Abstract
The identity of any cell type is determined by the specific pattern of gene expression. We show here that the ability of oligodendrocyte progenitors to acquire the identity of myelin-expressing cells or choose alternative fates is dependent on the activity of histone deacetylases. Using gene expression profiling, electrophysiological recordings, transplantation studies, and pharmacological inhibition, we demonstrate that specified NG2+ oligodendrocyte progenitors are plastic cells, whose decision to initiate an oligodendrocytic rather than astrocytic or neuronal program of gene expression requires the establishment of an epigenetic identity that is initiated by histone deacetylation.
Keywords: gene, oligodendrocyte, lineage, histone modification, neurogenesis, gliogenesis
Introduction
Cell replacement strategies using multipotential progenitors and stem cells are of great therapeutic significance for a wide variety of clinical disorders. For this reason, elucidating the mechanisms that regulate cell fate decisions after birth is of critical importance. In this manuscript, we use oligodendrocyte progenitors (OPCs) to address this question in the postnatal brain. OPCs were originally identified as the precursors of oligodendrocytes, the myelin-forming cells of the brain and spinal cord. Recent studies have shown that these cells have the ability to revert to stem-like cells (Kondo and Raff, 2000, 2004) and generate interneurons (Belachew et al., 2003; Aguirre and Gallo, 2004; Dayer et al., 2005). The mechanisms underlying the decision of an oligodendrocyte progenitor to become a myelinating oligodendrocyte or choose a distinct neural lineage are only partially understood. It is well accepted that the identity of distinct cell types is affected by extracellular factors, whose signaling pathways modulate the expression of critical transcription factors. However, distinct cell types need to retain their own unique “memory” while responding to the extracellular environment (Shen et al., 2007). This memory is engraved in the cell's genome and is achieved by a series of posttranslational modifications of histones and DNA methylation (Buszczak and Spradling, 2006). Among the posttranslational modifications, acetylation of lysine residues in the N-terminal tail of nucleosomal histones (mediated by histone acetyl transferases) is associated with transcriptionally competent chromatin (Ashraf and Ip, 1998; Strahl and Allis, 2000). Histone deacetylation, in contrast, is catalyzed by histone deacetylases (HDACs) and is functionally associated with transcriptional repression (Csordas, 1990; Rice and Allis, 2001). Because persistent histone acetylation in progenitors prevents the establishment of the mature oligodendrocytic phenotype (Shen et al., 2005), we asked whether this posttranslational modification is also responsible for alternative fate choice of these cells.
Materials and Methods
Immunocytochemistry of primary cultures of oligodendrocyte progenitors.
Oligodendrocyte progenitors were isolated from the cortex of neonatal rats using a positive selection strategy with antibodies against A2B5+, followed by incubation with anti-Ig magnetic microbeads (50 nm in size) and separation with a high-gradient magnetic field generated by the permanent magnet in the MACS Separator unit (Miltenyi Biotec, Auburn, CA). The immunoselected progenitors were cultured on Permanox chambers as described previously (Liu et al., 2003) and characterized using immunocytochemistry. The A2B5+ immunoselected population was composed of 99 ± 0.5% NG2+ cells and 94 ± 1% PDGF receptor α-positive cells and thereby represented a pure population of oligodendrocyte progenitors (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). These cells were induced to differentiate into oligodendrocytes by removing the mitogens from the medium and culturing them in the presence or absence of the HDAC inhibitors trichostatin A (TSA; 10 ng/ml) or valproic acid (VPA; 1 mm). The ability of oligodendrocyte progenitors to generate distinct cell types was further tested by culturing them in stem cell differentiation (SCD) medium (DMEM/F-12 plus 1 mm glutamine, 1% FBS, 25 mm glucose, and B27 supplement). For immunofluorescence, cells were incubated with antibodies against vimentin (Developmental Studies Hybridoma Bank, Iowa City, IA), nestin (Developmental Studies Hybridoma Bank), TuJ1 (Covance, Berkeley, CA), GFAP (Dako, Carpinteria, CA), and NPY (Sigma, St. Louis, MO) for 1 h at room temperature, followed by incubation with the appropriate secondary antibodies. Immunoreactive cells were analyzed by fluorescence microscopy (DM-RA; Leica, Bannockburn, IL), and the images were captured using a Hamamatsu (Hamamatsu, Japan) CCD camera.
In vivo treatment with HDAC pharmacological inhibitors.
Postnatal day 2 (P2) rats were injected subcutaneously with either PBS (n = 6) or VPA (300 mg/kg weight; n = 6) for 7 d. Each animal received a total of 13 injections, administered every 12 h. On postnatal day 9, the injected animals were anesthetized and perfused intracardially with 4% paraformaldehyde (PFA) in 0.1 m PBS. The brains were removed, postfixed overnight in the same solution, cryopreserved, and then sectioned into 20 μm coronal sections. Only sections corresponding to the levels +0.5 mm to +0.26 mm from bregma were selected for immunohistochemical analysis.
Cell transplantation.
A2B5+/NG2+ oligodendrocyte progenitors were isolated from the cortex of neonatal rats expressing green fluorescent protein (GFP) from the actin promoter (Okabe et al., 1997; Ito et al., 2001), using anti-Ig magnetic microbeads. A total of 2 × 105 GFP+/NG2+ oligodendrocyte progenitors in 1 μl were transplanted using a micropipette attached to a Hamilton syringe, into anesthetized P2 recipients (stereotactic coordinates, right, 0.5 mm; anterior to bregma, 0.5 mm; depth, 1.5 mm). After transplantation, the recipients received systemic administration of either PBS or VPA (300 mg/kg body weight), twice a day for 7 d. At the end of treatment, pups were perfused with 4% PFA, and the brains were removed, postfixed in 4% PFA, and cryopreserved.
Immunohistochemistry of rat brain sections and cell counts.
Cryosections of 20 μm thickness were washed, blocked with 10% NGS in a phosphate-based buffer containing 0.1% gelatin, 1% bovine serum albumin, and 0.5% Triton X-100. The following primary antibodies were used for immunostaining: GFP (Millipore, Billerica, MA), GFAP (Dako), APC/CC1 (EMD Biosciences, San Diego, CA), NeuN (Millipore), and neurofilament media chain (NFM; Millipore). Confocal images were obtained using the Zeiss (Thornwood, NY) LSM 510 Meta confocal laser-scanning microscope. Optical sections (Z = 1.0 μm) of confocal epifluorescence images were sequentially acquired with LSM5 Image Browser software, using a 25× objective (numerical aperture 0.80). The same software was used to merge images that were further processed in Photoshop 7.0 (Adobe Systems, San Jose, CA). Transplanted GFP+ cells were counted in the corpus callosum and cingulum 1 week after grafting. The anatomical distribution of GFP+/CC1+, GFP+/NeuN+, and GFP+/GFAP+ cells was analyzed in Z-series confocal scanning images (20 μm thickness; step size, 2.0 μm for 25× between successive images of the same field). The immunoreactive cells were analyzed in the above-mentioned brain regions surrounding the injection site. An average of 20 total coronal tissue sections per brain (20 μm thickness) from at least three to four transplanted animals were analyzed. The total number of double immunoreactive cells was expressed as a percentage of the total number of GFP+ cells. Cell-counting data in tissue sections are expressed as averages ± SEM. Statistical analysis was performed by Student's t test.
Gene expression profiling studies and statistical analysis.
Total RNA was isolated from oligodendrocyte progenitor cells, differentiated oligodendrocytes, or cells treated with TSA using Trizol (Invitrogen, Carlsbad, CA) and cleaned up with RNeasy Mini kit (Qiagen, Hilden, Germany). RNA from each group (n = 3) was pooled in equimolar amount before used for experiments with GeneChip Rat Genome U34A Arrays (Affymetrix, Santa Clara, CA), which contain probes for detecting 8000 transcripts. After RNA isolation, all of the subsequent technical procedures, including quality control, measurement of RNA concentration, cDNA synthesis, biotin labeling of cRNA, hybridization, and scanning of the arrays, were performed using standard procedure at the Cancer Institute of New Jersey. Differential gene expression was determined by the regularized t test, which uses a Bayesian procedure (Baldi and Long, 2001). Briefly, the expression level of each gene is assumed to be from a normal distribution with μ and σ2. Using a conjugate prior, the mean of the posterior (MP) estimate of μ is the sample mean. The MP estimate of σ2 is σ2 = [ν0σ02 + (n − 1)s2]/(ν0 + n − 2), where n is the sample size, s2 is the sample variance, ν0 is the degrees of freedom of the prior (a value of 10 is used in the analysis), and σ02 is the mean of sample variances of genes in the neighborhood of the gene under consideration. The neighborhood is the 50 genes with sample means immediately above and below the sample mean of the gene under consideration; that is, the neighborhood consists of the 101 genes centered on the gene. After the MP estimates of μ and σ2 were obtained, the t test of unequal variances was used to calculate a nominal p value of differential expression. Then the nominal p values was adjusted by the Benjamini and Hochberg method (Benjamini and Hochberg, 1995) to control the false discovery rate.
Reverse transcription-PCR analysis.
For reverse transcription (RT)-PCR, 9 μg of total RNA was used in 40 μl of RT reaction, using an RT-PCR kit (Invitrogen). The RT-PCR was performed in a 20 μl reaction mixture containing 2 μl of cDNA as template and 0.1 μm of the specific oligonucleotide primer pair. Cycle parameters were 30 s at 94°C, 30 s at 55°C, and 1.0 min at 72°C for 25 cycles. The DNA primers used were as follows: vimentin, 5′ primer, 5′-TGACAATGCTTCTCTGGCAC-3′; 3′ primer, 5′-TCCAGCAGCTTCCTGTAGGT-3′; reelin, 5′ primer, 5′-CTCCTCGGAAAACTACAGCG-3′; 3′ primer, 5′-CACCTGGTTGTCCATGTGAG-3′; neuropeptide Y, 5′ primer, 5′-TGGACTGACCCTCGCTCTAT-3′; 3′ primer, 5′-AACGACAACAAGGGAAATGG-3′; S100, 5′ primer, 5′-TGGGCTTCCAGAGCTTTCTA-3′; 3′ primer, 5′-TTCTTTATTGAGGGCAACCG-3′; nestin, 5′ primer, 5′-TGCAGCCACTGAGGTATCTG-3′; 3′ primer, 5′-CAGTTCCCACTCCTGTGGTT-3′; GFAP, 5′ primer, 5′-GGTGGAGAGGGACAATCTCA-3′; 3′ primer, 5′-TCCTTAATGACCTCGCCATC-3′; rat ceramide-galactosyltransferase (CGT), 5′ primer, 5′-GGAGTGCTGTTGGAATAGCAA-3′; 3′ primer, 5′-CGTACTCCTAGAACACAGACTT-3′; rat β-actin, 5′ primer, 5′-TGGAATCCTGTGGCATCC-3′; 3′ primer, 5′-TCGTACTCCTGCTTGCTG-3′.
Electrophysiological techniques.
Whole-cell patch-clamp recordings were performed on cells cultured in SCD medium for 4 d or cells pretreated with TSA for 1 d in SCD followed by 3 d in SCD medium alone. The external bath solution contained (in mm) 1.67 CaCl2, 1 MgCl2, 5.36 KCl, 137 NaCl, 17 glucose, 10 HEPES, and 13.15 sucrose. The pipette solution contained (in mm) 105 K-methanesulfonate, 17.5 KCl, 10 HEPES, 0.2 EGTA, 8 NaCl, 2 Mg-ATP, 2 Na2-ATP, 0.3 Na3-GTP, and 20 phosphocreatinine. The pH of the pipette solution was set to 7.3 with KOH. Pipette resistances were 3–5 MΩ. Potentials were recorded under current-clamp mode, digitized at 2.5 kHz, and filtered at 5 kHz. Signals were digitized with a CED Power1401 interface. For cell labeling experiments, 8-hydroxypyrene (1 mm; Sigma) was added to the recording pipette solution. After establishing a recording, hyperpolarizing current was injected to maintain a membrane potential of −80 mV. Depolarizing and hyperpolarizing current pulses were used to assess excitability. Data acquisition parameters and voltage pulse generation were controlled by custom software.
Results
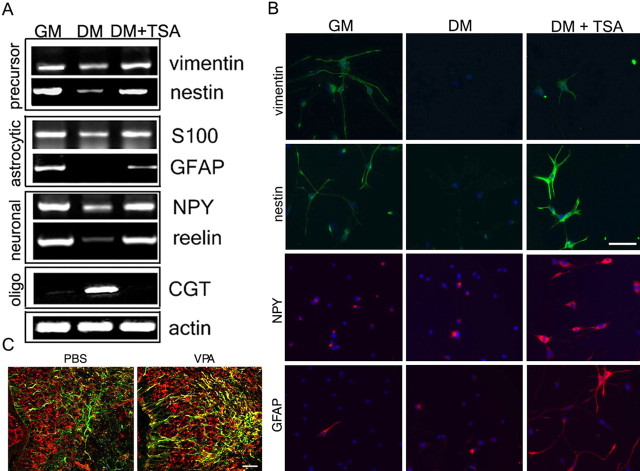
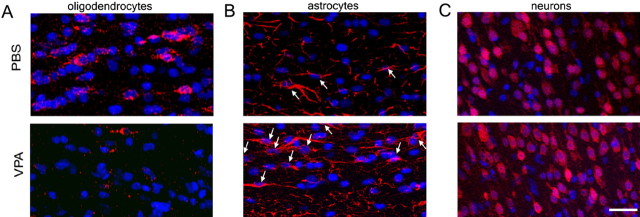
The mechanisms responsible for the acquisition of lineage-specific identity during postnatal development are only partially understood. We have previously shown that the decision of neonatal cortical progenitors to acquire oligodendrocytic identity is dependent on histone deacetylase activity (Shen et al., 2005, 2007). Because histone acetylation is functionally related to transcriptionally competent chromatin, we used gene expression profiling with Affymetrix microarrays to identify genes whose levels in differentiating oligodendrocyte progenitors were affected by this posttranslational modification of nucleosomes (supplemental Table 1, available at www.jneurosci.org as supplemental material). Among the genes whose expression was upregulated by persistent histone acetylation in oligodendrocyte progenitors, we identified early precursor (i.e., nestin and vimentin), astrocytic (i.e., tenascin and S-100), and neuronal (i.e., reelin, synaptotagmin, and neuropeptide Y) markers. These results were validated by RT-PCR (Fig. 1A) and suggested that the early stages of progenitor differentiation into oligodendrocytes were characterized by the downregulation of genes characteristic of other lineages modulated by histone deacetylation. Therefore, pharmacological inhibition of HDAC activity in cultured progenitors prevented differentiation into oligodendrocytes while favoring the expression of precursor, astrocytic, and neuronal markers (Fig. 1B). A similar effect was detected in vivo, after systemic administration of the HDAC inhibitor VPA, using a protocol that was previously reported to effectively increase global histone acetylation (Shen et al., 2005, 2007). The most striking consequence of maintaining high levels of global histone acetylation in the brain of VPA-treated animals (supplemental Fig. S2, available at www.jneurosci.org as supplemental material) was the persistence of immature cells characterized by nestin+ and GFAP+ immunoreactivity compared with PBS-injected controls (Fig. 1C). To define whether persistent histone acetylation in oligodendrocyte progenitors was associated with the persistence of an immature stage or with the choice of alternative lineages, we used immunohistochemistry (Fig. 2). The inability of HDAC to deacetylate histones in the progenitors of VPA-treated animals was associated with a 64.2 ± 7.6% reduction of oligodendrocyte generation and a corresponding 75 ± 8.7% increase of astrocytes and 23.7 ± 7.2% increase of neurons in the brain of VPA-treated animals relative to controls (Fig. 2). These data suggested that blocking histone deacetylation precluded the establishment of the oligodendrocytic lineage identity of progenitors and favored the differentiation of these cells along alternative lineages (Fig. 2).
Figure 1.
Oligodendrocyte progenitors express neuronal and astrocytic genes whose levels are regulated by global histone acetylation. A, RT-PCR of transcripts amplified from RNA isolated from rat immunoselected cortical oligodendrocyte progenitors cultured for 24 h in growth medium (GM) or in differentiation medium in the absence (DM) or presence of HDAC inhibitors (DM+TSA). Precursor, neuronal, and astrocytic genes were downregulated during oligodendrocyte differentiation and upregulated in response to HDAC inhibitors (i.e., TSA), whereas myelin genes (i.e., CGT) showed the opposite behavior. B, Immunofluorescence of progenitors in the same culture conditions, stained with antibodies specific for precursor (green; nestin and vimentin), neuronal (red; NPY), and astrocytic (red; GFAP) markers. 4′6′-Diamino-2-phenylindole dihydrochloride (DAPI; blue) was used as a nuclear stain. Scale bar, 100 μm. C, In vivo, in untreated rat pups (PBS), the expression of nestin (red) and GFAP (green) was rarely overlapping (yellow). In contrast, in VPA-treated animals, colabeling with nestin and GFAP cells (yellow) was observed at a much higher frequency. Scale bar, 50 μm.
Figure 2.
The lineage choice of endogenous oligodendrocyte progenitors can be manipulated by systemic administration of valproic acid. Postnatal day 2 rats were injected with PBS or VPA twice per day for 7 d. Oligodendrocytes, astrocytes, or neurons were identified in the P9 corpus callosum (A, B) and cortex (C), using antibodies against CC1 (A; red), GFAP (B; red; white arrows), or NeuN (C; red). DAPI (blue) was used as a nuclear stain. Scale bar, 20 μm.
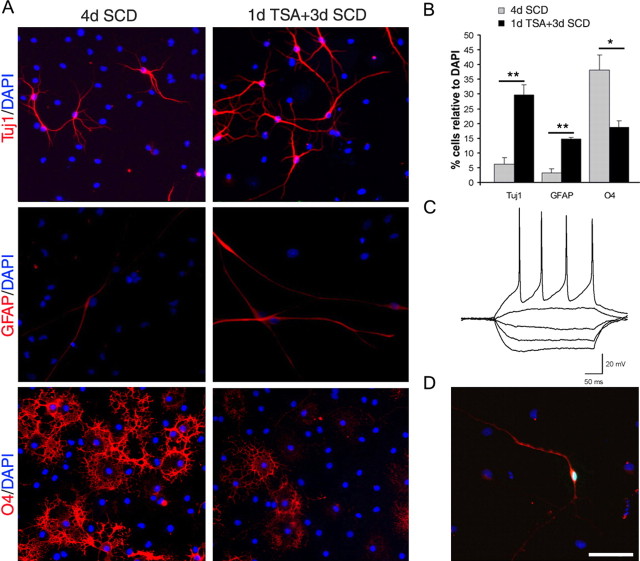
To better characterize the effect of HDAC inhibition on cell fate choice in the absence of systemic variables, we exposed primary cultures of immunoselected progenitors to the HDAC inhibitor TSA (Fig. 3A) for 1 d and then cultured the cells in conditions that had been previously defined as permissive for the differentiation along the three neural lineages (Belachew et al., 2003). In agreement with the in vivo results, the pretreatment with histone deacetylase inhibitors affected the epigenetic memory of oligodendrocyte progenitors and reduced by 50% the percentage of cells progressing into multipolar O4+ cells (Fig. 3A,B). The decreased percentage of oligodendrocyte lineage cells was associated with a threefold increase in the percentage of GFAP+ cells and a sixfold increase of TuJ1+ immunoreactive bipolar cells (Fig. 3A). To determine whether the cells in the TSA-treated cultures were functional neurons, we performed whole-cell patch-clamp recordings (Fig. 3C). Depolarizing current injections elicited action potentials in 86.6% (13 of 15 bipolar cells tested) of the cultures that were pretreated with TSA and confirmed that oligodendrocyte progenitors exposed to HDAC inhibitors had the functional properties of neurons. The neuronal identity of the cells was further assessed by filling the recorded cells with 8-hydroxypyrene injection and showing TuJ1 immunoreactivity after immunocytochemistry (Fig. 3D). Comparable morphological and electrophysiological results were obtained in cultures after VPA treatment (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). Together, these data indicate that the frequency of generation of functional neurons from OPCs can be dramatically increased by manipulation of the global acetylation levels of nucleosomal histones.
Figure 3.
Oligodendrocyte progenitors are multipotential cells whose fate choice is dependent on HDAC activity. A, Immunofluorescence of oligodendrocyte progenitors cultured for 4 d in SCD medium or pretreated with TSA for 1 d and then cultured in SCD medium for 3 d in the absence of TSA. Neurons were identified by TuJ1 immunoreactivity (red), astrocytes were identified by GFAP (red), and oligodendrocyte lineage cells were identified by O4 (red). DAPI (blue) was used as a nuclear stain. B, Bar graph showing the percentage of neurons, astrocytes, or O4+ oligodendrocytes generated in untreated (gray) or TSA-pretreated (black) cultures of oligodendrocyte progenitors. C, Example of current-clamp recording of a neuron-like cell from the TSA-pretreated condition. Action potentials in response to depolarizing currents were elicited in 13 of 15 cells. D, The bipolar cell recorded in C was labeled with 8-hydroxypyrene (green) while recording and then fixed and stained for TuJ1 (red). Scale bar, A, D, 100 μm. *p < 0.05; **p < 0.005.
To further determine whether in vivo astrocytes and neurons arise from redirected OPCs, we performed experiments to compare the fate of transplanted OPCs in experimental conditions affecting the global levels of histone acetylation. Positively immunoselected GFP+/A2B5+/NG2+ OPCs were isolated from the cortex of neonatal β-actin-egfp rats (Okabe et al., 1997; Ito et al., 2001) and transplanted into two groups of pups. One group received systemic administration of VPA, to enhance acetylation, whereas the control group received PBS. Seven days later, the animals were killed, and the brains were processed for immunofluorescence (Fig. 4). In PBS-treated recipients, 80 ± 5.4% of GFP+/NG2+ progenitors migrated to the corpus callosum and generated CC1+ oligodendrocytes, whereas only a small percentage of the remaining cells differentiated into GFP+/NeuN+ neurons (6 ± 3.5%) or GFP+/GFAP+ (6 ± 4%) astrocytes (Fig. 4A–C). In VPA-treated recipients, in contrast, only 14 ± 5.1% of GFP+/NG2+ transplanted cells differentiated into CC1+ oligodendrocytes, whereas the majority gave rise to GFAP+ astrocytes (29 ± 3.5%) and NeuN+ (43 ± 3.1%) neurons (Fig. 4A–C). Additional evidence for the neuronal identity of the GFP+ transplanted cells in VPA-treated animals was provided by the evidence of colocalization of GFP and NFM immunoreactivity (Fig. 4D). The mutually exclusive expression pattern of GFAP and NFM in the brain of VPA-treated animals suggested that global histone acetylation favored the differentiation of oligodendrocyte progenitors into neurons or astrocytes, rather than simply increasing the coexpression of astrocytic and neuronal markers within the same cells (Fig. 4E).
Figure 4.
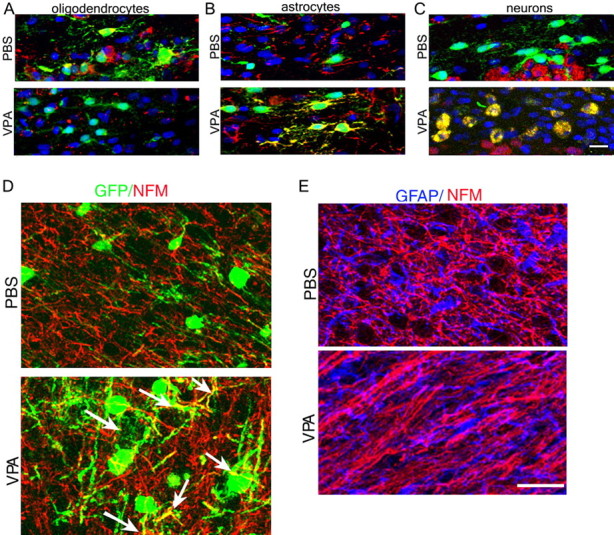
The lineage choice of transplanted immunoselected A2B5+/GFP+/NG2+ oligodendrocyte progenitors is affected by VPA. Confocal imaging of the medial corpus callosum in P10 rats transplanted with A2B5+/NG2+ progenitors isolated from GFP+ rats is shown. A–C, Note that in PBS-treated controls, the majority of the transplanted GFP+ cells (green) were also CC1+ (A, top, red) but not GFAP+ (B, top, red) or NeuN+ (C, top, red). DAPI (blue) was used as a nuclear stain. In contrast, in VPA-treated recipients, the majority of the transplanted cells were GFAP+ (B, bottom, yellow) or NeuN+ (C, bottom, yellow) but not CC1+ (A, bottom). D, Colabeling of GFP (green) and NFM (red) indicated the presence of double immunoreactivity (arrows) only after VPA treatment. E, Colabeling of GFAP and NFM showed their mutually exclusive expression in the cortex of VPA-treated animals and PBS controls. Scale bars: (in C) A–C, 25 μm; (in E) D, E, 80 μm.
Therefore, we conclude that persistent histone acetylation in OPCs alters their lineage choice decision, by suppressing the acquisition of the oligodendrocytic identity and favoring a conformation of chromatin that is consistent with the establishment of a neuronal or astrocytic phenotype.
Discussion
The concept of a molecular memory stored in the chromatin of the cells is a well characterized phenomenon in the study of imprinting and inheritance of parental traits (Zhou et al., 2005; Bantignies and Cavalli, 2006; Levenson and Sweatt, 2006). In the oligodendrocyte lineage, we and others have shown previously that global chromatin changes occur during developmental myelination (Shen et al., 2005) and that chromatin rearrangements are critical for the differentiation of oligodendrocytes (Marin-Husstege et al., 2002; Kondo and Raff, 2004; Hsieh et al., 2004).
In this study, we have combined gene-profiling studies of primary cultures of oligodendrocyte progenitors with electrophysiological recordings and transplantation studies to ask whether inhibitors of histone deacetylases “erase” the “oligodendrocyte epigenetic memory” in progenitors, thereby affecting their ability to choose among different lineages. The experimental results shown here suggest that the oligodendrocyte identity of OPCs is critically dependent on HDAC enzymatic activity. When HDAC activity is high, the oligodendrocytic epigenetic memory of specified progenitors is established by repressing neuronal and astrocytic genes as the cells progress to mature oligodendrocytes. However, when HDAC activity is inhibited, progenitors are unable to establish an oligodendrocyte-specific program of gene expression, and they are reprogrammed into a multipotential state that is responsive to neurogenic or astrogliogenic signals.
Therefore, this manuscript identifies global histone deacetylation as the epigenetic mechanism responsible for the definition of an epigenetic memory of oligodendrocyte lineage in OPCs. In agreement with this interpretation, in vitro experiments on cultured progenitors and in vivo experiments in developing rodents or transplanted recipients reveal that HDAC inhibition erases the oligodendrocyte memory of these cells, while allowing the acquisition of a pattern of gene expression consistent with neuronal and astrocytic lineage. More importantly, we show here that this effect translated into the ability of these “forgetful” OPCs to form functional neurons, rather than simply coexpress neuronal and glial markers within the same cell. The fact that histone acetylation enhances the expression of neuronal and astrocytic genes is consistent with previous work (Hsieh et al., 2004; Song and Ghosh, 2004; Ballas et al., 2001, 2005). Global histone acetylation is detected in precursor cells during the early stages of brain development that are associated with neurogenesis and astrogliogenesis. In contrast, histone deacetylation prevails in oligodendrocyte progenitors during the later stages of postnatal development and coincides with the onset of myelination (Shen et al., 2005). Therefore, we suggest that the establishment of an oligodendrocyte memory by using histone deacetylation is the likely result of an adaptive mechanism that precursor cells have selected to allow oligodendrocyte differentiation to remain silent during the neurogenic and astrogliogenic period.
The novel implication of our results is that erasing the epigenetic memory in oligodendrocyte progenitors, using HDAC inhibitors, is associated with the generation of functionally competent neurons. In addition to the developmental relevance, these findings might have significant therapeutic implications, considering that pharmacological compounds approved for the treatment of epilepsy could be used as stimuli favoring neurogenesis from glial progenitors.
Footnotes
This work was supported in part by National Multiple Sclerosis Society (NMSS) postdoctoral fellowship FA1534A1 (A.L.) and by NMSS Grant RG-3957 (P.C.-B.) and National Institutes of Health–National Institute of Neurological Disorders and Stroke Grants R01 42925 (P.C.-B.) and NS041310 (M.R.P.).
References
- Aguirre A, Gallo V. Postnatal neurogenesis and gliogenesis in the olfactory bulb from NG2-expressing progenitors of the subventricular zone. J Neurosci. 2004;24:10530–10541. doi: 10.1523/JNEUROSCI.3572-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, Ip YT. Transcriptional control: repression by local chromatin modification. Curr Biol. 1998;8:683–686. doi: 10.1016/s0960-9822(98)70435-x. [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr Opin Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Belachew S, Chittajallu R, Aguirre AA, Yuan X, Kirby M, Anderson S, Gallo V. Postnatal NG2 proteoglycan-expressing progenitor cells are intrinsically multipotent and generate functional neurons. J Cell Biol. 2003;161:169–186. doi: 10.1083/jcb.200210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- Buszczak M, Spradling AC. Searching chromatin for stem cell identity. Cell. 2006;125:233–236. doi: 10.1016/j.cell.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Csordas A. On the biological role of histone acetylation. Biochem J. 1990;265:23–38. doi: 10.1042/bj2650023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayer AG, Cleaver KM, Abouantoun T, Cameron HA. New GABAergic interneurons in the adult neocortex and striatum are generated from different precursors. J Cell Biol. 2005;168:415–427. doi: 10.1083/jcb.200407053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Suzuki A, Imai E, Okabe M, Hori M. Bone marrow is a reservoir of repopulating mesangial cells during glomerular remodeling. J Am Soc Nephrol. 2001;12:2625–2635. doi: 10.1681/ASN.V12122625. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science. 2000;289:1754–1757. doi: 10.1126/science.289.5485.1754. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004;18:2963–2972. doi: 10.1101/gad.309404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms: a common theme in vertebrate and invertebrate memory formation. Cell Mol Life Sci. 2006;63:1009–1016. doi: 10.1007/s00018-006-6026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Muggironi M, Marin-Husstege M, Casaccia-Bonnefil P. Oligodendrocyte process outgrowth in vitro is modulated by epigenetic regulation of cytoskeletal severing proteins. Glia. 2003;44:264–274. doi: 10.1002/glia.10290. [DOI] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil PA. A molecular insight of Hes-5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice' as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;3:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Liu A, Li J, Wolubah C, Casaccia-Bonnefil P. Epigenetic memory loss in aging oligodendrocytes in the corpus callosum. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2006.10.026. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MR, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Zhou GL, Xin L, Liu DP, Liang C. Remembering the cell fate during cellular differentiation. J Cell Biochem. 2005;96:962–970. doi: 10.1002/jcb.20572. [DOI] [PubMed] [Google Scholar]