Abstract
Rice (Oryza sativa L.) is a semiaquatic plant that forms adventitious root primordia at the stem nodes as part of normal development. Upon flooding, roots emerge, preceded by local death of epidermal cells above the root primordia. Cell death is strictly confined to cells above root primordia and is induced by ethylene and H2O2. These pro-death signals regulate expression of the transcription factor genes ANT-like, OsARF2, OsARF3 and Hox9 which have a proposed function in cell type specification, boundary formation, or organ polarity. It is hypothesized that local induction of cell death is dependent on epidermal cell identity as defined at the molecular level.
Key words: rice, ethylene, hydrogen peroxide, epidermal cell death, cell identity genes
Rice is a semiaquatic plant that forms adventitious root primordia at the stem nodes.1 Adventitious roots emerge from the nodes when plants become submerged.2 Adventitious root growth is preceded by death of epidermal cells that cover the root primordia.3 Both, adventitious root growth and epidermal cell death are promoted by ethylene.2,3 H2O2 acts as a mediator of defined cell death responses in plants4–6 and was shown to also participate in epidermal cell death in rice.7 Recent work identified the transcriptomes of rice epidermal cells above adventitious roots prior to and after induction of cell death and revealed that these cells have a unique molecular identity.7 Among the differentially expressed genes, several encoded transcription factors with a predicted function in determining cell identity.
Transcription Factors that are Differentially Expressed in Dying Cells are Predicted to Determine Cell Identity and Boundary Formation
Rice stem sections were isolated and treated for 4 h without effector, with 150 µM ethephon, as an ethylene source or with 3% H2O2. Treatments with ethephon or H2O2 were previously shown to induce cell death specifically in epidermal cells above adventitious roots (Fig. 1).7 Microarray analysis revealed that dying and non-dying epidermal cells possessed different transcriptomes prior to cell death. 2,642 genes were differentially expressed in epidermal cells above root primordia prior to induction of cell death compared to other epidermal cells. Using a differential microarray approach, 61 genes were identified in epidermal cells that undergo cell death which were regulated more than twofold in response to ethylene or H2O2 treatment. Of these, 6 encoded known transcriptional regulators (Table 1).
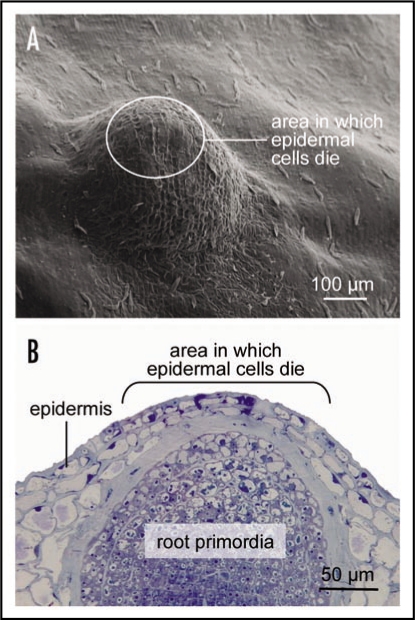
Figure 1.
(A) Scanning electron microscopic view of nodal tissue with an underlying adventitious root primordia. The waxy surface structures differ between epidermal cells above the root primordia which can undergo cell death and other epidermal cells. (B) Light micrograph of a cross section through the node showing a root primodia. Indicated in (A and B) are the approximate areas in which epidermal cells will undergo cell death when triggered by an appropriate signal. Approximately 2,600 genes are differentially expressed in these epidermal cells prior to cell death induction.
Table 1.
Transcription factor genes that are regulated by ethylene and H2O2 with a minimum fold change (FCh) of 2 at p < 0.001,7
| Gene name | FCh E | FCh H2O2 | Putative functions | |
| Os02g0614300 | ↑ | 2.3 | 2.5 | ANT-like AP2/ERF family transcription factor |
| Os01g0693400 | ↓ | 0.4 | 0.5 | RAV-like AP2/ERF family transcription factor |
| Os01g0192300 | ↓ | 0.4 | 0.3 | myb family transcription factor |
| Os01g0670800 | ↓ | 0.5 | 0.3 | OsARF2 transcription factor |
| Os01g0753500 | ↓ | 0.3 | 0.4 | OsARF3 transcription factor |
| Os10g0480200 | ↓ | 0.4 | 0.4 | Hox9 homeodomain leucine zipper transcription factor |
| Os01g0855900 | ↓ | 0.4 | 0.4 | CDC6, contains FAR1 DNA-binding domain |
Homologous transcription factors were previously shown to be involved in boundary formation, organ polarity, or cell type specification in other plants. The ANT-like factor of the AP2/ERF transcription factor family (Os02g0614300) of rice that was upregulated 2.3-fold after cell death induction is related to Arabidopsis ANT which is involved in lateral organ initiation and growth. A role for AINTEGUMENTA (ANT) in the establishment of adaxialabaxial polarity in Arabidopsis leaves and floral organs was proposed.8 OsARF2 (previously described as OsETTIN2; Os01g0670800) and OsARF3 (Os01g0753500) belong to the auxin response factor (ARF) family which consists of 25 predicted members in rice.9,10 Both were downregulated by cell death inducing signals. Most ARFs have a DNA binding domain and an AUX/IAA protein binding domain and are pivotal in auxin signaling. OsARF2 and OsARF3 are closely related members of class IIb of the ARF family which lacks the C-terminal AUX/IAA interaction domain. OsARF2 and OsARF3 were predicted to act as repressors,10 but have not been functionally characterized so far. Their closest Arabidopsis homolog AtARF3 (At2g33860) was shown to play a role in patterning of reproductive organs through cell type specification. It was suggested that AtARF3 participates either in establishing apical and basal boundaries in the gynoecium primordium or to provide positional information.11 AtARF3 mRNA is a target of small interferring TAS3 ta-siRNAs.12 In leaves, derepression of AtARF3 expression through a mutated siRNA binding site resulted in precocious development of trichomes on the lower (abaxial) leaf side, a phenotype that is reminiscent of the rld1 (rolled leaf protein1) mutant of maize.13 The maize class III homeodomain leucine zipper (HD-ZIPIII) RLD1 protein determines adaxial (upper) leaf cell fate specification.13 In plants, expression of HD-ZIPIII genes including RLD1 is controlled through miRNA. Maize RLD1 transcripts are degraded on the lower leaf side through miRNA166. Constitutive expression of RLD1 on both leaf sides due to a base mutation in miRNA166 results in partial relief of leaf polarity resulting in an upward rolled leaf.14 Rice Hox9 (Os10g0480200) which was downregulated in epidermal cells that undergo cell death is a HD-ZIPIII transcription factor and is in fact most closely related (86% identity/93% similarity) to RLD1 from maize.
The functions of the RAV-like AP2/ERF family transcription factor (Os01g0693400), the myb family transcription factor (Os01g0192300), and CDC6 (Os01g0855900) which contains a FAR1 domain have yet to be described.
Conclusions
Epidermal cells above adventitious roots have the unique ability to respond to appropriate signals with the execution of cell death.7 Cell identity determines the response to abiotic stress in Arabidopsis.15 In rice, the pro-death signals ethylene and H2O2 exerted transcriptional regulation of genes that specify the uniqueness of cells or organs and separate them from adjacent tissues. It is tempting to speculate that regulation of ANT-like, OsARF2, OsARF3 and Hox9 in epidermal cells above adventitious roots is required for the ability to execute a cell death program only in those epidermal cells that need to give way to an emerging root. It will be exciting to identify the signals and signal sources that initiate epidermal cell specification. The underlying root primordia can be considered as prime suspect in this process.
Footnotes
Previously published online as a Plant Signaling & Behavior E-publication: http://www.landesbioscience.com/journals/psb/article/7957
References
- 1.Bleecker AB, Schuette JL, Kende H. Anatomical analysis of growth and developmental patterns in the internode of deepwater rice. Planta. 1986;169:490–497. doi: 10.1007/BF00392097. [DOI] [PubMed] [Google Scholar]
- 2.Lorbiecke R, Sauter M. Adventitious root growth and cell cycle induction in deepwater rice. Plant Physiol. 1999;119:21–29. doi: 10.1104/pp.119.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mergemann H, Sauter M. Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol. 2000;124:609–614. doi: 10.1104/pp.124.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moeder W, Barry CS, Tauriainen AA, Betz C, Tuomainen J, Utriainen M, et al. Ethylene synthesis regulated by biphasic induction of 1-aminocyclopropane-1-carboxylic acid synthase and 1-aminocyclopropane-1-carboxylic acid oxidase genes is required for hydrogen peroxide accumulation and cell death in ozone-exposed tomato. Plant Physiol. 2002;130:1918–1926. doi: 10.1104/pp.009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overmyer K, Brosché M, Kangasjärvi J. Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 2003;8:335–342. doi: 10.1016/S1360-1385(03)00135-3. [DOI] [PubMed] [Google Scholar]
- 6.Bouchez O, Huard C, Lorrain S, Roby D, Balagué C. Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1. Plant Physiol. 2007;145:465–477. doi: 10.1104/pp.107.106302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steffens B, Sauter M. Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell. 2009 doi: 10.1105/tpc.108.061887. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nole-Wilson S, Krizek BA. AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiol. 2006;141:977–987. doi: 10.1104/pp.106.076604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato Y, Nishimura A, Ito M, Ashikari M, Hirano HY, Matsuoka M. Auxin response factor family in rice. Genes Genet Syst. 2001;76:373–380. doi: 10.1266/ggs.76.373. [DOI] [PubMed] [Google Scholar]
- 10.Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, et al. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa) Gene. 2007;394:13–24. doi: 10.1016/j.gene.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Sessions A, Nemhauser JL, McCall A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- 12.Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 13.Juarez MT, Twigg RW, Timmermans MCP. Specification of adaxial cell fate during maize leaf development. Development. 2004;131:4533–4544. doi: 10.1242/dev.01328. [DOI] [PubMed] [Google Scholar]
- 14.Nelson JM, Lane B, Freeling M. Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf's dorsoventral axis. Development. 2002;129:4581–4589. doi: 10.1242/dev.129.19.4581. [DOI] [PubMed] [Google Scholar]
- 15.Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]