Abstract
Prior to and/or accompanying lipolytic degradation of triacylglycerols (TAGs) during seed germination in oilseeds, certain enzymatic and non-enzymatic signaling molecules are expressed on the oil body membranes. These include certain proteases, lipoxygenase, phospholipase A2 and lipase. Although enough biochemical investigations have demonstrated their activities, recent developments in the in situ localization of these signaling molecules in germinating oilseeds, have enhanced our understanding in this field. This is evident from the temporal and spatial changes observed in the expression pattern of some of these molecules. Present review aims at providing an up-to-date account of these recent developments in the author's and other laboratories, which are largely based on fluorescence microscopic and confocal laser scanning microscopic (CLSM) imaging of the molecular changes using specific fluorescent probes. A model for the molecular events associated with oil body mobilization is also being presented.
Key words: confocal laser scanning microscopy, fluorescence microscopy, lipolysis, oil bodies, oilseeds, seed germination
Introduction
Oilseeds store triacylglycerols (TAGs) as food reserves for utilization during seed germination and seedling growth. TAGs are confined to small, spherical organelles called ‘oil bodies’ which are enclosed in a phospholipid monolayer embedded with some unique proteins.1–5 Oil bodies, averaging 0.5–2 µm in diameter, are remarkably stable both in cells and in isolated preparations, as a consequence of the steric hindrance and electronegative repulsion provided by the membrane proteins.6 The most thoroughly characterized proteins embedded in the oil body membranes are the oleosins2,7 and two other minor proteins—caleosins1,8 and steroleosins.3,4 Oleosins are alkaline plant proteins with molecular mass ranging from 15 to 30 kDa.9 Each oleosin molecule comprises of three structural domains: (i) an N-terminal amphipathic domain; (ii) a central, hydrophobic oil-body anchoring domain with a proline knot motif,6,10 and (iii) a C-terminal amphipathic α-helical domain. The central, 70 amino acid long anchoring domain of oleosins is highly conserved in diverse plant species. Presence of oleosins in seed oil bodies is consistent with their role in oil body biogenesis, their stabilization and maintenance of an appropriate size and surface/volume ratio for gradual oil body mobilization.4,11 Caleosins are a recently discovered class of calcium and lipid-binding oil body surface proteins.1,8 Different caleosin isoforms are present on the specific sub-domains of the ER and lipid bodies.12 Like oleosins, caleosins comprise of three domains: an N-terminal with a single Ca2+-binding EF hand domain, a central hydrophobic region and a C-terminal region with several protein kinase phosphorylation sites.13 They are believed to play a role in lipid trafficking and membrane expansion.14 They may also be involved in the docking of peroxisomes to lipid bodies and degradation of storage lipids during seed germination.5,8,15,16 Steroleosins, with a conserved NADPH-binding sub-domain and a divergent sterol-binding sub-domain,4 represent a class of dehydrogenase/reductase, involved in signal transduction events for a specialized function related to seed oil bodies.3 A steroleosin molecule consists of two structural domains: an N-terminal, oil body anchoring domain and sterol binding dehydrogenase domain. Unlike oleosins and caleosins, steroleosins are unable to individually stabilize the oil bodies.17
Upon lipolysis, TAGs stored in oil bodies provide energy and membrane building blocks during early stages of seedling growth in oilseeds.18 This process occurs by the sequential and/or collective action of many hydrolytic enzymes, such as phospholipases, lipoxygenase and lipase, at different stages of seed germination.19–22 Lipase is the principle enzyme involved in oil body mobilization during seed germination, which catalyzes the cleavage of carboxyl ester bonds of TAGs, releasing free fatty acids (FFAs) and glycerol. According to Feussner et al.23 lipoxygenase activity leads to oxygenation of storage TAGs to their corresponding hydroperoxy derivatives in some oilseeds. These derivatives are subsequently cleaved by lipases. But before lipase or lipoxygenase gets access across the oil body membrane for their action on the TAG matrix a controlled proteolytic degradation of oleosin isoforms and lipolytic degradation of phospholipids constituting the oil body membrane, is quite expected. Tzen and Huang,24 observed that trypsin treatment renders oil bodies susceptible to the action of phospholipase A2 or C. Matsui et al.25 observed that trypsin-assisted digestion of oil bodies isolated from the cotyledons of cucumber seedlings renders them more susceptible to lipoxygenase action. Both these reports led to the hypothesis that proteolysis of oil body membrane proteins is a prerequisite for oil body mobilization. Work in the author's laboratory has shown the association of the activities of phospholipase A2 (PLA2) and a cytosolic protease with oil bodies during early stages of seed germination in sunflower.22,26 Lipase is synthesized in the cytosol and gets associated with oil body membrane during seed germination.2,27 Lipase activity has also been shown to be initially localized at its storage sites in the protein storage vacuoles (PSVs) prior to seed germination, and its transfer to oil body surface during early phase of seed germination in sunflower.20 The entire process of oleosin processing and TAG mobilization is greatly enhanced in light, relative to seed germination in dark.28 The probable association of a thiol protease with oil bodies has also been demonstrated by us, leading to gradual degradation of oleosins during seedling growth.29 Maximum protease activity coincides with the phase of active TAG hydrolysis. As a consequence of proteolytic mobilization of oil body membrane proteins and subsequent TAG degradation, the remaining oil body membrane and its contents subsequently tend to fuse with the vacuolar membranes.
The present review aims at bringing together the currently available information on recently exploited novel fluorescence imaging methods for understanding the temporal and spatial distribution and roles of various signaling molecules during seed germination in oilseeds.
Fluorescent Probes for the Localization of Oil Body-Associated Signaling Molecules during Lipolysis
Fluorescence microscopy and confocal image analysis offer a sensitive approach for a rapid and accurate detection of enzyme-catalyzed events on/across the cellular and subcellular membranes. In order to understand the biogenesis and/or degradation of oil bodies, it is necessary to know the course of development and localization of PSVs, and temporal and spatial localization of various enzymes involved in signaling the lipid synthesis and degradation. Nile red and acetomethoxy derivative of 2′,7′-bis′(2-carboxyethyl)-5-(6)′carboxyfluorescien (BCECF-AM) are the probes for detection of the sites of neutral lipid accumulation (principally oil bodies) and PSVs, respectively, during seed development and germination. Some enzymatic proteins are also synthesized during the formation of lipid bodies in maturing seeds while a new set of proteins may be detected on the surface of oil bodies at the time of lipid mobilization during seed germination.23 In this context, specific probes have been exploited in the recent past in the author's laboratory. These include ‘glycerol-derived’ resorufin ester, PED6 and fluorescein mercuric acetate (FMA), for the localization of lipase, PLA2 and thiol protease activities, respectively.
Detection of oil bodies using nile red.
Nile red is phenoxazone probe which fluoresces upon interaction with lipids. It is an uncharged heterocyclic molecule and acts as a hydrophobic probe for lipids. Its fluorescence maximum varies depending on the relative hydrophobicity of the surrounding environment. Nile red-treated samples emit fluorescence ranging from green, golden, yellow, orange-red, red and yellow-gold depending on the nature of phospholipids or other lipids.30 This novel property of nile red makes it a sensitive fluorescent probe for the analysis of tissue lipids.30,31 The excitation and emission maxima of nile red in lipid suspensions also appear to depend on the concentration of the dye. At low concentrations of nile red, both the core hydrophobic and amphipathic components of lipids interact with nile red, giving a shorter wavelength emission maximum of approx. 580 nm. With increasing concentration of the dye, the hydrophobic core may become saturated and the emission maxima tend to shift towards higher wavelengths i.e., 620–650 nm.
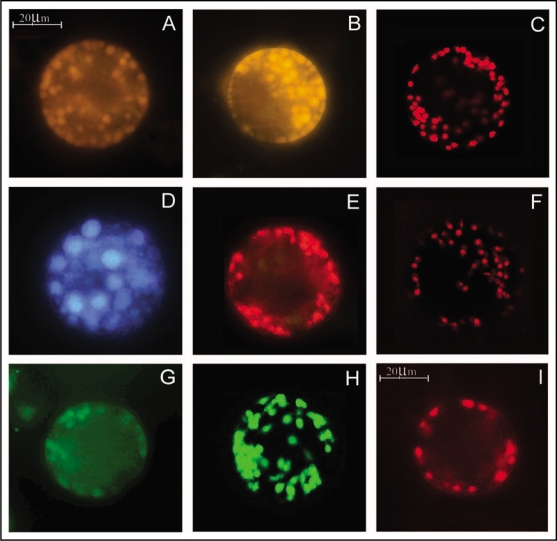
Using these unique properties of nile red, oil bodies and neutral lipids are detectable in the sections32 and protoplasts20,29 from the cotyledons of oilseeds. Variable fluorescence (yellow-green, yellow or red) from nile red treated oil bodies in protoplasts isolated from different seedling tissues (cotyledons, hypocotyls and roots), has been observed in sunflower29 (Fig. 1A) and safflower (Bhatla SC, unpublished; Fig. 1B). This signifies a change in the composition of neutral lipids present in these tissues. Recently, Nile red has also been exploited for confocal imaging of oil bodies in the protoplasts from sunflower seedling cotyledons29 (Fig. 1C) and also in the sections of embryos of sunflower (Bhatla SC, unpublished) and Arabidopsis.32
Figure 1.
Fluorescence microscopic and CLSM detection of signaling molecules in protoplasts from sunflower and safflower cotyledons. (A–C) Fluorescence of variable colors from oil bodies in response to nile red treatment indicating difference in neutral lipid composition. (A) Protoplast from a mature seed cotyledon of sunflower. (B) Fluorescence microscopic image of a protoplast from the cotyledons of imbibed seeds of safflower. (C) A CLSM image of a protoplast from sunflower seedling cotyledon (4 d old). (D) Fluorescence from PSVs in a protoplast from the cotyledons of mature sunflower seed, after treatment with BCECF-AM. (E) Sites of thiol protease activity from oil bodies, as observed by red fluorescence upon FMA treatment of protoplast from 4 d old sunflower seedling cotyledons. (F) A CLSM image of FMA-treated protoplast from sunflower seedling cotyledon (4 d old). (G) Epifluorescence image of a protoplast treated with PED6 for localizing PLA2 activity on the oil bodies. (H) A CLSM image of a sunflower seedling protoplast (4 d old) treated with PED6. (I) Fluorescence microscopic image of lipase activity sites in sunflower protoplast from 4 d old seedling cotyledon after treating with resorufin ester. All protoplasts obtained were from mature seeds or dark-grown seedlings to avoid autofluorescence from chloroplasts.
Localization of protein storage vacuoles.
Storage tissues of many fruits and seed cotyledons contain PSVs. They contain a variety of globulin storage proteins, lectins, acid phosphatases and cysteine and aspartic proteases. The H+-ATPase and H+-pyrophosphatase activities on the tonoplast are responsible for hormone—(principally GA)—induced acidification of PSVs during the phase of reserve mobilization. In the absence of GA action, pH of the lumen of PSVs in the aleurone cells of barley has been reported to be near neutral and it becomes acidic with GA treatment.33 Acidification of lumen of PSVs increases protease activity in it since aspartic and cysteine proteases have pH optima near 4.34 Thus, pH of the PSVs' lumen plays a critical role in regulating the various metabolic activities operating within and through them.
Keeping in view the above stated biochemical features of PSVs, the in situ localization of PSVs in plant cells and protoplasts is largely based on measuring the fluorescence from pH sensitive and aspartic and/or cysteine protease-specific probes loaded as cell permeant derivatives or by microinjection.34 ZFR-CMAC (7-amino-4-chloromethyl coumarin benzyloxycarbonoyl-L-phenylalanyl-L-arginine amide hydrochloride) is a non-fluorescent protease substrate used to identify regions of proteolytic activity in plant protoplasts.34 It releases fluorescent CMAC when the peptide bond between the CMAC fluorophore and arginine residue is cleaved. This probe can be loaded into the cells/protoplasts non-invasively and is not cytotoxic. Its transport into the PSVs is facilitated by conjugation with glutathione in the cytosol.
BCECF and Lysosensor yellow/blue are the two commonly used pH-sensitive probes to localize PSVs and determine their lumen pH status. Lysosensor yellow/blue (pKa = 4.2) emits yellow/green fluorescence at pH 3.5, blue/green fluorescence at pH 4.5, pale blue fluorescence at pH 5.5 and dim-blue fluorescence near neutral pH. BCECF-AM is a non-fluorescent lipophilic acetoxymethyl ester that readily enters the cells. Inside the cell, the ester gets cleaved by esterases producing fluorescent BCECF anions which get actively sequestered into the vacuoles. BCECF anions emit blue fluorescence upon excitation at 482 nm (em: 520 nm). BCECF-AM is widely used in cellular and pharmacological studies without significant toxicity. Leakage of BCECF out of the cells is negligible, thereby greatly reducing the background fluorescence. Accumulation of both glutathione-conjugated fluorescent probe (ZFR-CMAC) and pH-sensitive probes (e.g., BCECF-AM) can be blocked by using probenacid, an inhibitor of organic anion transport.
It is necessary to monitor the development and mobilization of PSVs in germinating oilseeds not only because they are a store house of various proteins and enzymes but also because PSVs have been reported to store lipase prior to its migration to oil body surface during seed germination in oilseeds. Using BCECF-AM as a probe for fluorescence microscopy of protoplasts, changes in the size and number of PSVs have been reported in sunflower, cotton and peanut.35 Protoplasts from the young sunflower seedling cotyledons show a number of PSVs of variable sizes (Fig. 1D). With advancing age of the seedlings, PSVs coalesce into a larger vegetative vacuole which are depleted of their contents.35
Localization of thiol-protease activity.
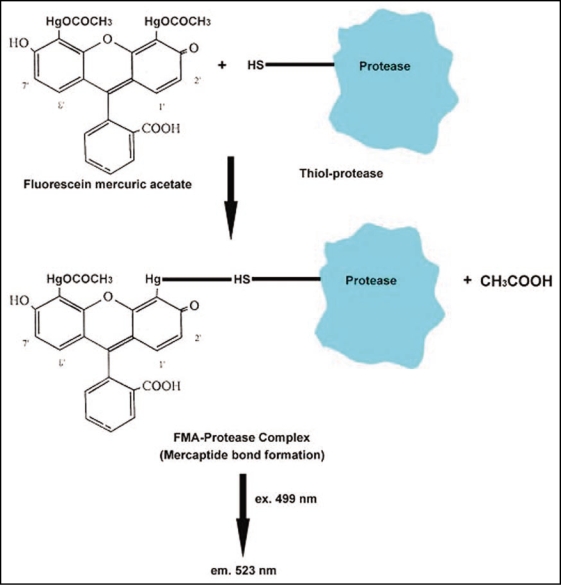
Spatial distribution of protein degradation enzymes can enhance specificity and timing of proteolysis, generate asymmetry and maintain subcellular compartmentation of protease activity. FMA is known to be a highly fluorescent reagent which reacts with thiol-groups. It is also a SH-protease inhibitor and its fluorescence is quenched upon binding with proteins containing thiol groups.36 This reagent is, therefore, useful for the analysis of intracellularly localized thiol groups.37 FMA is believed to catalyze conformational changes in the SH-proteases by forming a mercaptide bond with a cysteine sulphahydryl group (Fig. 2). It has also been explored as a possible reagent for ultrastructural detection of SH-proteases in animal systems.38
Figure 2.
Mechanism of binding of FMA with thiol protease, leading to detection of FMA-protease complex by fluorescence microscopy and CLSM.
A simple and highly sensitive fluorometric method for the determination of SH-proteases using FMA, was recently developed by Morenkova and Nagler.39 This method allows the determination of the amount of SH-groups up to 0.1 nmol/mg proteins. FMA has been used in the author's laboratory to localize thiol protease activity from sunflower seedling cotyledon by in vivo labeling of viable protoplasts.29 It involves the uptake and binding of FMA at the intracellular thiol-protease activity sites in protoplasts, leading to fluorescence emission at 523 nm, following excitation at 499 nm (Fig. 2). The observations provide evidence for the expression of the said thiol-protease activity on the oil body surface, leading to gradual proteolysis of oleosins during seed germination (Fig. 1E and F). Biochemical investigations in the author's laboratory have demonstrated the activity of a 65 kDa, cytosolic protease from sunflower seedling cotyledons, coinciding with the degradation of oleosins during seed germination.22 Further investigations from this laboratory have also indicated the probable association of a thiolprotease with oil bodies, leading to gradual degradation of oleosins during seedling growth.29
Detection of PLA2 activity.
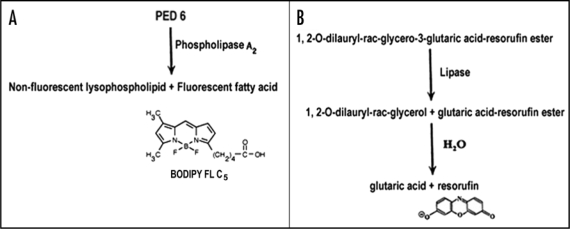
Phospholipases play a pivotal role in releasing a diverse array of lipid-derived products by degrading membrane phospholipids. PLA2 is an acyl hydrolase that catalyzes phospholipid hydrolysis at the sn-2 position, thereby generating a fatty acid and a lysophospholipid. A transient, temporal and tissue specific expression of PLA2 activity is associated with oil bodies during seed germination in cucumber.26 Hendrickson et al.40 developed PED6-a PLA2 specific substrate analog. PED6 incorporates a BODIPY FL-dye-labeled-sn-2-acyl chain and a dinitrophenyl quencher group. Cleavage of this PED6-labeled acyl chain by PLA2 results in the formation of non-fluorescent lysophospholipid and fluorescent fatty acid tagged to the fluorophore-Bodipy FL (Fig. 3A). This product can be localized by fluorescence upon excitation at the appropriate wavelength (505 nm). In animal system, PED6 has been used for in vivo assay of intestinal PLA2 metabolism in zebra fish larvae.41 This probe is reported to have affinity for both cytosolic and secretory PLA2 activities.42 Thus, this method will prove useful to localize intracellular PLA2 activity of both kinds in plant cells as well.
Figure 3.
(A) Mechanism of localization of intracellular PLA2 activity sites, using a synthetic substrate (PED6). (B) Mechanism of intracellular lipase action on plant cells/protoplasts incubated with a ‘glycerol-derived’ resorufin ester (synthetic substrate for lipase), leading to the release of fluorescent resorufin at the sites of lipase activity.
Recently, using PED6, a novel protocol has been developed in the author's laboratory for in situ localization of development-associated changes in the expression of PLA2 activity on the surface of oil bodies in protoplasts from sunflower seedling cotyledons.21 Intracellular PED6-binding sites fluoresce bright green upon excitation at 505 nm. This work reports the first use of a fluorogenic substrate to localize PLA2 activity in plant protoplasts. The principal intracellular sites exhibiting PLA2 activity in the oilseed cotyledon protoplasts are the oil bodies and the enzyme is apparently expressed or activated on the surface of oil bodies during early phase of seedling development, prior to lipase action (Fig. 1G and H). PED6 fluorescence is also evident in the cytoplasm surrounding the oil bodies, along with oil bodies themselves. This indicates the possibility of preferential accumulation of PLA2 on/around the oil bodies for the degradation of the oil body membranes at a specific stage of seed germination. A PLA2 might possibly be causing alteration or hydrolysis of the oil body membrane phospholipids, rendering the oil bodies susceptible to the action of lipases, leading to TAG mobilization from the oil body matrix during seed germination. The observation of PED6 affinity sites (PLA2 activity) in the protoplasts from sunflower seedling cotyledons has been made in the author's laboratory, using epifluorescence microscopy and confocal laser scanning microscopy (CLSM).21
Fluorometric and fluorescence microscopic methods for localizing lipase activity.
The principal enzyme associated with oil body mobilization during seed germination is lipase (triacylglycerol acylhydrolase, E.C 3.1.1.3), which catalyzes the hydrolysis of carboxyl ester bonds of TAGs, releasing FFAs and glycerol. Most lipases have optimum activity for the primary ester groups of TAGs. While some lipases remove fatty acyl groups from either the C1 or C3 positions, others remove both C1 and C3 acyl groups. The first fluorometric determination of lipase activity was undertaken by the lipase-catalyzed hydrolysis of fluorescein esters.43 Ishiyama et al.44 used a synthetic substrate—a resorufin derivative (7-isobutyloxycarbonyloxy-3 H-phenoxazin-3-one) for the fluorometric determination of viability in human leukemia cells (HL 60) and mouse lymphocytes (P 388). This resorufin derivative is non-fluorescent with an absorption maximum below 500 nm but produces an intense fluorescence at 590 nm when cleaved by endogenous esterases at pH 7.4. Resorufin (in primary alcohols) exists in dissociated form as free anion (R−), with absorption maxima at 580 nm and prominent emission at 600 nm.45 The emission corresponds to that of resorufin and fluorescence is due to the deprotonated anion of resorufin.
Resorufin derivatives permeate the membranes of viable cells because of their lipophilicity and get hydrolyzed by intracellular esterases to produce fluorescence. Beisson et al.46,47 assayed lipase activity from the crude tissue extracts of Arabidopsis seedlings, using three sensitive methods (radioactivity, fluorescence and colorimetry). Specific activity of the enzyme (lipase) was found to be similar with all the three methods. Unlike the radioactivity assay, however, the fluorescence assay can be monitored continuously. Hydrolysis of a synthetic, “glycerol-derived” lipase substrate-(1,2,o-diluaryl-rac-glycero-3-glutaric acid-resorufin ester) involves a series of steps, starting with a lipase-catalyzed formation of 1,2-o-dilauryl glycerol and glutaric acid-resorufin ester, followed by the spontaneous intramolecularly-assisted hydrolysis of the latter product to release fluorescent resorufin (Fig. 3B). Time course of the reaction corresponds to the release of resorufin, monitored at 572 nm.48 Sensitive monitoring of lipase activity in live cells has also been accomplished using the fluorescent substrate-1,2-dioleolyl-3-(pyren-1-yl) decanoyl-rac-glycerol (Marker Gene Technologies, Inc., USA). Upon enzymatic cleavage, the fluorescent fatty acid, pyrenedecanoic acid, is released at the site of lipase action. The fluorescence of the product shifts to a longer wavelength and can be distinguished from that of the substrate, because it forms eximers inside the membrane (ex: 390 nm, em: 470 nm).
Measurement of lipase activity is sometimes difficult in tissue homogenates of oilseeds due to lipase inhibitors released during tissue homogenization.49,50 In order to overcome this problem, a novel method has been devised in the author's laboratory for in vivo localization of lipase activity in plant cells.35 The non-destructive nature of this methodology overcomes the problem of lipase inhibitors released during tissue homogenization. Using this protocol, lipase activity is detectable in protoplasts from oilseeds. A ‘glycerol-derived’ resorufin ester (1,2,o-dilauryl-rac-glycero-3-glutaric acid-resorufin ester), used for assaying lipase activity in mammalian, fungal and plant tissue extracts.46,47,51,52 was adopted for localizing lipase activity in protoplasts from sunflower seedling cotyledons by fluorescence microscopy (Fig. 1I).20,35 The resorufin ester, being lipophilic, permeates plasma membranes of plant protoplasts. Uncleaved lipase substrate, i.e., resorufin ester, is non-fluorescent in solution. The resorufin ester is hydrolyzed by intracellular lipase, producing fluorescence due to resorufin formation at its intracellular locations (λexmax: 569 nm, λemmax: 584 nm).
Using this probe (resorufin ester), fluorescence due to lipase activity on the oil bodies in protoplasts from the cotyledons of germinating seeds of peanut and cotton has also been reported from the author's laboratory.35 Thus, resorufin acts as a marker of lipasespecific intracellular locations. Excitation and fluorescence emission of resorufin also exhibit a slight shift in λmax for excitation and emission when analyzed in intact protoplasts or in ethanolic solutions and also a pH-dependent shift.20 Investigations from the author's laboratory were further employed to investigate the spatial and temporal changes in lipase activity sites during oil body mobilization in protoplasts from sunflower seedling cotyledons during seed germination.20 Subcellular distribution of lipase activity in the seedling cotyledons was observed to shift from PSVs to oil bodies, with advancing stages of seedling development.
A Model for Signaling Events Accompanying Lipolysis in Oilseeds during Germination
A model depicting the expression of the probable signaling molecules on the oil body membranes in light and dark-grown seedlings, is presented in Figure 4. It is based on the current knowledge and the author's recent findings on sunflower as a model system for localization of the various oil body-associated molecules, using specific fluorescent probes. The model shows that an imbibed seed oil body possesses the required components of oil body membrane proteins i.e., oleosins, caleosins and steroleosins, on the monolayer of phospholipids. This is followed by the action of protease, lipoxygenase and lipase which are transiently expressed on the oil body surface by migration from other intracellular locations. Imaging of these molecular events by fluorescence microscopy has provided evidence for an earlier expression of protease than any other enzyme. Subsequent events (PLA2,lipoxygenase and lipase action) seem to be occurring soon thereafter more or less simultaneously, but the role of caleosins and steroleosins in this phase of lipolysis still needs to be investigated. Dark-grown seedlings exhibit a lower activity of these enzymes then those grown in light. Investigations on the photomodulation component of the above-stated lipolysis-associated signaling molecules are also likely to bring to light significant findings.
Figure 4.
A model depicting the biomolecular events on the surface of oil bodies from imbibed seeds and young seedling cotyledons (light and dark-grown) during seed germination in sunflower.
Future Scope
Significant progress has been made in the development of fluorescent probes and protocols for the temporal and spatial localization of oil bodies, PSVs, PLA2, thiol proteases and lipase activity during seed germination in oilseeds. Further work is, however, required to be undertaken for developing protocols for imaging the pattern of expression of lipoxygenase, steroleosins and caleosins. Current investigations in the author's laboratory are focusing on developing the protocols for imaging the possible changes in the expression pattern of calcium channel proteins as well as co-localization of physiologically and biochemically compatible fluorescent probes, for deciphering the various signaling molecules associated with oilseed biogenesis and germination. Such and similar future investigations are likely to highlight more signaling molecules having a possible role during lipolysis in oilseeds. Lastly, all the probes under present and future investigations need to be further investigated for exploitation using suitable laser lines of CLSM. Protein-protein interaction also needs to be investigated on the oil body membranes, using techniques like fluorescence resonance energy transfer (FRET), coupled with multiphoton fluorescence lifetime imaging. This is likely to provide crucial information on the oil body membrane protein interaction with various signaling proteins such as proteases.
Acknowledgements
Thanks are due to Alexander von Humboldt Foundation (Germany) for fluorescence microscope facility installed in the laboratory of S.C.B. Confocal images from the author's laboratory were made possible on the CLSM facility in the Department of Botany, Delhi University, under the FIST programme of the Department of Science and Technology, New Delhi. Senior research fellowship from CSIR, New Delhi to V.K., is gratefully acknowledged.
Abbreviations
- BCECF-AM
2′,7′-bis'(2-carboxyethyl)-5-(6)′-carboxyfluorescein acetomethoxy derivative
- CLSM
confocal laser scanning microscope
- FFAs
free fatty acids
- FMA
fluorescein mercuric acetate
- FRET
fluorescence resonance energy transfer
- PED6
N-((6-(2,4-dinitrophenyl)amino)hexanoyl)-2-(4,4difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine,triethylammonium salt
- PLA2
phospholipase A2
- PSVs
protein storage vacuoles
- TAGs
triacylglycero
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7799
References
- 1.Frandsen GI, Mundy J, Tzen JTC. Oil bodies and their associated proteins, oleosin and caleosin. Physiol Plant. 2001;112:301–307. doi: 10.1034/j.1399-3054.2001.1120301.x. [DOI] [PubMed] [Google Scholar]
- 2.Huang AHC. Oil bodies and oleosins in seeds. Ann Rev Plant Physiol Plant Mol Biol. 1992;43:177–200. [Google Scholar]
- 3.Lin LJ, Tai SSK, Peng CC, Tzen JTC. Steroleosin, a sterol-binding dehydrogenase in seed oil bodies. Plant Physiol. 2002;128:1200–1211. doi: 10.1104/pp.010982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin LJ, Tzen JTC. Two distinct steroleosins are present in seed oil bodies. Plant Physiol Biochem. 2004;42:601–608. doi: 10.1016/j.plaphy.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Murphy DJ. Lipid associated proteins. In: Murphy DJ, editor. Plant Lipids: Biology, Utilization and manipulation. USA: CRC Press; 2005. pp. 226–269. [Google Scholar]
- 6.Jaing PL, Wang CS, Hsu CM, Jauh GY, Tzen JTC. Stable oil bodies sheltered by a unique oleosin in lily pollen. Plant Cell Physiol. 2007;48:812–821. doi: 10.1093/pcp/pcm051. [DOI] [PubMed] [Google Scholar]
- 7.Murphy DJ, Li M, Keddie JS, Smith LJ, Clark DC. Expression and characterization of the N-terminal domain of an oleosin protein from sunflower. J Biol Chem. 1993;268:17504–17512. [PubMed] [Google Scholar]
- 8.Chen JCF, Tsai CCY, Tzen JTC. Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol. 1999;40:1079–1086. doi: 10.1093/oxfordjournals.pcp.a029490. [DOI] [PubMed] [Google Scholar]
- 9.Murphy DJ, Hernendez-Pinzon I, Patel K. Roles of lipid bodies and lipid-body proteins in seeds and other tissues. J Plant Physiol. 2001;158:471–478. [Google Scholar]
- 10.Lacey DJ, Wellner N, Beaudoin F, Napier JA, Shewry PR. Secondary structure of oleosins in oil bodies isolated from seeds of safflower (Carthamus tinctorius L.) and sunflower (Helianthus annuus L.) Biochem J. 1998;334:469–477. doi: 10.1042/bj3340469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capuano F, Beaudoin F, Johnathan A, Napier A, Shewry PR. Properties and exploitation of oleosins. Biotechnol Adv. 2007;25:203–206. doi: 10.1016/j.biotechadv.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Murphy DJ, Hernendez-Pinzon I, Patel K, Hope RG, Mc Lauchlan J. New insights into the mechanisms of lipid-body biogenesis in plants and other organisms. Biochem Soc Trans. 2000;28:710–711. [PubMed] [Google Scholar]
- 13.Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM. Role of the proline knot motif in oleosin-endoplasmic reticulum topology and oil body targeting. Plant Cell. 1997;9:1481–1493. doi: 10.1105/tpc.9.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin LJ, Liao PC, Yang HH, Tzen JTC. Determination and analyses of the N-termini of oil-body proteins, steroleosin, caleosin and oleosin. Plant Physiol Biochem. 2005;43:770–776. doi: 10.1016/j.plaphy.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Naested H, Frandsen GI, Jauh G-Y, Hernandez-Pinzon I, Neilsen HB, Murphy DJ, et al. Caleosins: Ca2+ binding proteins associated with lipid bodies. Plant Mol Biol. 2002;44:463–476. doi: 10.1023/a:1026564411918. [DOI] [PubMed] [Google Scholar]
- 16.Poxleitner M, Rogers SW, Samuels AL, Browse J, Rogers JC. A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J. 2006;47:917–933. doi: 10.1111/j.1365-313X.2006.02845.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen MC, Chan CL, Lee TT, Huang SH, Tzen JT. Constitution of stable artificial oil bodies with triacylglycerol, phospholipid and caleosin. J Agri Food Chem. 2004;52:3982–3987. doi: 10.1021/jf035533g. [DOI] [PubMed] [Google Scholar]
- 18.Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. 1999;24:109–115. doi: 10.1016/s0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- 19.Eastmond PJ. SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell. 2006;18:665–675. doi: 10.1105/tpc.105.040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A, Bhatla SC. Spatial and temporal changes in the lipase activity sites during oil body mobilization in protoplasts from sunflower seedling cotyledons. Plant Growth Regul. 2005;46:11–17. [Google Scholar]
- 21.Gupta A, Bhatla SC. Preferential phospholipase A2 activity on the oil bodies in cotyledons during seed germination in Helianthus annuus L. cv. Morden. Plant Sci. 2007;172:535–543. [Google Scholar]
- 22.Sadeghipour HR, Bhatla SC. Differential sensitivity of oleosins to proteolysis during oil body mobilization in sunflower seedlings. Plant Cell Physiol. 2002;43:1117–1126. doi: 10.1093/pcp/pcf142. [DOI] [PubMed] [Google Scholar]
- 23.Feussner I, Kuhn H, Wasterneck C. Lipoxygenase-dependant degradation of storage lipids. Trends Plant Sci. 2001;6:268–273. doi: 10.1016/s1360-1385(01)01950-1. [DOI] [PubMed] [Google Scholar]
- 24.Tzen JTC, Huang AHC. Surface structure and properties of plant seed oil bodies. J Cell Biol. 1992;117:327–335. doi: 10.1083/jcb.117.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsui K, Hijiya K, Tabuchi Y, Kajiwara T. Cucumber cotyledon lipoxygenase during postgerminative growth: Its expression and action on lipid bodies. Plant Physiol. 1999;119:1279–1287. doi: 10.1104/pp.119.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.May C, Preisig-Muller R, Hohne M, Gnau P, Kindl H. A phospholipase A2 is transiently synthesized during seed germination and localized on lipid bodies. Biochim Biophys Acta. 1998;1393:267–276. doi: 10.1016/s0005-2760(98)00081-2. [DOI] [PubMed] [Google Scholar]
- 27.Beisson F, Ferte N, Bruley S, Voultoury R, Verger R, Arondel V. Oil bodies as substrate for lipolytic enzymes. Biochim Biophys Acta. 2001;1531:47–58. doi: 10.1016/s1388-1981(01)00086-5. [DOI] [PubMed] [Google Scholar]
- 28.Sadeghipour HR, Bhatla SC. Light-enhanced oil body mobilization in sunflower seedlings accompanies faster protease action on oleosins. Plant Physiol Biochem. 2003;41:309–316. [Google Scholar]
- 29.Vandana S, Bhatla SC. Evidence for the probable oil body association of a thiol-protease, leading to oleosin degradation in sunflower seedling cotyledons. Plant Physiol Biochem. 2006;44:714–723. doi: 10.1016/j.plaphy.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 30.Greenspan P, Fowler SD. Spectrofluorometric studies of the lipid probe, Nile red. J Lipid Res. 1985;26:781–789. [PubMed] [Google Scholar]
- 31.Fowler SD, Greenspan P. Application of nile red, a fluorescent hydrophobic probe, for the detection of neutral lipid deposits in tissue sections: comparison with oil red O. J Histochem Cytochem. 1985;33:833–836. doi: 10.1177/33.8.4020099. [DOI] [PubMed] [Google Scholar]
- 32.Siloto RMP, Findlay K, Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. The accumulation of oleosins determines the size of seed oil bodies in Arabidopsis. Plant Cell. 2006;18:1961–1974. doi: 10.1105/tpc.106.041269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson SJ, Jones RL. Gibberellic acid induces vacuolar acidification in barley aleurone. Plant Cell. 1996;8:2211–2221. doi: 10.1105/tpc.8.12.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanson SJ, Bethke PC, Jones RL. Barley aleurone cells contain two types of vacuoles: characterization of lytic organelles by use of fluorescent probes. Plant Cell. 1998;10:685–698. doi: 10.1105/tpc.10.5.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta A, Sadeghipour HR, Bhatla SC. Subcellular detection of lipase activity in plant protoplasts using fluorescence microscopy. Plant Growth Regul. 2003;41:259–264. [Google Scholar]
- 36.Molla A, Yamamoto T, Maeda H. Characterization of 73 kDa thiol-protease from Serratia marcescens and its effect on plasma proteins. J Biochem. 1988;104:612–616. doi: 10.1093/oxfordjournals.jbchem.a122521. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi S, Maeda A. Use of fluorescein mercuric acetate as a probe in studies of thiol-containing protein. J Bochem. 1977;81:971–976. doi: 10.1093/oxfordjournals.jbchem.a131563. [DOI] [PubMed] [Google Scholar]
- 38.Biggiogera M, Pellicciari C. Fluorescein mercuric acetate as a dye for fluorescence and electron microscopic detection of—SH group in mouse sperm protamines: a critical evaluation. Acta Histochem. 1988;83:1–10. doi: 10.1016/S0065-1281(88)80064-3. [DOI] [PubMed] [Google Scholar]
- 39.Morenkova SA, Nagler LG. Fluorometric method for determination of keratin SH-groups in human epidermis. Biomed Khim. 2005;51:220–223. [PubMed] [Google Scholar]
- 40.Hendrickson HS, Hendrickson EK, Johnson ID, Farber SA. Intramolecularly quenched BODIPY-labeled phospholipids analogs in phospholipase A2 and platelet-activating factor acetyl hydrolase assays and in vivo fluorescence imaging. Anal Biochem. 1999;276:27–35. doi: 10.1006/abio.1999.4280. [DOI] [PubMed] [Google Scholar]
- 41.Farber SA, Pack M, Ho S-Y, Johnson ID, Wagner DS, Dosch R, et al. Genetic analysis of digestive physiology using fluorescent phospholipids reporters. Science. 2001;292:1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- 42.Haugland RP. Handbook of Fluorescent Probes and Research Products. 9th Edn. USA: Molecular probes; 2002. p. 736. [Google Scholar]
- 43.Kramer DN, Guilbault GG. A substrate for the fluorometric determination of lipase activity. Anal Chem. 1963;35:588–589. [Google Scholar]
- 44.Ishiyama M, Furusama H, Shiga M, Ohseto F, Sasamoto K. A resorufin derivative as a fluorogenic indicator for cell viability. Anal Sci. 1999;15:1025–1028. [Google Scholar]
- 45.Rasimas JP, Blanchard GJ. A time resolved spectroscopic study of solution phase ionic association and dissociation. J Phys Chem. 1996;100:11526–11533. [Google Scholar]
- 46.Beisson F, Arondel V, Verger R. Assaying Arabidopsis lipase activity. Biochem Soc Trans. 2000;28:773–775. doi: 10.1042/0300-5127:0280773. [DOI] [PubMed] [Google Scholar]
- 47.Beisson F, Tiss A, Riviere C, Verger R. Methods for lipase detection and assay: a critical review. Eur J Lipid Sci Technol. 2000;102:133–153. [Google Scholar]
- 48.Bothner C, Chanez R, Wei J, Strupp C, Phung Q, Schneemann A, et al. Monitoring enzymes and catalysis with mass spectrometry. J Biol Chem. 2000;275:13455–13459. doi: 10.1074/jbc.275.18.13455. [DOI] [PubMed] [Google Scholar]
- 49.Chapman JRGW. A proteinaceous competitive inhibitor of lipase isolated from Helianthus annuus seeds. Phytochem. 1987;26:3127–3131. [Google Scholar]
- 50.Wang SM, Huang AHC. Inhibitors of lipase activities in soybean and other oilseeds. Plant Physiol. 1984;76:929–934. doi: 10.1104/pp.76.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ncube J, Aldercreutz PJ, Mattiasson B. Purification of Rape (Brassica napus) seedling lipase and its use in organic media. Biotechnol Appl Biochem. 1993;17:327–336. [Google Scholar]
- 52.Teissere M, Borel M, Cailol B, Nari J, Gardies AM, Noat G. Purification and characterization of a fatty acid acyl-ester hydrolase for post germinated sunflower seeds. Biochim Biophys Acta. 1995;1255:105–112. doi: 10.1016/0005-2760(94)00222-k. [DOI] [PubMed] [Google Scholar]