Abstract
Despite progress in the management of breast cancer, the molecular underpinnings of clinically aggressive subtypes of the disease are not well-understood. Here, we show that activation of Notch developmental signaling in estrogen receptor (ER)-negative breast cancer cells results in direct transcriptional up-regulation of the apoptosis inhibitor and cell cycle regulator survivin. This response is associated with increased expression of survivin at mitosis, enhanced cell proliferation, and heightened viability at cell division. Conversely, targeting Notch signaling with a peptidyl γ-secretase inhibitor suppressed survivin levels, induced apoptosis, abolished colony formation in soft agar, and inhibited localized and metastatic tumor growth in mice, without organ or systemic toxicity. In contrast, ER+ breast cancer cells, or various normal cell types, were insensitive to Notch stimulation. Therefore, ER- breast cancer cells become dependent on Notch-survivin signaling for their maintenance, in vivo. Therapeutic targeting of this pathway may be explored for individualized treatment of patients with clinically aggressive, ER-xs breast cancer.
Introduction
Despite the success of “targeted” agents (1), including estrogen ablation (2), breast cancer remains a potentially deadly disease, marked by extraordinary molecular and clinical heterogeneity (3). Gene expression signatures have been useful to catalog the different molecular subtypes of breast cancer, and in some cases, this information carries important prognostic (4, 5), and predictive (6) value to identify patients at risk of recurrent disease (7). However, the molecular pathways underlying clinically aggressive variants of breast cancer, typically estrogen receptor (ER)-negative disease, have not been identified (8), and “individualized” therapy for these patients based on molecular disease profile is not presently available.
Developmental signaling pathways, such as Notch, are critical for tissue specification and organ morphogenesis (9), and are frequently deregulated in cancer (10). Although it remains to be seen to what extent these mechanisms are truly disease “drivers,” mutations in Notch-1 (11) cause oncogene expression in a subset of T-cell acute lymphoblastic leukemias (12), and deregulated Notch activity may influence cellular transformation (13), cell cycle dynamics (14), progenitor/stem cell maintenance (15), and disease outcome (16) in breast cancer. Notch signaling initiates with the binding of extracellular ligands, Jagged-1, Jagged-2 or Delta-1, or Delta-4 to Notch receptors at the cell surface. In turn, this results in proteolytic cleavage of Notch intracellular domain (NIC) by the enzyme γ-secretase, nuclear import of NIC, and recruitment of transcriptional modulators, including RBP-Jκ, to drive de novo expression of target genes (17). Although it is clear that Notch and other developmental pathways control multiple downstream networks of cell proliferation, cell survival, and progenitor/stem cell maintenance (9), only a handful of target genes capable of orchestrating such pleiotropic responses has been identified, and their contribution to the cancer phenotype has remained largely elusive.
As a unique member of the Inhibitor of Apoptosis (IAP) gene family (18), survivin has emerged as a pivotal cancer gene with multiple roles in the regulation of mitosis, suppression of cell death, and enhanced adaptation to cellular stress (19). The sharp differential expression of survivin in cancer, as opposed to normal tissues, is largely controlled at the level of transcription, and several oncogenic pathways, including developmental signaling by Wnt/β-catenin (20), have been shown to promote survivin gene expression. There is also evidence that survivin may be a critical gene in breast cancer, linked to aggressive disease (6), resistance to apoptosis (21), and modulation of ErbB2 signaling (22).
In this study, we mapped novel molecular circuitries of breast cancer pathogenesis. We found that survivin is a novel target of Notch signaling, and this occurs preferentially in ER- breast cancer cells. Therapeutic targeting of a Notch-survivin axis produces strong anticancer activity, and is well-tolerated in vivo, opening new opportunities for “individualized” therapy in clinically aggressive breast cancer subtypes.
Materials and Methods
Cell lines and viral transductions
Breast adenocarcinoma cell lines MCF-7, MDA-MB-231, and cervical carcinoma HeLa cells were obtained from the American Type Culture Collection (ATCC) and maintained in culture as recommended by the supplier. Breast adenocarcinoma cell lines T47D, Sum149, and HBL100 were generously provided by Dr. Arthur M. Mercurio (University of Massachusetts Medical School, Worcester, MA). Primary human mammary epithelial cells (HMEC) were obtained from Lonza. Primary human foreskin fibroblasts (HFF), human gingival fibroblasts (HGF), human epithelial fibroblasts (WS-1), and human intestinal epithelial cells (INT; all from ATCC) were cultured per manufacturer's instructions and used at early passages before the onset of senescence. A NIC construct has been characterized previously (23). For adenoviral transduction, various cell types (1 × 105) were incubated with replication-defective adenovirus pAd-NIC, pAd-GFP, or pAd-Control at multiplicity of infection of 50 for 4 to 8 h at 37°C, harvested at increasing time intervals, and processed for individual experiments.
Semiquantitative reverse transcription-PCR
Total RNA was harvested using the RNeasy kit (Qiagen). One microgram of total RNA was reversed transcribed in the presence of SuperScript II polymerase plus random primer (Invitrogen). Survivin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was amplified (28 cycles) and separated on 1.5% agarose gels. Band intensity normalized to GAPDH expression was quantified using Labworks 4.6 (UVP BioImaging Systems).
Western blotting and antibodies
Cellular extracts were prepared in a lysis buffer containing 20 mmol/L Tris (pH 7.5), 0.5% sodium deoxycholate, 1% Triton, 0.1% SDS, 150 mmol/L NaCl, 1 mmol/L EDTA, 50 mmol/L NaF, 1 mmol/L Na3VO4, plus protease inhibitors (Roche Applied Science). Lysates were separated by SDS gel electrophoresis and transferred to Immobilon membranes (Millipore). The antibodies used were as follows: survivin (1:1,000) from NOVUS Biologicals; XIAP(1:500), green fluorescent protein (GFP; 1:500), cytochrome c (1:1,000), and Cox-4 (1:5,000) from BD Biosciences; Bcl-2 (1:500) from Santa Cruz Biotechnology; V5 epitope for detection of NIC or Jagged-1 (1:5,000) from Invitrogen; NIC (1:1,000) from Rockland; and β-actin (1:5,000) from Sigma.
Analysis of survivin gene expression
A cDNA construct comprising the first 830 bp of the mouse survivin promoter upstream of the translational initiation codon fused to GFP (ms-830-GFP) was characterized previously (24). HeLa cells were cotransfected with ms-830-GFP plus Notch-1 or Jagged-1 cDNA by Lipofectamine. After 24 h, cells were analyzed for GFP expression by Western blotting, or fluorescence microscopy with image acquisition on an Olympus IX71 microscope outfitted with an Olympus Regina camera. A putative RBP-Jκ binding site at position -355 in ms-830-GFP was mutated using the QuikChange Site-Directed Mutagenesis kit with primer: 5′-GAACCTGCAGAGCACATGttACTTGCAGCGGACATGC-3′. The mutant construct (mutant ms-830-GFP) was confirmed by DNA sequencing, transfected in HeLa cells, and analyzed for GFP expression after Notch stimulation. For electrophoretic mobility shift assay (EMSA), MDA-MB-231 nuclear extracts were prepared using the CelLytic NuCLEAR extraction kit (Sigma) according to the manufacturer's instructions. cDNA sequences corresponding to the -355 region of ms-830-GFP (5′-GGAAGAACCTGCAGAGCACATGGGACTTGCAGCGGACATGCT-3′), a random survivin promoter region (5′-TGCAACGCCAACCTGGGCTGTGTT CGGGGCATGCCCAGCCTG-3′), or an RBP-Jκ binding region in the human Hes promoter (5′-GTTACTGTGGGAAAGAAAGTC-3′) were synthesized. Probes (3.5 pmol) were made double stranded by annealing at equimolar concentrations, and 5′ end labeled with 1 μL of [γ-32P] ATP (3,000 Ci/mmol) and 10 U of T4 polynucleotide kinase (NEB) for 10 min at 37°C. MDA-MB-231 nuclear extracts were incubated with the reaction mixture (10 μL) containing radiolabeled DNA probes, 2 μg of poly(deoxyinosinic-deoxycytidylic) acid (Sigma), 10 mmol/L Tris (pH 7.5), 1 mmol/L MgCl2, 50 mmol/L NaCl, 4% glycerol, 0.5 mmol/L EDTA, 0.5 mmol/L DTT, 50 μg bovine serum albumin, and various unlabeled competing oligonucleotides. After 30 min of incubation at 22°C, samples were separated by electrophoresis on 4% nondenaturing polyacrylamide gels, and bands were visualized by autoradiography.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using the ChIP-IT Express kit (Active Motif) per the manufacturer's protocols. MDA-MB-231 cells were infected with pAd-NIC before fixation with 1% formaldehyde for 10 min. Cells were washed, lysed, and sonicated to reduce DNA lengths to a range of 300 to 600 bp. The chromatin/DNA complexes were incubated with antibodies to RNA Pol II (Active Motif), RBP-Jκ (Santa Cruz Biotechnology), or IgG (Active Motif) for 18 h at 4°C. The immune complexes were precipitated, eluted, reverse crosslinked, and treated with proteinase K. The resulting DNA samples were amplified with primers to the RNA Polymerase II site in the GAPDH promoter ( forward, 5′-TACTAGCGGTTTTACGGGCG-3′; reverse, 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′), the -305 bp putative RBP-Jκ binding site in the human survivin promoter (forward, 5′-ACCACGCCCAGCTAATTTTTG-3′; reverse, 5′-CCTCGACTGCTTTCAAAGAACG-3′), and the RBP-Jκ binding site in the Hes promoter (forward, 5′-CGTGTCTCCTCCTCCCATTG-3′; reverse, 5′-CCAGGACCAAGGAGAGAGGT-3′).
Cell cycle analysis
Cells (1.5 × 105) were synchronized at the G1-S transition by treatment with 2 mmol/L thymidine for 18 h at 37° C. In some experiments, cells were released after 18 h, infected with pAd-NIC or pAd-GFP for 8 h, and resynchronized with 1 mmol/L thymidine for another 18 h. Aliquots of synchronized cultures were collected at 0, 4, 8, 10, 12, and 14 h after the second thymidine release, and analyzed for DNA content by propidium iodide staining and flow cytometry, or alternatively, by Western blotting. For single-color cell cycle analysis, cells were fixed in 70% ethanol for 18 h at -20°C, and analyzed by flow cytometry. Cell cycle populations were gated and quantified using FlowJo software (TreeStar). A pAd-T34A survivin dominant-negative mutant was characterized previously (25). In some experiments, MDA-MB-231 cells were infected with the combinations pAd-NIC/pAd-Control, pAd-T34A/pAd-Control, or pAd-NIC/pAd-T34A for 24 h; synchronized with 2 mmol/L thymidine for 24 h; and released into S phase. Cultures were collected after various time intervals and analyzed for DNA content by propidium iodide staining, or, alternatively, by Western blotting.
Cell death assays
A γ-secretase inhibitor (GSI) z-Leu-Leu-Nle-CHO was purchased from Calbiochem and dissolved in DMSO. In metabolic activity assays, cells were treated with GSI or DMSO for 24 h, incubated with 50 μg/mL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) for 2 h, and analyzed at A595. In other experiments, cells treated with GSI or DMSO were spun onto slides, DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI), and cells with chromatin condensation/fragmentation were scored by fluorescence microscopy. For experiments of cytochrome c release, GSI-treated cells were harvested, fractionated into cytosolic extracts using the ApoAlert Cell Fractionation kit (Clontech), and analyzed for time-dependent release of cytochrome c by Western blotting. Caspase activity was determined using the Capase-3/7 In Situ Assay kit (Millipore) by multiparametric flow cytometry, in the presence or absence of a broad spectrum caspase inhibitor zVAD-fmk (American Peptides; 20 μmol/L).
Analysis of tumorigenesis
For soft agar colony formation, 1 × 104 cells were treated with GSI (0.125-10 μmol/L) for 24 h, suspended in 2 mL of DMEM/0.35% bactoagar, and plated onto 6-well tissue culture plates containing 2 mL DMEM/0.75% bactoagar as a bottom layer. After a 2-wk incubation at 37°C, 5% CO2, colonies were stained with 0.005% crystal violet and counted under high-power field.
All experiments involving animals were approved by an Institutional Animal Care and Use Committee. For xenograft tumor studies, 5 × 106 MDA-MB-231 cells were injected into each flank of 6- to 8-wk-old female CB17 severe combined immunodeficient (SCID)/beige mice (Taconic). When tumors reached 150-175 mm3 in volume, mice were randomized (3 animals per group, 6 tumors per group, 2 independent experiments) and treated daily with i.p. injections of 3 mg/kg GSI or vehicle control. For experiments in vivo, z-Leu-Leu-Nle-CHO (American Peptides) was synthesized to >99% purity and dissolved in 25% Cremophor/PBS (Sigma). Mice were weighed daily, monitored for signs of pain and distress, and tumor size were measured in three dimensions with a caliper using the equation L × W2/2 (mm3). All animals were euthanized when tumor volume exceeded 1,500 mm3. At the end of the experiment, liver, spleen, lung, kidney, pancreas, and colon were harvested, fixed in formalin, and embedded in paraffin blocks for histology. For quantification of lung micrometastases, serial lung sections were cut 20 μm apart, stained with H&E, and analyzed by light microscopy.
Statistical analysis
Data were analyzed using the unpaired Student's t test on a GraphPad software program (Prism 4.0). All statistical tests were two sided. A P value of 0.05 was considered to be statistically significant.
Results
Notch-1 induction of survivin in ER- breast cancer
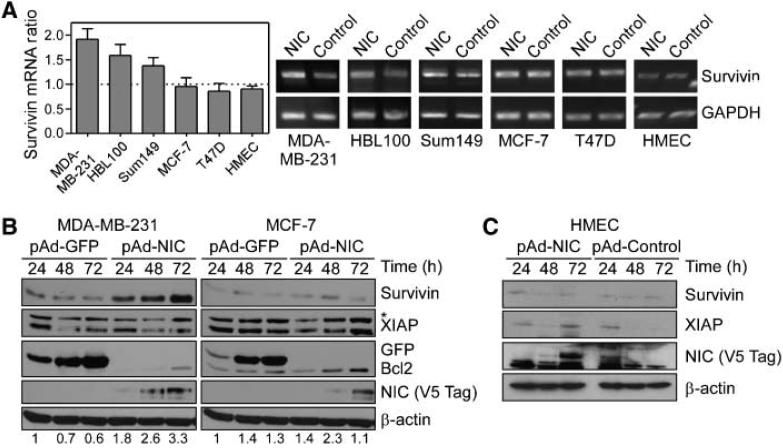
Preliminary meta-analysis of published microarray data revealed that Notch-1 and survivin cosegregated with ER- breast cancer cases and correlated with abbreviated overall survival.3 Consistent with this, transduction of ER- breast adenocarcinoma MDA-MB-231, HBL100, and Sum149 cells (26) with pAd-NIC, the active intracellular domain of Notch, resulted in a 1.5- to 2-fold increased survivin mRNA expression, compared with pAd-GFP-treated cultures (Fig. 1A). In contrast, Notch stimulation of ER+ breast cancer cells MCF-7 and T47D, or normal HMECs, did not modulate survivin mRNA levels (Fig. 1A). In time course experiments, pAd-NIC increased expression of survivin in ER- MDA-MB-231 cells, but not ER+ MCF-7 cells, by Western blotting (Fig. 1B). Conversely, Bcl-2 was comparably up-regulated in both cell types (27), whereas another survival factor, XIAP, was not significantly affected (Fig. 1B). Even a prolonged, 72-h stimulation of normal primary HMECs with pAd-NIC did not modulate survivin or XIAP expression (Fig. 1C).
Figure 1.
Regulation of survivin expression by Notch. A, semiquantitative reverse transcription-PCR. A panel of ER- (MDA-MB-231, HBL100, and Sum149), ER+ (MCF-7 and T47D), or primary HMEC were transduced with pAd-NIC or pAd-Control, and assayed for survivin mRNA levels. B, Western blotting. Transduced MDA-MB-231 or MCF-7 cells were harvested at the indicated time intervals and analyzed by Western blotting. C, analysis of normal cells. Transduced primary HMEC were analyzed by Western blotting at the indicated time intervals. *, nonspecific. Numbers at the bottom of each panel correspond to normalized densitometric quantification of survivin protein bands.
Notch regulation of survivin gene expression
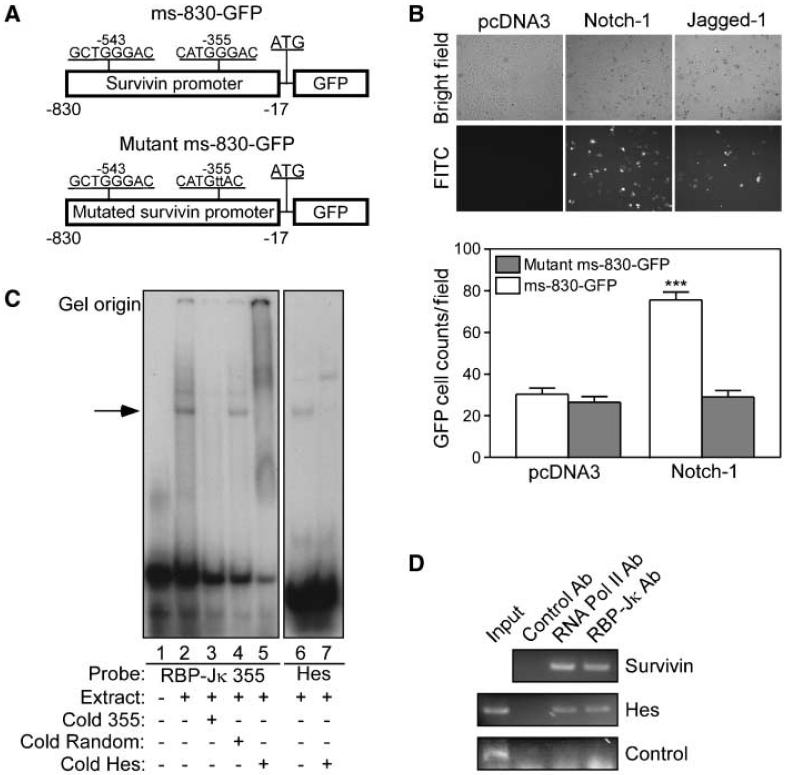
To determine whether Notch directly stimulated survivin gene expression, we next used an 830-bp fragment of the survivin promoter fused to GFP, i.e., ms-830-GFP (24), containing two putative RBP-Jκ sites (17) at -543 and -355 (Fig. 2A). Cotransfection of Notch-1 or Jagged-1 together with wild-type ms-830-GFP resulted in strong expression of GFP, by Western blotting (data not shown), and quantitative fluorescence microscopy (Fig. 2B), compared with pcDNA3-expressing cells (Fig. 2B, top). Mutation of the proximal RBP-Jκ site at -355 in ms-830-GFP (Fig. 2A) completely abolished Notch-1 induction of GFP to background levels of control cultures (Fig. 2B, bottom). By EMSA, a radiolabeled probe of the -355 RBP-Jκ site formed DNA-protein complexes in MDA-MB-231 nuclear extracts (Fig. 2C). This interaction was competed out by equimolar concentrations of unlabeled probe, or a canonical Hes RBP-Jκ sequence but not by an unrelated competitor (Fig. 2C). Similarly, a radiolabeled Hes RBP-Jκ probe bound to MDA-MB-231 nuclear extracts, in a reaction also competed out by molar excess of unlabeled probe (Fig. 2C). In contrast, a probe of the -543 survivin RBP-Jκ site did not form DNA-protein complexes in MDA-MB-231 extracts (data not shown) and was not further investigated. Consistent with these data, ChIP experiments using MDA-MB-231 cells transduced with pAd-NIC revealed that RNA polymerase II and RBP-Jκ were physically associated with the human survivin promoter in vivo at the putative RBP-Jκ binding site at -305 (Fig. 2D).
Figure 2.
Notch regulation of survivin gene expression. A, maps of survivin promoter constructs (ms-830-GFP). Two putative RBP-Jκ sites at -355 and -543 in the mouse survivin gene are indicated.B, fluorescence microscopy. HeLa cells cotransfected with ms-830-GFP plus Notch-1, Jagged-1, or pcDNA3 were analyzed by fluorescence microscopy (top), and GFP-positive cells were quantified (bottom). Columns, mean (n = 10 high-power fields with ~200 cells per field) from two independent experiments; bars, SE. ***, P < 0.0001. C, EMSA. Nuclear extracts from MDA-MB-231 cells were incubated with 32P-γ ATP-labeled probe duplicating the survivin RBP-Jκ site at -355 (left), in the presence or absence of unlabeled competitors. An RBP-Jκ probe from the Hes promoter was used as a control (right). Arrow, position of a DNA-protein complex. D, ChIP assay. Chromatin from MDA-MB-231 cells transduced with pAd-NIC was incubated with antibodies to RNA polymerase II (RNA Pol II), RBP-Jκ, or IgG, and the immunoprecipitated DNA was amplified with primers for the putative RBP-Jκ site in the human survivin promoter at -305, the RBP-Jκ site in the Hes promoter, or a nonspecific promoter region.
Notch-1 regulation of cell proliferation
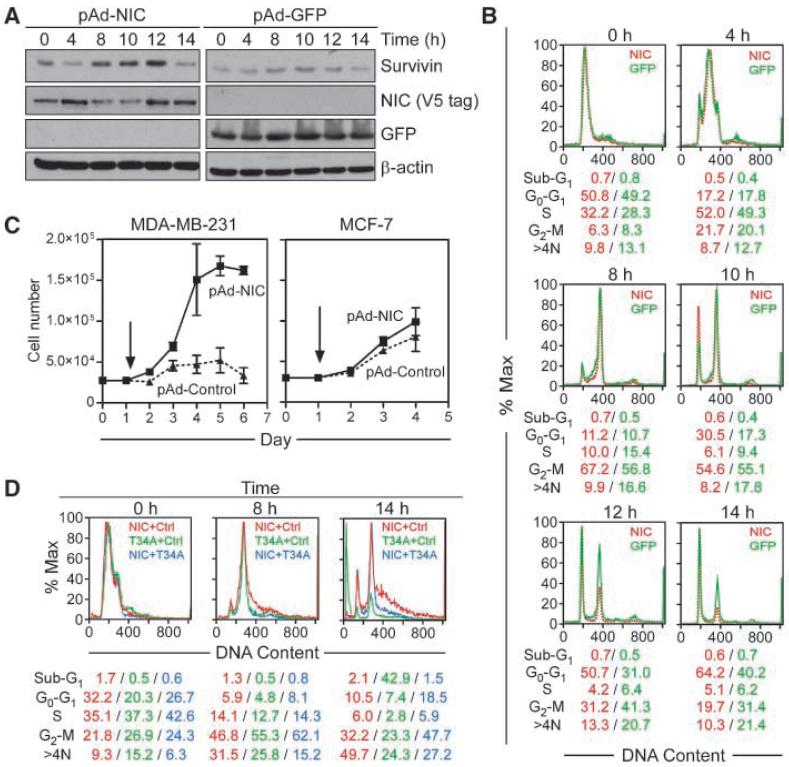
To determine the kinetics of Notch-1 induction of survivin, we next studied cell cycle transitions in synchronized MDA-MB-231 cells after transduction with pAd-NIC. In synchronized cultures expressing pAd-GFP, endogenous survivin was expressed in a cell cycle-dependent manner, peaking at mitosis, 8- to 12-hours after thymidine release (Fig. 3A), consistent with previous observations (28). Activation of Notch-1 in these cells up-regulated survivin with the same kinetics, in a reaction that was also maximal at mitosis (Fig. 3A). Quantification of cell cycle phases revealed that GFP-expressing cells approached S phase 4 hours after thymidine release, entered mitosis after 8 hours, and completed cell division between 10 and 12 hours, with reaccumulation in the next G1 phase by 14 hours (Fig. 3B). In contrast, Notch activation resulted in accelerated mitotic transitions, with 30% of NIC-expressing cells exiting mitosis 10 hours after release, compared with 17% of GFP-transduced cells, and 64% of Notch-1-stimulated cells re-entering G1 after 14 hours, versus 40% of control cultures (Fig. 3B). Accelerated mitotic progression in Notch-stimulated MDA-MB-231 cells was associated with increased cell proliferation over a 6-day interval, compared with pAd-GFP-expressing cultures (Fig. 3C). In contrast, pAd-NIC or pAd-GFP did not induce changes in cell proliferation in ER+ MCF-7 cultures (Fig. 3C).
Figure 3.
Effect of activated Notch-1 on cell cycle progression. A, thymidine synchronization. Transduced MDA-MB-231 cells were synchronized with 2 mmol/L thymidine and analyzed by Western blotting at the indicated time intervals after release. B, cell cycle analysis. Synchronized and transduced MDA-MB-231 cells were analyzed for DNA content by propidium iodide staining and flow cytometry at the indicated time intervals. The percentage of cells in each cell cycle phase is indicated. C, cell proliferation. Transduced MDA-MB-231 or MCF-7 cells were counted at the indicated time intervals, starting at day 1 (arrow). Columns, mean (n = 3); bars, SE. D, rescue from mitotic cell death. MDA-MB-231 cells were transduced with combinations of pAd-NIC, pAd-Control, and pAd-T34A; synchronized; and analyzed for DNA content by propidium iodide staining and flow cytometry. The percentage of cells in each cell cycle phase is indicated.
We next asked whether Notch-1 induction of survivin was important for cell viability during this proliferative response. For these experiments, we transduced MDA-MB-231 cells with a dominant-negative survivin mutant (pAd-T34A), which interferes with the mitotic function of survivin (28). Expression of pAd-T34A had no effect on synchronized, interphase cultures, but caused acute loss of cell viability as cells progressed through mitosis (Fig. 3D), in agreement with previous observations (28). Notch stimulation reversed pAd-T34A-induced cell death and preserved cell viability at mitosis (Fig. 3D). In control experiments, pAd-NIC had no effect on MDA-MB-231 cell viability (Fig. 3D). The higher number of cells with >4N DNA content after pAd-NIC transduction may reflect mitotic slippage and accumulation of chromosomal abnormalities due to accelerated mitotic transition.
Effect of Notch targeting on breast cancer cell viability
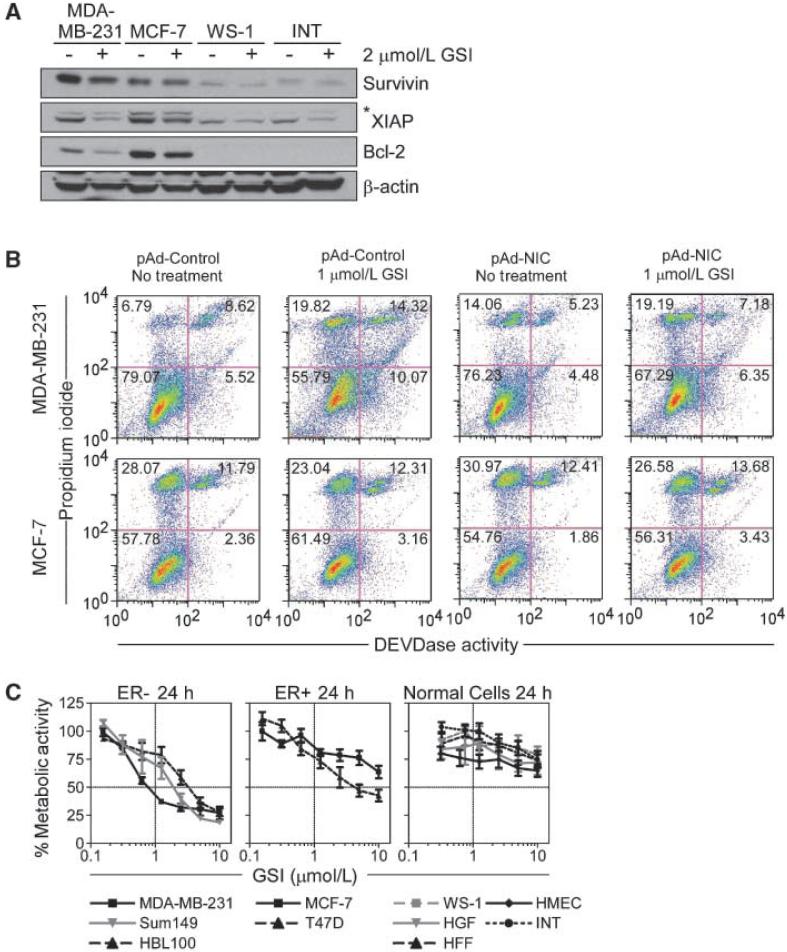
To test the effect of Notch signaling in breast cancer, we used a GSI z-Leu-Leu-Nle-CHO, which inhibits receptor cleavage and NIC generation (17). GSI treatment reduced endogenous survivin levels in MDA-MB-231 cells, and this was associated with parallel suppression of Bcl-2 and XIAP (Fig. 4A). In contrast, GSI minimally affected the expression of these antiapoptotic molecules in MCF-7 cells (Fig. 4A). In addition, GSI had no effect on survivin levels in WS-1 fibroblasts or INT colonic epithelial cells, whereas XIAP was modestly reduced, and Bcl-2 was undetectable in these cells (Fig. 4A). Treatment of MDA-MB-231 cells with a second, structurally independent GSI molecule, z-Ile-Leu-CHO, reduced Notch-1 expression and attenuated survivin levels in MDA-MB-231 cells (data not shown).
Figure 4.
Effect of Notch inhibition on breast cancer cell viability. A, Western blotting. The indicated breast cancer or normal cell types were treated with DMSO (-) or 2 μmol/L GSI (+), harvested after 24 h, and analyzed by Western blotting. B, NIC rescue of GSI-induced cell death. Transduced MDA-MB-231 (top) or MCF-7 (bottom) cells were treated with 1 μmol/L GSI or DMSO for 24 h and analyzed for apoptosis by DEVDase activity. The percentage of cells in each quadrant is indicated. C, cell viability. The indicated breast cancer (left and middle) or normal (right) cell types were treated with increasing concentrations of GSI (0.125-10 μmol/L) for 24 h, and analyzed for cell viability by MTT. Points, mean (n = 4); bars, SE. IC50 values for GSI-induced cell killing are as follows: MDA-MB-231, 1.196 μmol/L; HBL100, 3.416 μmol/L; Sum149, 2.030 μmol/L; MCF-7, >10 μmol/L; T47D, 5.709 μmol/L; and normal cell lines, >10 μmol/L.
Transduction of MDA-MB-231 cells with pAd control or pAd-NIC did not significantly affect cell viability, as determined by DEVDase, i.e., caspase, activity and multiparametric flow cytometry (Fig. 4B, top). GSI treatment of these cells resulted in a 2-fold increase in the fraction of apoptotic cells, in a reaction nearly completely reversed by pAd-NIC (Fig. 4B, top). In contrast, GSI did not significantly induce caspase activity in MCF-7 cultures, and transduction of these cells with pAd-NIC or pAd-Control had no further effect on cell viability (Fig. 4B, bottom). Consistent with these data, GSI induced concentration-dependent killing of multiple ER- breast cancer cell types, including MDA-MB-231, HBL100, Sum149 (Fig. 4C, left) with IC50 of <3.5 μmol/L. Comparatively, ER+ cell types, MCF-7 and T47D, were less sensitive to GSI-induced killing, with IC50 concentrations of >10 and 5.907 μmol/L, respectively (Fig. 4C, middle). In addition, comparable concentrations of GSI did not affect the viability of three primary fibroblast cell lines, (WS-1, HFF, and HGF), normal INT epithelial cells, or primary HMECs (Fig. 4C, right).
GSI induction of apoptosis in breast cancer cells
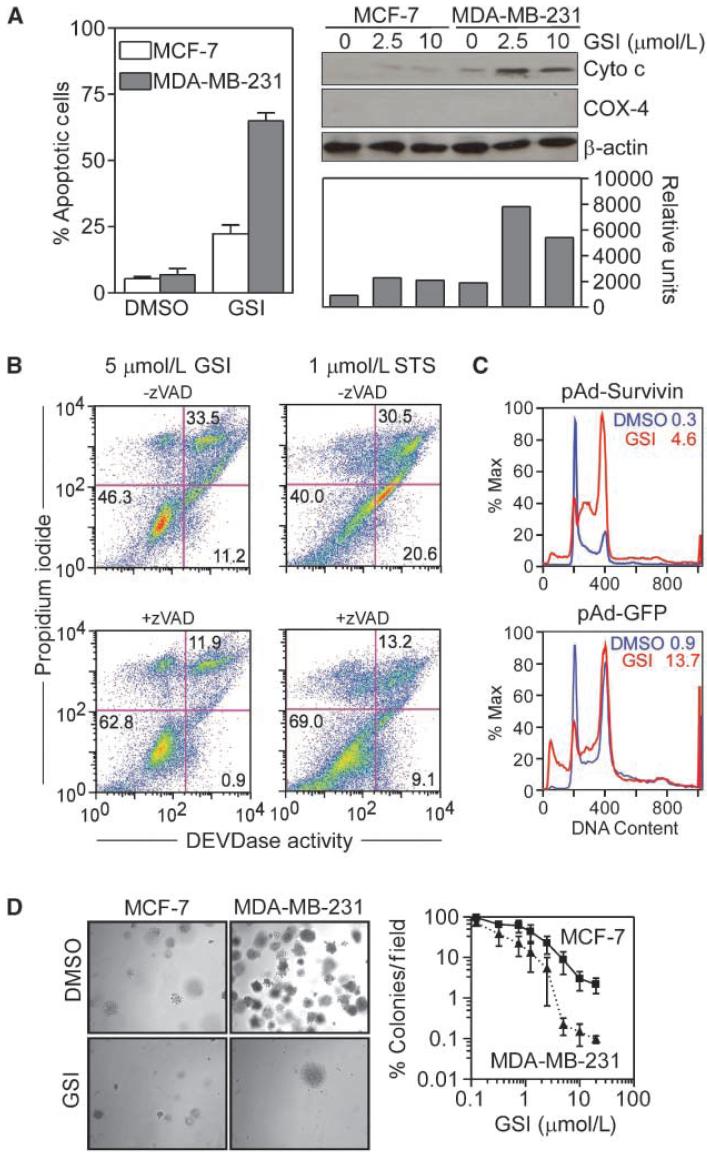
MDA-MB-231 cells exposed to GSI exhibited morphologic hallmarks of apoptosis, including chromatin condensation and fragmentation (Fig. 5A, left), and concentration-dependent release of cytochrome c in the cytosol (Fig. 5A, right). GSI-induced cell death under these conditions was fully reversed by a broad spectrum caspase inhibitor, zVAD, comparable with its inhibition of staurosporine (STS)-induced apoptosis (Fig. 5B). In contrast, GSI did not affect cell viability or cytochrome c release in MCF-7 cultures (Fig. 5A). To test whether loss of survivin after Notch inhibition contributed to apoptosis, we transduced MDA-MB-231 cells with pAd-GFP or pAd-Survivin, and quantified GSI-induced cell death. In GFP-expressing cells, GSI induced a sustained G2-M arrest and apoptosis (Fig. 5C). Expression of survivin in these cells did not significantly affect the mitotic arrest imposed by Notch inhibition but completely reversed GSI-induced cell death, whereas pAd-GFP was ineffective (Fig. 5C).
Figure 5.
Induction of apoptosis by Notch inhibition. A, cell death. MCF-7 or MDA-MB-231 cells were treated with GSI (2.5 μmol/L), harvested after 24 h, and analyzed for nuclear morphology of apoptosis by DAPI staining and fluorescence microscopy (left; columns, mean of 5-10 fields with an average of 60-30 cells per field; bars, SE) or release of mitochondrial cytochrome c in the cytosol by Western blotting (right). Bottom right, normalized densitometric quantification of cytochrome c protein bands. Cox-4 was used as a control mitochondrial marker. B, caspase requirement. MDA-MB-231 cells were treated with 5 μmol/L GSI or 1 μmol/L STS, and analyzed for DEVDase activity after 24 h, in the presence or absence of zVAD. The percentage of cells in each quadrant is indicated. C, survivin rescue. Transduced MDA-MB-231 cells were treated with 1 μmol/L GSI for 24 h, and analyzed for DNA content by propidium iodide staining and flow cytometry. The percentage of cells with hypodiploid (sub-G1) DNA content is indicated. D, colony formation assay. MCF-7 or MDA-MB-231 cells were treated with increasing concentrations of GSI (0.125-20 μmol/L) for 24 h, plated onto semisolid medium, and colonies (left) were stained with 0.005% crystal violet, and quantified by light microscopy (right) after 2 wk. Points, mean (n = 3); bars, SE. Six fields per GSI concentration were quantified per condition (right).
Targeting Notch signaling for breast cancer therapy
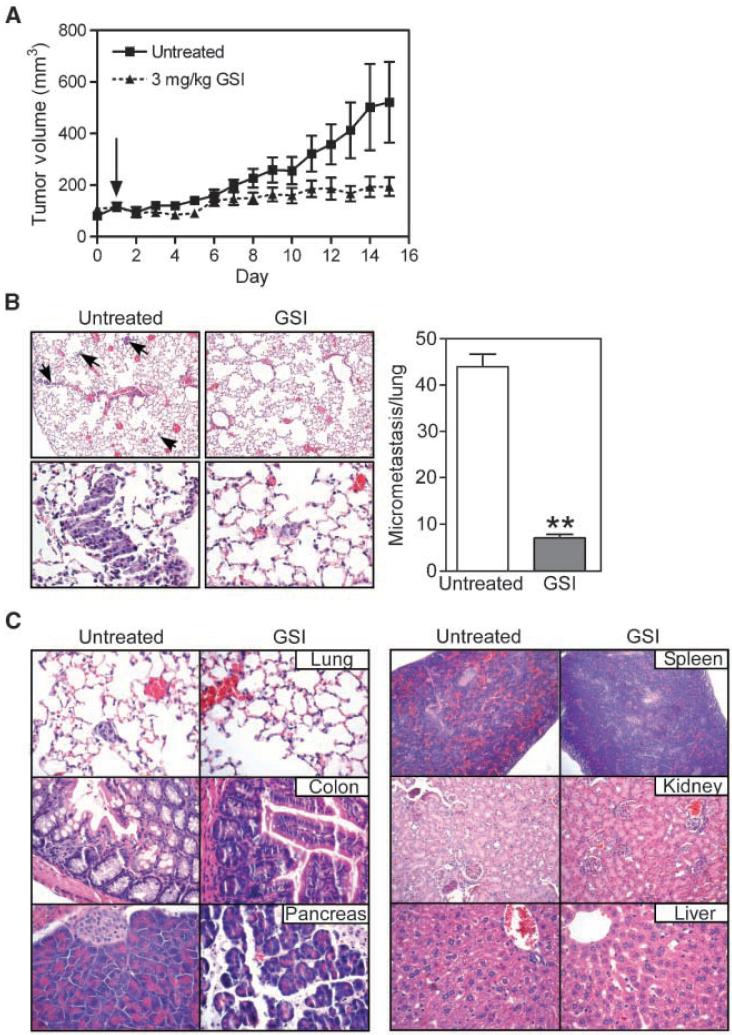
GSI treatment of MDA-MB-231 cells abolished colony formation in soft agar (Fig. 5D, left), whereas comparable concentrations of GSI were three orders of magnitude less effective in MCF-7 cells (Fig. 5D, right). Next, we grew MDA-MB-231 cells as superficial tumors in immunocompromised mice and treated the animals with vehicle or systemic GSI (3 mg/kg/daily i.p.). In control animals, MDA-MB-231 tumors grew at a steady exponential rate over a 2-week time interval (Fig. 6A). In contrast, GSI treatment significantly inhibited tumor growth over a comparable time interval (Fig. 6A). In addition, lungs from mice treated with vehicle exhibited a high density of epithelial micrometastases (29) by H&E staining of serial lung sections (Fig. 6B, left). In contrast, GSI treatment inhibited formation of lung metastases, in vivo (Fig. 6B, right).
Figure 6.
Anticancer activity of Notch inhibition. A, kinetics of tumor growth. CB17 SCID/beige mice were injected s.c. with 5 × 106 MDA-MB-231 cells into each flank. When tumors reached 150 to 175 mm3, mice (3 animals per group, 6 tumors per group) were treated with 3 mg/kg GSI daily or vehicle as i.p. injections. Arrow, start of GSI treatment. Points, mean of tumor volume at each indicated time point; bars, SE. B, effect of Notch inhibition on lung metastases. Lung sections from vehicle- or GSI-treated animals were analyzed by H&E staining (left), and lung micrometastases were quantified in serial tissue sections (20 μm apart) by light microscopy (right; columns, mean from 5 independent fields; bars, SE; **, P < 0.0001). Magnifications, ×10 (top) and ×40 (bottom). C, tissue histology. The indicated organs were collected from mice treated with vehicle (untreated) or GSI at the end of the experiment (14 d), paraffin embedded, and analyzed by H&E staining.
Safety of Notch targeting for cancer therapy
Mice in the GSI group did not exhibit signs of systemic toxicity, and had no significant weight loss, compared with vehicle-treated animals (data not shown). Histologic examination of lung, colon, pancreas, spleen, kidney, and liver was largely indistinguishable in control or GSI-treated mice (Fig. 6C). Specifically, lung sections from GSI or control group showed a conserved architecture with a normal alveolar septum, absence of lymphatic dilation, or interstitial inflammation (Fig. 6C). With respect to the gastrointestinal tract, GSI treatment did not cause goblet cell hyperplasia of colonic epithelium, and resulted in only mild fascitis of pancreatic tissue, compared with vehicle-treated control (Fig. 6C). The liver architecture of hepatic nodules was fully preserved in GSI-treated animals, without steatosis or inflammatory infiltration, and extramedullary hematopoiesis was comparably observed in control- or GSI-treated animals (Fig. 6C).
Discussion
In this study, we have shown that Notch signaling increases cell proliferation and promotes resistance to apoptosis preferentially in ER- breast cancer (30), a disease subtype often marked by aggressive clinical behavior (31). A critical effector of this pathway was identified as survivin, a pleiotropic regulator of cell proliferation and inhibitor of apoptosis overexpressed in many cancers (19). Conversely, inhibition of Notch signaling with a peptidyl GSI down-regulated survivin, inhibited cell proliferation, increased apoptosis, and suppressed the growth of localized and metastatic ER- breast cancer in mice, without overt toxicity.
Recently, there has been considerable interest in elucidating how developmental signaling pathways, including Notch (17), may aberrantly contribute to tumorigenesis. Mounting evidence suggests that Notch deregulation may engender critical tumor hallmarks, including oncogene expression (12, 32), angiogenesis (33), stem cell maintenance (34), deregulated cell cycle progression (14), and antiapoptotic mechanisms (35). In this context, survivin fits well the pleiotropic requirements of a Notch effector gene for its essential roles in mitosis (36), preservation of stem/progenitor cell homeostasis (37), inhibition of apoptosis (38), and regulation of angiogenesis (39). Mechanistically, Notch stimulation resulted in direct activation of survivin gene transcription through at least one RPB-Jκ site (17) in the survivin promoter. This is reminiscent of how another developmental signaling program, i.e., Wnt/β-catenin, directly induces survivin gene transcription in cancer (20). Consistent with the known tissue specificity of Notch signaling, which has been variously associated with malignant transformation (11), or tumor suppression (40), a Notch-survivin axis was preferentially operative in ER- versus ER+ breast cancer cells, and not at all detected in various normal cell types.
Three potential mechanisms may be envisioned for a role of survivin as a Notch target in clinically aggressive breast cancer (31). First, Notch-induced heightened survivin levels at mitosis may deregulate multiple mitotic checkpoints (36), and ultimately contribute to genetic instability and aneuploidy, in vivo. Second, this pathway may directly promote drug and radiation resistance. Higher survivin levels have been consistently linked to inhibition of apoptosis induced by DNA damaging agents (41), as well taxanes (28), two mainstay therapeutic regimens in breast cancer. Third, survivin may operate as a Notch-regulated cytoprotective and/or mitotic factor to promote long-term persistence of breast cancer “stem cells” (42), potentially contributing to ductal carcinoma in situ (15), an idea consistent with the presence of survivin in “stemness” gene signatures (43), and its role in hematopoietic stem cell viability (37).
Although the introduction of “targeted” therapies (1), including antihormonal strategies (2), has significantly improved the survival of breast cancer patients, the prognosis of metastatic disease remains grim. This often involves ER- subsets of the disease, for instance triple-negative (ER, progesterone receptor, and HER-2), basal-like breast cancer, which is characterized by high recurrence rates (7). Here, the increased sensitivity of ER- versus ER+ breast cancer cells to therapeutic Notch inhibition, in vitro and in preclinical models, suggests that these cells may become dependent or “addicted” (44) to Notch signaling. Although the importance of “oncogene addiction” in long-term maintenance of the neoplastic phenotype, in vivo, is debated, the notable clinical responses observed in subsets of cancer patients after targeting of specific cellular pathways support their pivotal role(s) as disease “drivers” (45). A similar paradigm has been proposed for Notch (46), and GSIs have been pursued as therapy for potential “Notch-addicted” tumors, especially T-cell leukemias (47). Despite its rationale, this approach has not been without concerns. GSIs are not entirely specific for Notch, as they also affect other transmembrane proteins, including E-cadherin, the epidermal growth factor receptor, and CD44 (48), and global inhibition of Notch receptors has been associated with serious toxicity, especially aberrant differentiation of intestinal epithelium, and T-cell development, in vivo (49). The data presented here suggest a more encouraging scenario, as a prototype peptidyl GSI was safely administered systemically to mice with effective anticancer activity but negligible systemic or organ toxicity. Although this bodes well for potential further (pre)clinical development of similar compounds, it remains to be seen whether long-term GSI treatment will eventually result in pharmacologic resistance, as observed in leukemias (11).
In summary, we have uncovered a novel Notch-survivin signaling axis, preferentially exploited in ER- breast cancer cells. Although this pathway may contribute to worse clinical outcome, a potential “addiction” of ER- breast cancer cells to a Notch-survivin axis may open new prospects for individualized therapy of these recurrence-prone patients.
Acknowledgments
Grant support: NIH grants HL54131, CA90917, and CA78810; (D.C. Altieri), HL32348, HL35627, and RR15555 (I. Prudovsky); and CA132622 (C.W. Lee).
We thank Drs. T. Maciag for inspiring this work, A. Mercurio and B. Osborne for reagents and advice, D. Garlick for histologic evaluation, and M. Kelliher for helpful discussion.
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
C.W. Lee, K. Simin, Q. Liu, M. Guha, C.C. Hsieh, D.C. Altieri. Unpublished observations
References
- 1.Hudis CA. Trastuzumab-mechanism of action and use in clinical practice. N Engl J Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 2.Jordan VC. SERMs: meeting the promise of multifunctional medicines. J Natl Cancer Inst. 2007;99:350–6. doi: 10.1093/jnci/djk062. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 5.Liu R, Wang X, Chen GY, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–26. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 6.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 7.Sotiriou C, Piccart MJ. Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nat Rev Cancer. 2007;7:545–53. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 8.Nahta R, Yu D, Hung MC, Hortobagyi GN, Esteva FJ. Mechanisms of disease: understanding resistance to HER2-targeted therapy in human breast cancer. Nat Clin Pract Oncol. 2006;3:269–80. doi: 10.1038/ncponc0509. [DOI] [PubMed] [Google Scholar]
- 9.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–54. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 11.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 12.Sharma VM, Calvo JA, Draheim KM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26:8022–31. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–25. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 14.Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21:5925–34. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farnie G, Clarke RB, Spence K, et al. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst. 2007;99:616–27. doi: 10.1093/jnci/djk133. [DOI] [PubMed] [Google Scholar]
- 16.Reedijk M, Odorcic S, Chang L, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–7. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 17.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–89. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 18.Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–94. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 20.Kim PJ, Plescia J, Clevers H, Fearon ER, Altieri DC. Survivin and molecular pathogenesis of colorectal cancer. Lancet. 2003;362:205–9. doi: 10.1016/S0140-6736(03)13910-4. [DOI] [PubMed] [Google Scholar]
- 21.Gritsko T, Williams A, Turkson J, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12:11–9. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 22.Xia W, Bisi J, Strum J, et al. Regulation of survivin by ErbB2 signaling: therapeutic implications for ErbB2-overexpressing breast cancers. Cancer Res. 2006;66:1640–7. doi: 10.1158/0008-5472.CAN-05-2000. [DOI] [PubMed] [Google Scholar]
- 23.Small D, Kovalenko D, Kacer D, et al. Soluble Jagged 1 represses the function of its transmembrane form to induce the formation of the Src-dependent chord-like phenotype. J Biol Chem. 2001;276:32022–30. doi: 10.1074/jbc.M100933200. [DOI] [PubMed] [Google Scholar]
- 24.Xia F, Altieri DC. Mitosis-independent survivin gene expression in vivo and regulation by p53. Cancer Res. 2006;66:3392–5. doi: 10.1158/0008-5472.CAN-05-4537. [DOI] [PubMed] [Google Scholar]
- 25.Mesri M, Wall NR, Li J, Kim RW, Altieri DC. Cancer gene therapy using a survivin mutant adenovirus. J Clin Invest. 2001;108:981–90. doi: 10.1172/JCI12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Zhang Y, Li Y, et al. Down-regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol Cancer Ther. 2006;5:483–93. doi: 10.1158/1535-7163.MCT-05-0299. [DOI] [PubMed] [Google Scholar]
- 28.O'Connor DS, Wall NR, Porter AC, Altieri DC. A p34(cdc2) survival checkpoint in cancer. Cancer Cell. 2002;2:43–54. doi: 10.1016/s1535-6108(02)00084-3. [DOI] [PubMed] [Google Scholar]
- 29.Fraker LD, Halter SA, Forbes JT. Growth inhibition by retinol of a human breast carcinoma cell line in vitro and in athymic mice. Cancer Res. 1984;44:5757–63. [PubMed] [Google Scholar]
- 30.Yehiely F, Moyano JV, Evans JR, Nielsen TO, Cryns VL. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol Med. 2006;12:537–44. doi: 10.1016/j.molmed.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 31.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–72. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Es JH, van Gijn ME, Riccio O, et al. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–63. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 35.Beverly LJ, Felsher DW, Capobianco AJ. Suppression of p53 by Notch in lymphomagenesis: implications for initiation and regression. Cancer Res. 2005;65:7159–68. doi: 10.1158/0008-5472.CAN-05-1664. [DOI] [PubMed] [Google Scholar]
- 36.Lens SM, Vader G, Medema RH. The case for Survivin as mitotic regulator. Curr Opin Cell Biol. 2006;18:616–22. doi: 10.1016/j.ceb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Leung CG, Xu Y, Mularski B, et al. Requirements for survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. J Exp Med. 2007;204:1603–11. doi: 10.1084/jem.20062395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dohi T, Xia F, Altieri DC. Compartmentalized phosphorylation of IAP by protein kinase A regulates cytoprotection. Mol Cell. 2007;27:17–28. doi: 10.1016/j.molcel.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh RP, Dhanalakshmi S, Agarwal C, Agarwal R. Silibinin strongly inhibits growth and survival of human endothelial cells via cell cycle arrest and downregulation of survivin, Akt and NF-κB: implications for angioprevention and antiangiogenic therapy. Oncogene. 2005;24:1188–202. doi: 10.1038/sj.onc.1208276. [DOI] [PubMed] [Google Scholar]
- 40.Nicolas M, Wolfer A, Raj K, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–21. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 41.Ghosh JC, Dohi T, Raskett CM, Kowalik TF, Altieri DC. Activated checkpoint kinase 2 provides a survival signal for tumor cells. Cancer Res. 2006;66:11576–9. doi: 10.1158/0008-5472.CAN-06-3095. [DOI] [PubMed] [Google Scholar]
- 42.Liu S, Dontu G, Wicha MS. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 2005;7:86–95. doi: 10.1186/bcr1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taubert H, Wurl P, Greither T, et al. Stem cell-associated genes are extremely poor prognostic factors for soft-tissue sarcoma patients. Oncogene. 26:7170–4. doi: 10.1038/sj.onc.1210530. Epub 2007 May 21. [DOI] [PubMed] [Google Scholar]
- 44.Weinstein IB, Joe AK. Mechanisms of disease: oncogene addiction-a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3:448–57. doi: 10.1038/ncponc0558. [DOI] [PubMed] [Google Scholar]
- 45.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 46.Roy M, Pear WS, Aster JC. The multifaceted role of Notch in cancer. Curr Opin Genet Dev. 2007;17:52–9. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Shih Ie M, Wang TL. Notch signaling, γ-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–82. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 48.Fortini ME. γ-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat Rev Mol Cell Biol. 2002;3:673–84. doi: 10.1038/nrm910. [DOI] [PubMed] [Google Scholar]
- 49.Wong GT, Manfra D, Poulet FM, et al. Chronic treatment with the γ-secretase inhibitor LY-411,575 inhibits β-amyloid peptide production and alters lymphopoiesis and intestinal cell differentiation. J Biol Chem. 2004;279:12876–82. doi: 10.1074/jbc.M311652200. [DOI] [PubMed] [Google Scholar]