PubMed] [Google Scholar]
- 49.Sun L, Trausch-Azar JS, Muglia LJ, Schwartz AL. Glucocorticoids differentially regulate degradation of MyoD and Id1 by N-terminal ubiquitination to promote muscle protein catabolism. Proc Natl Acad Sci USA 105: 3339–3344, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuckermann JP, Kleiman A, McPherson KG, Reichardt HM. Molecular mechanisms of glucocorticoids in the control of inflammation and lymphocyte apoptosis. Crit Rev Clin Lab Sci 42: 71–104, 2005. [DOI] [PubMed] [Google Scholar]
- 51.Vander Kooi BT, Onuma H, Oeser JK, Svitek CA, Allen SR, Vander Kooi CW, Chazin WJ, O'Brien RM. The glucose-6-phosphatase catalytic subunit gene promoter contains both positive and negative glucocorticoid response elements. Mol Endocrinol 19: 3001–3022, 2005. [DOI] [PubMed] [Google Scholar]
- 52.Vander Kooi BT, Streeper RS, Svitek CA, Oeser JK, Powell DR, O'Brien RM. The three insulin response sequences in the glucose-6-phosphatase catalytic subunit gene promoter are functionally distinct. J Biol Chem 278: 11782–11793, 2003. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Hu Z, Hu J, Du J, Mitch WE. Insulin resistance accelerates muscle protein degradation: Activation of the ubiquitin-proteasome pathway by defects in muscle cell signaling. Endocrinology 147: 4160–4168, 2006. [DOI] [PubMed] [Google Scholar]
- 54.Weinstein SP, Paquin T, Pritsker A, Haber RS. Glucocorticoid-induced insulin resistance: dexamethasone inhibits the activation of glucose transport in rat skeletal muscle by both insulin- and non-insulin-related stimuli. Diabetes 44: 441–445, 1995. [DOI] [PubMed] [Google Scholar]
- 55.Whiteman EL, Cho H, Birnbaum MJ. Role of Akt/protein kinase B in metabolism. Trends Endocrinol Metab 13: 444–451, 2002. [DOI] [PubMed] [Google Scholar]
- 56.Wright AP, Zilliacus J, McEwan IJ, Dahlman-Wright K, Almlof T, Carlstedt-Duke J, Gustafsson JA. Structure and function of the glucocorticoid receptor. J Steroid Biochem Mol Biol 47: 11–19, 1993. [DOI] [PubMed] [Google Scholar]
- 57.Yang H, Mammen J, Wei W, Menconi M, Evenson A, Fareed M, Petkova V, Hasselgren PO. Expression and activity of C/EBPbeta and delta are upregulated by dexamethasone in skeletal muscle. J Cell Physiol 204: 219–226, 2005. [DOI] [PubMed] [Google Scholar]
- 58.Yang H, Menconi MJ, Wei W, Petkova V, Hasselgren PO. Dexamethasone upregulates the expression of the nuclear cofactor p300 and its interaction with C/EBPbeta in cultured myotubes. J Cell Biochem 94: 1058–1067, 2005. [DOI] [PubMed] [Google Scholar]
- 59.Zhao HH, Herrera RE, Coronado-Heinsohn E, Yang MC, Ludes-Meyers JH, Seybold-Tilson KJ, Nawaz Z, Yee D, Barr FG, Diab SG, Brown PH, Fuqua SA, Osborne CK. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J Biol Chem 276: 27907–27912, 2001. [DOI] [PubMed] [Google Scholar]
- 60.Zhao X, Gan L, Pan H, Kan D, Majeski M, Adam SA, Unterman TG. Multiple elements regulate nuclear/cytoplasmic shuttling of FOXO1: characterization of phosphorylation- and 14-3-3-dependent and -independent mechanisms. Biochem J 378: 839–849, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]