Abstract
Cryptochromes (CRYs) are photoreceptors mediating developmental responses to blue light throughout the life of plants. Function and signal transduction of CRYs in photomorphogenesis have been well characterized in Arabidopsis. Studies on rice CRYs demonstrate that monocots CRYs may function similarly to their Arabidopsis counterparts. However, there is inconsistency in subcellular localization of CRYs in different species and little has been known about the effects of environmental cues on CRYs except for light. We recently reported that TaCRY1a of monocot wheat displays a light-responsive nucleocytoplasmic shuttling pattern similar to Arabidopsis CRY1 but differs from AtCRY1 and OsCRY1 by containing nuclear localization domains in both its N and C termini and the sequence for nuclear export in its N-terminal domain. TaCRY1a and TaCRY2 are transcriptionally regulated by osmotic stress/ABA and overexpression of TaCRY1a-GFP and TaCRY2-GFP led to higher sensitivity to high salinity, osmotic stress and ABA treatment. Mining wheat EST database provided additional clues for CRY's involvement in pathways apart from photomorphogenesis.
Key words: cryptochrome, signal transduction, stress, subcellular localization, wheat
Blue light receptor cryptochorome proteins (CRYs) in model plant species have been extensively investigated for their roles in photomorphogenesis and photoperiodic timing. Arabidopsis CRY1 (AtCRY1) is the primary receptor mediating blue light regulation of seedling de-etiolation and circadian clock;1–3 AtCRY2 works redundantly with AtCRY1 under relatively low light and plays a key role in the control of photoperiodic flowering.4,5 Rice CRYs function similarly to their Arabidopsis counterparts and their corresponding N-terminal domains are functionally interchangeable.6–8 This indicates conserved CRY signaling in monocots and dicots. However, the underlying mechanisms may not be identical. For example, AtCRY2 is a constitutively nuclear localized protein and AtCRY1 shuttles between nucleus and cytoplasm in a light-dependant manner;9–11 while rice CRY1 (OsCRY1) distributes both in nucleus and cytoplasm irrespective of the light condition.6 This raises an interest to explore the subcellular localization of CRYs in more plant species and its relation to the functions.
In a recent paper,12 we reported the cloning and characterization of two CRY genes, TaCRY1a and TaCRY2, from hexaploid wheat, an important monocot cereal crop. We presented evidence that TaCRY2, like AtCRY2,9,10,13 is a nuclear protein degrading when exposed to light. Similar to AtCRY1, TaCRY1a is a light-dependent nucleocytoplasmic shuttling protein. But they differ in that TaCRY1a possesses nuclear localization domains in both its N and C termini and carries sequence for nuclear export in its N-terminal domain. Through examining sub-cellular distributions of various TaCRY1a segment-GFP fusions, we determined that the N-terminal segment (AA136-260) is important for both nuclear targeting and blue light responsive nuclear export. Interestingly, even though the AA136-260 segment alone is capable for nuclear import and export, a complete N-terminal domain is necessary for nuclear export when the highly hydrophilic C-terminus of TaCRY1a exists.12 These results, together with other reports, demonstrate that plant CRYs utilize complicated mechanisms to regulate their subcellular localization. A few studies have indicated that CRY homodimerization14 and partner binding8,15 might be required for correct subcellular localization of Arabidopsis and rice CRY1 proteins.
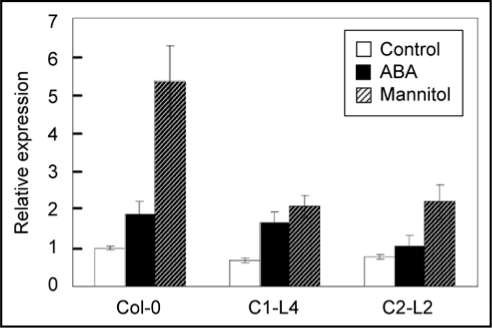
Growth and development of plants is the expression of interactions between genes and the environment. It has been known that light quality16,17 and circadian clock17,18 affect the expression of CRY genes. To investigate whether environmental stresses affect the behavior of CRYs, we examined the expression of TaCRY1a and TaCRY2 under various treatments. TaCRY1a and TaCRY2, especially the latter, are clearly transcriptionally regulated by osmotic stress and ABA in roots and germinating embryos. By analyzing the responses of transgenic Arabidopsis to osmotic stress, we found that overexpression of TaCRY1a or TaCRY2 resulted in decreased seed germination, impaired cotyledon opening, and less tolerance to ABA and high salinity at the vegetative growth stage, suggesting that the high level of CRY expression compromises plant resistance to osmotic stress. In the attempt to uncover the mechanisms by which CRY signals affect osmotic stress/ABA responses, we examined in the transgenic lines the expression of some marker genes for stress responses, and found that the upregulation of both RD29A and ADH1 under osmotic stress/ABA was impaired.12 Since their upregulation has been positively associated with stress tolerance,19,20 their decreased induction is in accordance with the increased susceptibility of these transgenic lines to osmotic stresses. Besides these two genes, a few ABA biosynthesis/metabolism-related genes in the transgenic lines were also examined for their expression under the osmotic stress/ABA treatment. ABA2, an ABA biosynthesis fine-tuning gene, was less induced under the ABA treatment in the line overexpressing TaCRY1a-GFP and under the mannitol treatment in lines overexpressing either TaCRY1a-GFP or TaCRY2-GFP (Fig. 1).
Figure 1.
Expression of ABA2 in Col-0 and transgenic Arabidopsis lines overexpressing TaCRY genes. Total RNA was extracted from 15-day-old T2 transgenic seedlings grown on MS plates (control), on MS plates for 12 days and then on MS plates supplemented with 300 mM mannitol or 10 µM ABA for three days. Error bars indicate SD. Col-0: wild type; C1-L4: TaCRY1a-GFP overexpressing line; C2-L2: TaCRY2-GFP overexpressing line.
Involvement of CRY proteins in stress responses may exist in a broader extent. By mining wheat EST database with 1,050,000 ESTs (Genbank 159th release) from tissues under various growth conditions, we found that the TaCRY2 transcript in roots has ∼tenfold increase under desiccation compared with that under normal growing conditions. In a 24267-clone cDNA library prepared with crown and leaf tissues after a long exposure to low temperature, the TaCRY1a ESTs increased 7-fold. TaCRY2 EST frequency also increased in leaves after septoria tritici infection. Thus, the crosstalk of CRY signalling with other pathways should be more extensively explored.
Acknowledgements
This project was partially supported by ‘863’ program (2006AA10A104), NSFC program (30430440 and 30025030), Outstanding Youth Funds of MOE, GCP Project (SP2-1), and ‘111’ project (Bo8025).
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7756
References
- 1.Lin C, Ahmad M, Cashmore AR. Arabidopsis Cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 1996;10:893–902. doi: 10.1046/j.1365-313x.1996.10050893.x. [DOI] [PubMed] [Google Scholar]
- 2.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 3.Somers DE, Devlin PF, Kay SA. Phytochromes and Cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 4.Lin C, Yang H, Guo H, Mockler TC, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor Cryptochrome 2. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of cry2. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto N, Hirano T, Iwasaki T, Yamamoto N. Functional analysis and intracellular localization of rice cryptochromes. Plant Physiol. 2003;133:1494–1503. doi: 10.1104/pp.103.025759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirose F, Shinomura T, Tanabata T, Shimada H, Takano M. Involvement of rice cryptochromes in de-etiolation responses and flowering. Plant Cell Physiol. 2006;47:915–925. doi: 10.1093/pcp/pcj064. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YC, Gong SF, Li QH, Sang Y, Yang HQ. Functional and signalling mechanism analysis of rice CRYPTOCHROME 1. Plant J. 2006;46:971–983. doi: 10.1111/j.1365-313X.2006.02753.x. [DOI] [PubMed] [Google Scholar]
- 9.Guo H, Duong H, Ma N, Lin C. The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J. 1999;19:279–287. doi: 10.1046/j.1365-313x.1999.00525.x. [DOI] [PubMed] [Google Scholar]
- 10.Kleiner O, Kircher S, Harter K, Batschauer A. Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J. 1999;19:289–296. doi: 10.1046/j.1365-313x.1999.00535.x. [DOI] [PubMed] [Google Scholar]
- 11.Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR. The C-termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 12.Xu P, Xiang Y, Zhu HL, Xu HB, Zhang ZZ, Zhang CQ, et al. Wheat cryptochromes: subcellular localization and involvement in photomorphogenesis and osmotic stress responses. Plant Physiol. 2009;149:760–774. doi: 10.1104/pp.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu XH, Klejnot J, Zhao XY, Shalitin D, Maymon M, Yang HY, et al. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell. 2007;19:3146–3156. doi: 10.1105/tpc.107.053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, et al. Arabidopsis cryptochrome 1 N-terminal domain-mediated homodimerization is required for its photoreceptor activity. Plant Cell. 2005;17:1569–1580. doi: 10.1105/tpc.104.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang HY, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- 16.Chatterjee M, Sharma P, Khurana JP. Cryptochrome 1 from Brassica napus is upregulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol. 2006;141:61–74. doi: 10.1104/pp.105.076323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platten JD, Foo E, Foucher F, Hecht V, Reid JB, Well JL. The Cryptochrome gene family in pea includes two differentially expressed CRY2 genes. Plant Mol Biol. 2005;59:683–696. doi: 10.1007/s11103-005-0828-z. [DOI] [PubMed] [Google Scholar]
- 18.Toth R, Kevei E, Hall A, Millar AJ, Nagy F, Kozma-Bognar L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conley TR, Peng HP, Shih MC. Mutations affecting induction of glycolytic and fermentative genes during germination and environmental stresses in Arabidopsis. Plant Physiol. 1999;119:599–608. doi: 10.1104/pp.119.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:17306–17311. doi: 10.1073/pnas.0407758101. [DOI] [PMC free article] [PubMed] [Google Scholar]