Abstract
Introduction
“Race-specific” PSA needs evaluation in men at high-risk for prostate cancer (PCA) for optimizing early detection. Baseline PSA and longitudinal prediction for PCA was examined by self-reported race and genetic West African (WA) ancestry in the Prostate Cancer Risk Assessment Program, a prospective high-risk cohort.
Materials and Methods
Eligibility criteria are age 35–69 years, FH of PCA, African American (AA) race, or BRCA1/2 mutations. Biopsies have been performed at low PSA values (<4.0 ng/mL). WA ancestry was discerned by genotyping 100 ancestry informative markers. Cox proportional hazards models evaluated baseline PSA, self-reported race, and genetic WA ancestry. Cox models were used for 3-year predictions for PCA.
Results
646 men (63% AA) were analyzed. Individual WA ancestry estimates varied widely among self-reported AA men. “Race-specific” differences in baseline PSA were not found by self-reported race or genetic WA ancestry. Among men with ≥ 1 follow-up visit (405 total, 54% AA), three-year prediction for PCA with a PSA of 1.5–4.0 ng/mL was higher in AA men with age in the model (p=0.025) compared to EA men. Hazard ratios of PSA for PCA were also higher by self-reported race (1.59 for AA vs. 1.32 for EA, p=0.04). There was a trend for increasing prediction for PCA with increasing genetic WA ancestry.
Conclusions
“Race-specific” PSA may need to be redefined as higher prediction for PCA at any given PSA in AA men. Large-scale studies are needed to confirm if genetic WA ancestry explains these findings to make progress in personalizing PCA early detection.
Keywords: genetics, population; mass screening; African Americans; prostatic neoplasms
INTRODUCTION
African American (AA) men and European American (EA) men with a family history (FH) of prostate cancer (PCA) are at substantially increased risk for the disease and may have clinically more aggressive disease (1–3). Early detection of PCA in these high-risk men presents the ideal situation in which to diagnose PCA while at a curable point, with screening likely leading to the detection of intermediate to high-grade PCA (4, 5). However, several issues remain regarding the optimal PSA level at which to recommend prostate biopsies in order to detect PCA as well as not to subject these men to unnecessary biopsies. High-risk men have been found to have PCA detected at low PSA values, even less than 2.5 ng/mL (5, 6). Determining which high-risk men should have prostate biopsies at low PSA values is crucial to early detection and avoiding unnecessary biopsies. An important concept regarding the PSA in AA men is the “race-specific PSA”. This concept is based on several previous reports that found that AA men had higher PSA values stage-for-stage or at baseline (7–10). Many of these studies were performed when PSA was being implemented for PCA screening in the late 1980’s-mid 1990’s. In addition, due to population admixture, self-reported AA men exhibit large variance in individual genetic West African (WA) ancestry (11, 12) which has not been explored in the context of the “race-specific PSA”. Furthermore, concerns have been raised regarding missing high-grade PCA if PSA is adjusted for race based on this concept (13). Thus issues of PCA prediction at lower PSA values and “race-specific” PSA deserve further study in a prospective high-risk cohort with characterization of genetic markers of WA ancestry in order to gain insight on how to use the PSA to make tailored diagnostic decisions in high-risk men.
Reports from previous studies regarding PSA characteristics in the screening setting are challenging to apply to younger, ethnically-diverse, high-risk men undergoing aggressive PCA screening. The Prostate Cancer Prevention Trial (PCPT) reported on PSA performance characteristics in the control arm when describing their risk assessment calculator (14). However, this population was predominantly EA (95.6%), were 55 years or older, and had prostate biopsies recommended for a PSA > 4.0 ng/mL. PSA characteristics were also reported from the more diverse population enrolled in PCA screening in the San Antonio Center of Biomarkers of Risk for Prostate Cancer (SABOR) (15). This cohort, while having 13.5% AA men and 37.2% Hispanics, reported on sensitivity/specificity characteristics of the PSA in relation to the PCPT risk calculator and not on point-by-point PSA predictions for PCA by race. AA men were also underrepresented in this cohort. The European Randomized Study of Screening for Prostate Cancer (ERSPC) enrolled over 21,000 men in Europe to be randomized to screening vs. no screening (16). Men in this study are older (over 55 years) and are predominantly EA. Prostate biopsies are not recommended at PSA values less than 3.0 ng/mL in the setting of a normal rectal examination, thus making these data difficult to apply to high-risk US men of diverse racial background.
The purpose of this study was to investigate if the baseline PSA and PSA predictive ability for PCA was substantially different in self-reported AA men compared to self-reported EA men with a FH of PCA undergoing aggressive screening in the Prostate Cancer Risk Assessment Program (PRAP) at Fox Chase Cancer Center (FCCC), a prospective, longitudinal PCA screening and research program for men at high-risk. PRAP has approximately 60% AA participation, a demographic not seen in other screening studies. Another distinguishing feature of the PRAP cohort is that recommendations for prostate biopsy have been made at PSA values as low as 1.5 ng/mL to study aggressive screening measures (6). Therefore, the predictive ability for PCA at low PSA values was able to be investigated. We investigated the concept of “race-specific PSA” by determining if baseline PSA differed by self-reported race or was associated with genetic markers of WA ancestry based on a panel of 100 previously validated ancestry informative markers (AIMs) in AA men (17). We further explored if differences in PSA predictive ability for PCA was due to variation in WA ancestry. By grouping self-reported AA men from lowest to highest genetic WA ancestry, we explored the ability to identify subsets of AA men in whom the PSA has a higher prediction for PCA to move forward with personalized PCA early detection.
MATERIALS AND METHODS
Prostate Cancer Risk Assessment Program (PRAP) cohort
The Prostate Cancer Risk Assessment Program (PRAP) at Fox Chase Cancer Center (FCCC) was established in 1996 to provide screening and perform research for men at high risk for PCA (18). Eligibility for PRAP include any man between ages 35–69 years with one first-degree relative with PCA, two second-degree relatives with PCA on the same side of the family, any AA man regardless of FH, or men with known mutations in BRCA1 or BRCA2. Recruitment is primarily through radio advertisements in the Philadelphia area, as well as through physician referrals, newsletters, and community events. Accrual to PRAP is ongoing and participants are followed longitudinally for PCA screening and early detection. Three community Partner hospitals also participate in PRAP.
PCA screening in PRAP
PCA screening procedures, biopsy criteria, PCA incidence, and PCA features have been described previously (5). Briefly, screening for PCA is performed on an annual basis. Screening tests include the total prostate specific antigen (PSA), percent free PSA (fPSA) (6), digital rectal examination (DRE) by a PRAP physician, and the PSA velocity (PSAv). If all screening parameters are within normal limits per PRAP criteria (see Biopsy criteria below), then the participant returns in one year for repeat screening.
Criteria for Biopsy
Until November 2005, the criteria for prostate biopsy were (1) PSA > 4 ng/mL, (2) PSA 2.0–4.0 ng/mL with fPSA less than 27%, (3) any abnormality on DRE, or (4) PSAv of 0.75 ng/mL/year. After November 2005, the criteria for biopsy were changed to (1) PSA > 2.0 ng/mL, (2) PSA 1.5–2.0 ng/mL with fPSA ≤25%, (3) any abnormality on DRE, or (4) PSAv of 0.75 ng/mL/year to investigate the detection of PCA at lower PSA values based on emerging guidelines recommending discussing aggressive PCA screening in high-risk men (19).
Biopsy Approach
All biopsies are transrectal ultrasound-guided 5-region patterned prostate biopsies (20, 21). The mean number of cores from these five regions was identical for AA and EA men (mean = 9.4 cores and median = 10.0 cores for both race groups). All pathology is evaluated at FCCC by the Department of Pathology or by the pathology departments at three Partner hospital sites.
Ascertainment of Self-Reported Race
Self -reported race is determined by phone during the eligibility interview. Participants are asked for their race/ethnicity and the majority will volunteer the information. However, if they are unclear, several categories are offered to them to identify their race/ethnicity as follows: White/EA, Black/AA/Caribbean, Hispanic/Latino, Southeast Asian, South Asian, Native American, Other, or Unknown. Participants are classified as AA or White/EA if either of these groups are chosen regardless of other race/ethnic indications. If both AA and White/EA are indicated, the participant is classified as AA. Twenty five participants indicated more than one race of whom 22 were classified as AA and 3 were classified as EA. Since these 25 participants represented only 3.9% of the entire cohort, we have included these men in the analysis.
Genetic Markers of Ancestry
One hundred carefully selected AIMs were genotyped for all samples. These autosomal markers have previously been identified and validated and can be used to extract continental ancestry information in African Americans (17). Genotyping methods are available by request.
Statistical Methods
Individual genetic ancestry (IA) was determined for each person using 100 AIMs for West African and European genetic ancestry (17). IA was estimated from the genotype data using the Bayesian Markov Chain-Monte Carlo (MCMC) method implemented in the program STRUCTURE 2.1 (22). STRUCTURE 2.1 was run under the admixture model using prior population information and independent allele frequencies. We ran the MCMC method using K=2 parental populations and a burn-in length of 30,000 for 70,000 repetitions. Each participant was then scored from 0–100% on individual ancestry estimates of WA ancestry.
Cox proportional hazards regressions were used for inferences about the relationship of time to diagnosis with race, age, and PSA covariates. Men who did not develop PCA were censored at their last available follow-up date. Harrell's Concordance Index was used as a measure of model fit (23). For calculation of p-values, continuous variables were entered as linear terms in models and all tests of statistical significance were two-sided. To display the relationship of PSA with time to diagnosis in the models, we used Cox models with age and PSA entered through the use of restricted cubic splines (23). Three knots were used in the restricted cubic splines (specified at 0.5 ng/mL, 2.0 ng/mL, and 4.0 ng/mL for PSA and 40 years, 50 years, and 60 years for age) to estimate three-year probabilities of being diagnosed with PCA. The estimated probabilities versus PSA level were then plotted. We estimated the baseline survivor function, as implemented in STATA 10.0, to estimate the three year predicted probabilities for PCA. Since age and PSA were continuous variables, such plots of the predicted probabilities provided more interpretable descriptions of the relationship of PSA with PCA diagnosis than multiple time to event curves stratified by PSA and age categories. We used restricted cubic spline models of the probabilities to display the average predicted probability across PSA. Differences between populations were tested by using interaction terms in regressions.
RESULTS
As of June 2007, 657 high-risk men were accrued to PRAP. This analysis includes 646 of these men with complete data for race, baseline PSA, and AIM genotypes. The demographics of this cohort by self-reported race are shown in Table 1. No differences exist in mean baseline age, PSA, percent free PSA, DRE findings, or biopsy history. In addition, the median age at entry was identical for self-reported AA and EA men at 49.0 years. Age-adjusted baseline PSA values were not significantly different between self-reported AA and EA men when testing for “race-specific PSA” effect (1.60 ng/mL vs. 1.67 ng/mL respectively, p=0.69).
Table 1.
Demographics and Prostate Cancer Characteristics by Self-reported Race of 646 PRAP Participants
| African American (n=408) | European American (n=238) | |||||
|---|---|---|---|---|---|---|
| N | Mean | Range | N | Mean | Range | |
| Age at entry (years) | 408 | 49.6 | 34–69 | 238 | 49.8 | 35–69 |
| Duration of follow-up (months) | 223 | 40.4 | 0.3–122.5 | 188 | 48.3 | 0.6–127.1 |
| PSA at baseline (ng/mL) | 408 | 1.6 | 0.1–27.2 | 238 | 1.7 | 0.1–22.5 |
| Percent Free PSA at baseline+ | 81 | 16.8 | 3.5–39.4 | 50 | 17.0 | 4.6–40.0 |
| DRE at baseline++ | ||||||
| Normal/BPH (%) | 371 | 210 | ||||
| (95.6) | (95.4) | |||||
| Abnormal (%) | 17 | 10 | ||||
| (4.4) | (4.6) | |||||
| Biopsy history (reported at baseline)§ | ||||||
| No Prior Biopsy/Unknown | 298 | 205 | ||||
| (92.3%) | (92.8%) | |||||
| Had Prior Negative Biopsy | 25 | 16 | ||||
| (7.7%) | (7.2%) | |||||
| Genetic West African Ancestry * | 408 | 0.751 | 0.016–0.995 | 238 | 0.058 | 0.004–0.751 |
| PCA diagnosis ** | 37 | - | 29 | - | ||
| (9.1%) | (12.2%) | |||||
| PSA prior to PCA dx (ng/mL) | 37 | 4.8 | 0.9–31.6 | 29 | 4.3 | 1.1–22.5 |
| Gleason Score | 37 | 6.2 | 5–8 | 29 | 6.2 | 5–7 |
Difference in percent free PSA by race at baseline was not statistically significant, t-test p=0.783. (Note: Percent free PSA is only performed for men with a PSA 2.0–4.0 ng/mL by the previous criteria or a PSA 1.5–2.0ng/mL by the current criteria in PRAP. Therefore, not all men have a percent free PSA performed at baseline.)
Difference in abnormal DRE by race at baseline was not statistically significant, Fisher’s Exact Test p=1.00 (Note: DRE information missing for 18 AA and 20 EA participants)
Difference in biopsy history by race was not statistically significant, Fisher’s Exact Test p= 0.870 (Note: Biopsy history missing for 85 AA men and 17 EA men.)
Highest possible is 1.00. This is based on genotyping 100 Ancestry Informative Markers.
Percent of the group; last diagnosis was 4/08
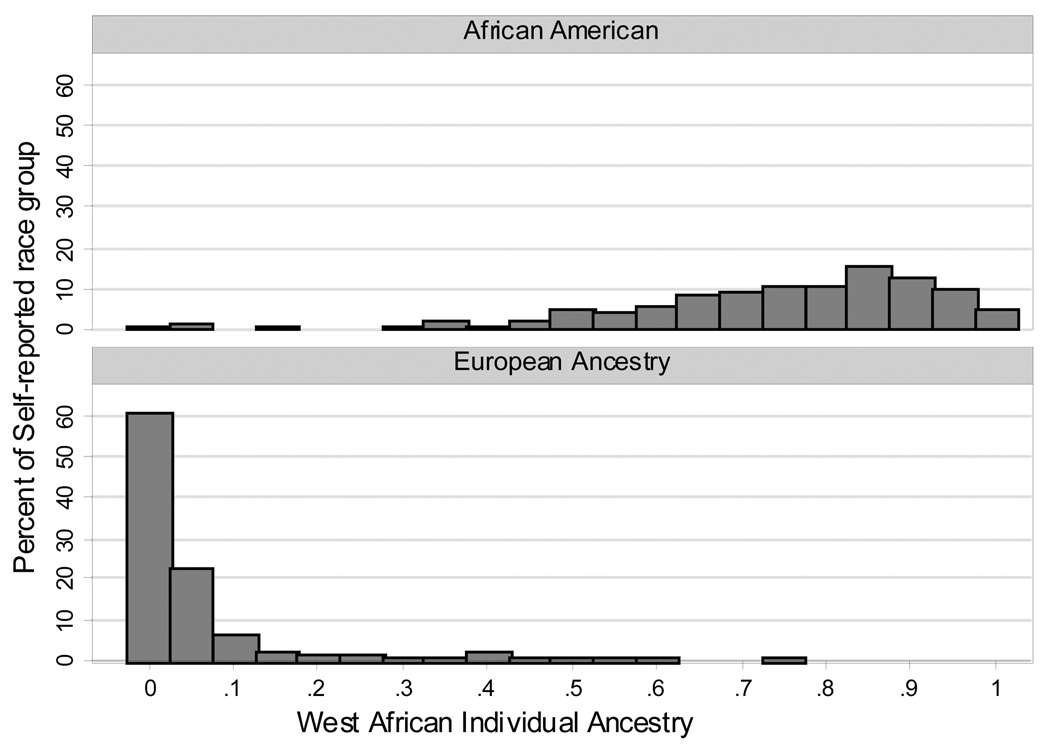
To further explore the concept of “race-specific PSA”, we investigated if the baseline PSA was higher for PRAP men with higher genetic West African (WA) ancestry. The distribution of WA ancestry by IA estimates grouped by self-reported race is shown in Figure 1 and the demographics of this cohort of 646 men is the same as in Table 1.
Figure 1. Distribution of Individual Ancestry Estimates by Self-Reported Race in 646 PRAP Participants.
As can be seen from Figure 1, genetic WA ancestry was significantly higher in self-reported AA men compared to EA men. The distribution of WA ancestry varied widely in self-reported AA men compared to EA men. We found no significant correlation between IA estimates of WA ancestry and baseline PSA in self-reported EA men, however for AA men there appeared to be a nominal yet non-significant correlation (Pearson Correlations: ; EA men 0.071, p=0.30; AA men −0.024, p=0.06).
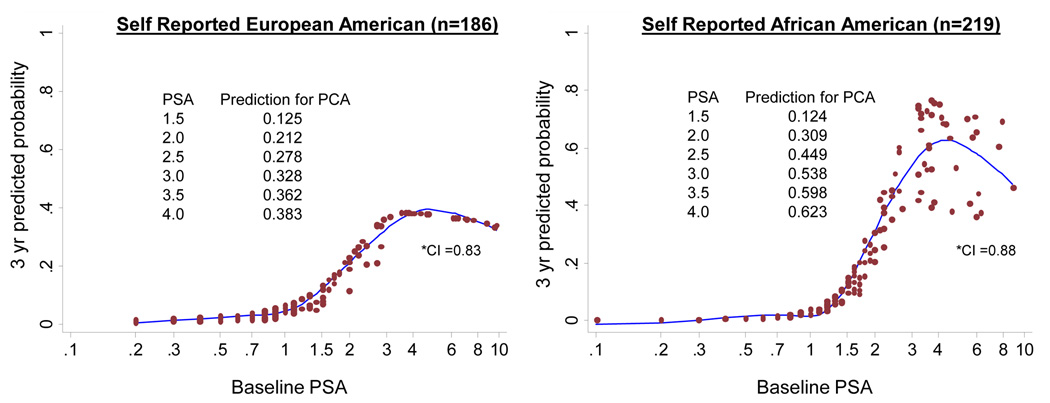
We next explored if the PSA prediction for PCA differed between AA and EA men by self-reported race using Cox models. This cohort included 411 out of the total 646 men; the remaining 235 men who were excluded were: 46 men who were not yet scheduled to return for their follow-up visit, and 189 men who did not return for any follow-up visits. Compared to the men included in the analysis, men lost to follow-up tended to be younger (mean age 46.9 year vs 50.9 years, t-test p-value <0.0001) and have lower baseline PSA values (mean PSA 1.25 ng/ml vs 1.86 ng/ml, t-test p-value = 0.001). AA men were more likely to be lost to follow-up than EA men (40% vs 19%, Chi-square test p-value <0.0001). Table 2 shows the demographics and PCA characteristics of this group of 411 PRAP participants. For the Cox model analysis, an additional six PRAP men with a baseline PSA over 10 ng/mL were removed to reduce the possibility that they would be influential points (PSA levels of excluded men were 13, 15, 15, 22, 23, and 27 ng/mL). Therefore the final Cox model analyses were performed on 405 PRAP men and the results in this paper are hence only generalizable to men with PSA<=10 ng/mL. Figure 2 shows the plots of the Cox models of the PSA prediction for PCA at 3-years with age in the model. This 3-year time frame was chosen for study as the mean duration of follow-up in PRAP has been approximately 40–48 months. As can be seen from Figure 2, the PSA had a noticeably higher prediction for PCA in the range of ~1.5–4.0 ng/mL in self-reported AA men compared to self-reported EA men. A statistically significant difference was seen by race in the association of baseline PSA to PCA development based on the Cox model when testing for interactions for race-PSA and race-age (p=0.025). When testing the model for the race-PSA interaction only, the interaction was still statistically significant (p=0.04).
Table 2.
Demographics and Prostate Cancer Characteristics by Self-reported Race in 411 PRAP Participants with At-Least One Follow-up Visit
| African American (n=223) | European American (n=188) | |||||
|---|---|---|---|---|---|---|
| N | Mean | Range | N | Mean | Range | |
| Age at entry (years) | 223 | 51.6 | 35–69 | 188 | 50.1 | 35–69 |
| Duration of follow-up (months) | 223 | 40.4 | 0.3–122.5 | 188 | 47.9 | 0.6–127.1 |
| PSA at baseline (ng/mL) | 223 | 2.0 | 0.1–27.2 | 188 | 1.7 | 0.2–22.5 |
| Genetic West African Ancestry* | 223 | 0.755 | 0.045–0.995 | 188 | 0.052 | 0.004–0.751 |
| PCA diagnosis ** | 37 | - | - | 29 | - | - |
| PSA prior to PCA dx (ng/mL) | 37 | 4.8 | 0.9–31.6 | 29 | 4.3 | 1.1–22.5 |
| Gleason Score | 37 | 6.2 | 5–8 | 29 | 6.2 | 5–7 |
Highest possible is 1.00. This is based on genotyping 100 Ancestry Informative Markers.
last diagnosis was 4/08.
Figure 2. Three-year Predicted Probability for Prostate Cancer of Baseline PSA by Self-Reported Race with Age in the Model.
Cox model plots for predicting 3-year probability for prostate cancer by baseline PSA. Interaction of self-reported race and PSA was significant at p=0.025.
*CI= Harrell’s Concordance Index (23)
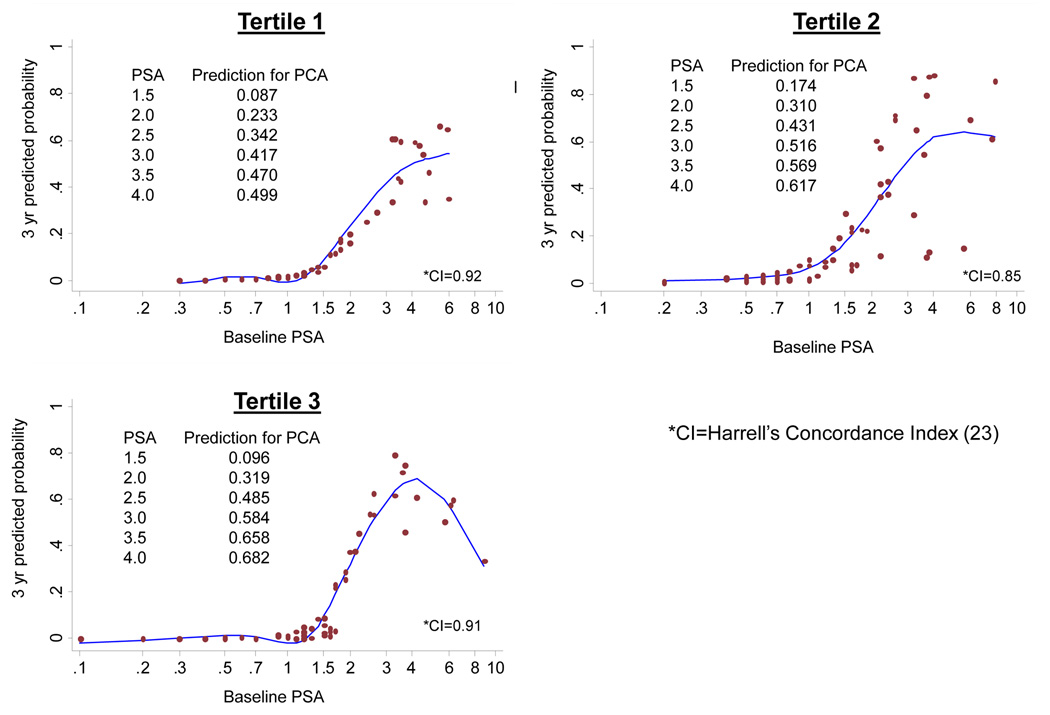
We next investigated if the higher prediction for PCA in self-reported AA men was due to the influence of genetic WA ancestry. We divided 219 self-reported AA men from Figure 2 into tertiles of lowest to highest IA estimates for WA ancestry. Each tertile included 73 AA men and the mean and range of WA ancestry were as follows: 0.561 (0.045–0.684) for Tertile 1, 0.785 (0.686–0.865) for Tertile 2, and 0.921 (0.866–0.995) for Tertile 3. Figure 3 shows the Cox model plots of 3-year prediction for PCA by baseline PSA in these tertiles of AA men with point estimates included on each figure. There is a trend for higher prediction for PCA at any given PSA in the range of 1.5–4.0 ng/mL with increasing genetic WA ancestry by IA estimates, although this was not statistically significant. Hazard ratios of PSA for PCA were significantly higher in self-reported AA men compared to self-reported EA men (1.59 [95% CI 1.38–1.84] vs. 1.30 [95% CI 1.12–1.51], p=0.04, respectively). By tertiles of WA ancestry, hazard ratios of PSA for PCA were as follows: tertile 1 = 2.19 (95% CI 1.49–3.22), tertile 2 = 1.46 (95% CI 1.19–1.79), and tertile 3 = 1.45 (95% CI 1.04–2.01). The joint test of equality of interaction terms in an interaction model was not statistically significant (p=0.150). The hazards estimated from the interaction models did not substantially differ from those reported above.
Figure 3. Predicted Probability for Prostate Cancer at 3-Years of the Baseline PSA with Increasing Genetic WA Ancestry in 219 AA Men in PRAP with Age in the Model.
DISCUSSION
Screening for PCA in high-risk men (AA men and in EA men with a FH of PCA) can be challenging. The potential benefits include early detection of intermediate or high-grade PCA with improved chances for cure with modern approaches. The risks include performing unnecessary prostate biopsies and subjecting these men to physical and psychological morbidities. Previous studies (including from our group) have shown that early detection strategies do detect intermediate to high-grade PCA and at lower PSA values (4, 5). In addition these high-risk men are at risk for developing PCA at a relatively young age, adding to the potential benefits of early detection (5). However, using the PSA for screening for PCA in high-risk men is challenging, with positive predictive values ranging from 38–45% in our cohort. Novel strategies are needed to personalize the early detection of PCA in high-risk men. PRAP is an ideal cohort in which to investigate PCA screening issues in high-risk men as there is 60% AA participation, participants are followed longitudinally, and prostate biopsies have been performed at lower PSA values for several years.
In this study, we investigated the concept of “race-specific PSA” with the high-risk, prospective, and longitudinal cohort in PRAP. The “race-specific PSA” concept implies that AA men have higher PSA at baseline and at diagnosis, and therefore, these men may need to have prostate biopsies recommended at higher PSA values compared to EA men when performing PCA screening. The concern here is missing high-grade PCA (13). Indeed, we did not find an association between higher baseline PSA and self-reported race in our cohort. We further explored the concept of “race-specific PSA” by investigating for any association of baseline PSA to genetic markers of WA ancestry and found no association, further showing evidence against a higher baseline PSA in AA men. Of importance is the wide range of genetic ancestry estimates of WA ancestry among self-reported AA men, indicating that each individual AA man seeking screening for PCA does not have the same genetic ancestral proportions and therefore may not have the same risk for PCA.
We did find that the PSA has a higher prediction for PCA at any given value between ~1.5–4.0 ng/mL in self-reported AA compared to EA men with a FH of PCA (all a higher risk group compared to the general population). We have the unique ability to study lower PSA values (<4.0 ng/mL) because of the aggressive screening approaches being studied in the PRAP high-risk cohort in a longitudinal fashion. These results have the opposite implication of the “race-specific PSA”, where the same PSA value has a higher risk for PCA at 3-years in AA men. Clinically, our data support aggressive screening measures in self-reported AA men based on higher predictions for PCA.
Given the wide variation in genetic WA ancestry among AA men and recent studies which reveal higher risk for PCA among AAs with higher genetic WA ancestry (11, 12), we explored if genetic WA ancestry was the contributing factor for higher PCA risk among our AA subjects. Based on tertiles of increasing genetic WA ancestry, we did find higher 3-year point estimates for predictions of PCA for PSA between ~1.5–4.0 ng/mL. The apparent paradox between these increasing point estimates for PCA shown in Figure 3 and the lower hazard ratios for PSA by tertile of genetic WA ancestry is likely related to the fact that the baseline hazard for PCA at low PSA levels increases for AA men in tertiles 2 and 3 compared with tertile 1 which is demonstrated by Cox modeling as follows: tertile 2 vs. tertile 1, HR=10.0, p=0.08; tertile 3 vs. tertile 1, HR=2.4, p=0.56. If such a higher baseline hazard is indeed found to be significant in a larger sample, it could negate previous beliefs that AA men inherently have a higher PSA than EA men. Furthermore, these hazard ratios of PSA for PCA by tertile of WA ancestry need to be interpreted cautiously since they were estimated from models in which baseline PSA was entered as a linear term rather than the flexible way in which PSA was modeled for Figure 3. Figure 3 gives a more accurate depiction of the flexible relationship of baseline PSA with tertile of WA ancestry in our sample. Given the trend in increasing PCA prediction at lower PSA values with increasing genetic WA ancestry among AA men, we have identified a potentially higher risk group of men based on markers of ancestry in which to further investigate aggressive screening approaches in larger studies. These genetic markers of ancestry may have clinical use in individualizing PCA screening among AA men, particularly since our data show a wide range of WA ancestry estimates among self-reported AA men. Other factors to explore regarding the difference in the PSA predictive ability for PCA between AA and EA men include genetic modifiers, diet, environment, and socioeconomic factors.
There are some limitations in this study. The overall follow-up rate among PRAP participants is 60%, which can hinder the interpretation of PCA development over time. This is a known challenge in prospective screening studies and highlights the need for efforts to enhance adherence to screening protocols. This follow-up rate of 60% is close to rates reported by other high-risk screening cohorts of 60–72% (4). We also do not perform prostate biopsies in all PRAP participants as have been performed in the control arm of the PCPT (24). Therefore there may be some men who have PCA in PRAP that is undetected at this time. However, our cohort represents a true clinical setting of aggressive PCA screening which would not include prostate biopsies for everyone. Another limitation is that at this time we were able to evaluate PCA prediction at 3-years. Longer follow-up in PRAP is needed and is planned to determine the prediction for PCA at 5 and 10 years. Finally the sample size limited the ability to firmly assess the association of estimates of WA genetic ancestry to predictions for PCA based on PSA among AA men. PRAP is continuously accruing participants, and we plan to analyze our findings in a larger cohort in the future. Follow-up on outcomes from PCA treatment (biochemical recurrence after radical prostatectomy vs. radiation therapy, distant recurrence, quality of life after treatment, and death from PCA) is also planned in the future with longer follow-up.
Overall, we find no evidence in support of the traditional “race-specific” PSA. We do find that the PSA has a higher prediction for PCA in AA men - a new concept of the “race-specific” effect. This finding may be explained by genetic WA ancestry which deserves further study. Our findings of the role of genetic WA ancestry in modifying the ability of the PSA to predict for PCA in AA men need to be confirmed on a larger scale in order to explore their role in personalizing PCA early detection in high-risk men.
Acknowledgements
We are grateful to all participants of the Prostate Cancer Risk Assessment Program.
Funding: (V. Giri) 98-PADOH-ME-98155; National Institutes of Health CORE Grant (CA06927): (R. Kittles) Department of Defense (DAMD W81XWH-07-1-0203 and DAMD W81XWH-06-1-0066).
REFERENCES
- 1.Cancer Facts and Figures. Atlanta: American Cancer Society; 2008. [Google Scholar]
- 2.Ahaghotu C, Baffoe-Bonnie A, Kittles R, et al. Clinical characteristics of African-American men with hereditary prostate cancer: the AAHPC Study. Prostate Cancer Prostatic Dis. 2004;7(2):165–169. doi: 10.1038/sj.pcan.4500719. [DOI] [PubMed] [Google Scholar]
- 3.Carter BS, Bova GS, Beaty TH, et al. Hereditary prostate cancer: epidemiologic and clinical features. J Urol. 1993;150(3):797–802. doi: 10.1016/s0022-5347(17)35617-3. [DOI] [PubMed] [Google Scholar]
- 4.Catalona WJ, Antenor JA, Roehl KA, Moul JW. Screening for prostate cancer in high risk populations. J Urol. 2002;168(5):1980–1983. doi: 10.1016/S0022-5347(05)64276-0. discussion 1983–4. [DOI] [PubMed] [Google Scholar]
- 5.Giri VN, Beebe-Dimmer J, Buyyounouski M, et al. Prostate cancer risk assessment program: a 10-year update of cancer detection. J Urol. 2007;178(5):1920–1924. doi: 10.1016/j.juro.2007.07.010. discussion 1924. [DOI] [PubMed] [Google Scholar]
- 6.Uzzo RG, Pinover WH, Horwitz EM, et al. Free prostate-specific antigen improves prostate cancer detection in a high-risk population of men with a normal total PSA and digitalrectal examination. Urology. 2003;61(4):754–759. doi: 10.1016/s0090-4295(02)02524-4. [DOI] [PubMed] [Google Scholar]
- 7.Henderson RJ, Eastham JA, Culkin DJ, et al. Prostate-specific antigen (PSA) and PSA density: racial differences in men without prostate cancer. J Natl Cancer Inst. 1997;89(2):134–138. doi: 10.1093/jnci/89.2.134. [DOI] [PubMed] [Google Scholar]
- 8.Vijayakumar S, Karrison T, Weichselbaum RR, Chan S, Quadri SF, Awan AM. Racial differences in prostate-specific antigen levels in patients with local-regional prostate cancer. Cancer Epidemiol Biomarkers Prev. 1992;1(7):541–545. [PubMed] [Google Scholar]
- 9.Moul JW. Use of prostate-specific antigen in black men: age-adjusted reference ranges for maximal cancer detection. J Natl Med Assoc. 1998;90(11 Suppl):S710–S712. [PMC free article] [PubMed] [Google Scholar]
- 10.Abdalla I, Ray P, Vaida F, Vijayakumar S. Racial differences in prostate-specific antigen levels and prostate-specific antigen densities in patients with prostate cancer. Am J Clin Oncol. 1999;22(6):537–541. doi: 10.1097/00000421-199912000-00001. [DOI] [PubMed] [Google Scholar]
- 11.Freedman ML, Haiman CA, Patterson N, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103(38):14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins C, Torres JB, Hooker S, Bonilla C, Hernandez W, Candreva A, et al. Confirmation study of prostate cancer risk variants at 8q24 in African Americans identifies a novel risk locus. Genome Res. 2007;17(12):1717–1722. doi: 10.1101/gr.6782707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed A, Ankerst DP, Pollock BH, Thompson IM, Parekh DJ. Current age and race adjusted prostate specific antigen threshold values delay diagnosis of high grade prostate cancer. J Urol. 2007;178(5):1929–1932. doi: 10.1016/j.juro.2007.07.045. discussion 1932. [DOI] [PubMed] [Google Scholar]
- 14.Thompson IM, Ankerst DP, Chi C, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98(8):529–534. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 15.Parekh DJ, Ankerst DP, Higgins BA, et al. External validation of the Prostate Cancer Prevention Trial risk calculator in a screened population. Urology. 2006;68(6):1152–1155. doi: 10.1016/j.urology.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Schroder FH, Bangma CH. The European Randomized Study of Screening for Prostate Cancer (ERSPC) Br J Urol. 1997;79(Suppl 1):68–71. doi: 10.1111/j.1464-410x.1997.tb00804.x. [DOI] [PubMed] [Google Scholar]
- 17.Tian C, Hinds DA, Shigeta R, Kittles R, Ballinger DG, Seldin MF. A genomewide single-nucleotide-polymorphism panel with high ancestry information for African American admixture mapping. Am J Hum Genet. 2006;79(4):640–649. doi: 10.1086/507954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruner DW, Baffoe-Bonnie A, Miller S, et al. Prostate cancer risk assessment program. A model for the early detection of prostate cancer. Oncology (Williston Park) 1999;13(3):325–334. discussion 337-9, 343-4 pas. [PubMed] [Google Scholar]
- 19.National Comprehensive Cancer Network; 2004. 2004. Practice Guidelines in Oncology: Prostate Cancer Early Detection. [DOI] [PubMed] [Google Scholar]
- 20.Eskew LA, Woodruff RD, Bare RL, McCullough DL. Prostate cancer diagnosed by the 5 region biopsy method is significant disease. J Urol. 1998;160(3 Pt 1):794–796. doi: 10.1016/S0022-5347(01)62789-7. [DOI] [PubMed] [Google Scholar]
- 21.Rosser CJ, Broberg J, Case D, Eskew LA, McCullough D. Detection of high-grade prostatic intraepithelial neoplasia with the five-region biopsy technique. Urology. 1999;54(5):853–856. doi: 10.1016/s0090-4295(99)00236-8. [DOI] [PubMed] [Google Scholar]
- 22.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrell F. Regression modeling strategies. New York; Springer; 2001. [Google Scholar]
- 24.Thompson IM, Goodman PJ, Tangen CM, et al. The influence of finasteride on the development of prostate cancer. N Engl J Med. 2003;349(3):215–224. doi: 10.1056/NEJMoa030660. [DOI] [PubMed] [Google Scholar]