Abstract
Human UDP-glucuronosyltransferase (UGT) 2B7 is the major isoform catalyzing the glucuronidation of a variety of endogenous compounds including bile acids. To determine the role of bile acids in the regulation of UGT2B7 expression, Caco-2 cells were incubated with the natural human farnesoid X receptor (hFXR) ligand, chenodeoxycholic acid, as well as the secondary bile acid, lithocholic acid, derived from chenodeoxycholic acid. Incubation of Caco-2 cells with lithocholic acid in the absence of exogenous hFXR resulted in a dose-dependent down-regulation of UGT2B7 mRNA levels, with an IC50 of 13 μM. Similar down-regulation was also observed with chenodeoxycholic acid; however, much higher concentrations were required. Transient transfection of Caco-2 cells with hFXR suppressed UGT2B7 mRNA expression both in the absence and presence of ligand. UGT2B7 promoter transfection experiments and deletion/mutation analysis showed that lithocholic acid-activated hFXR decreased UGT2B7 promoter activity via a negative hFXR response element (NFRE) located between nucleotides −148 and −134. Cotransfection with hFXR and/or human retinoid X receptor further enhanced the repression. Electrophoretic mobility shift assays additionally confirmed the role of NFRE in UGT2B7 down-regulation by lithocholic acid. These findings suggest that lithocholic acid, an activator of nuclear hFXR, acts as a negative regulator of UGT2B7 expression, indicating that hFXR may play an essential role in lithocholic acid homeostasis through negative regulation of this UGT that is involved in lithocholic acid biotransformation. Therefore, it is postulated that lithocholic acid toxicity may be due to down-regulation of genes involved in its detoxification, including UGT2B7, leading to limited excretion of lithocholic acid from the body.
Bile acids, in addition to their role in the emulsification of dietary lipids (Borgstrom et al., 1985), have been recognized as important signaling molecules and physiological ligands for several nuclear receptors (Chiang, 2002). The nuclear receptors that have been recognized as being involved in the regulation of biosynthesis, metabolism, and transport of bile acids are the human farnesoid orphan receptor (hFXR), the liver X receptor, the peroxisome proliferator-activated receptors (PPARs), the human pregnane orphan receptor (hPXR) (Chiang, 2002), and the human vitamin D receptor (hVDR) (Makishima et al., 2002). hFXR is the major bile acid receptor, and primary bile acids are its physiological ligands (Takeshita et al., 2002).
Growing evidence has been collected that bile acids are strongly involved in the regulation of their own synthesis and transport, as well as cholesterol homeostasis and lipid metabolism. It has been shown that bile acids activate hFXR, which regulates several target genes, including drug-metabolizing enzymes. Several examples of gene regulation via bile acid activation of nuclear receptors include cytochrome P450 7A1, which has been identified as a target gene of the bile acid-specific receptor, hFXR (Russell and Setchell, 1992). hFXR negatively regulates bile acid synthesis by decreasing transcription of cytochrome P450 7A1, and cholesterol 12α-hydroxylase, the enzyme responsible for cholic acid synthesis, is also under negative control by bile acids (Vlahcevic et al., 2000). In addition, hFXR activates transcription of ileal-bile acid binding protein (I-BABP), which is involved in the enterohepatic circulation of bile acid (Princen et al., 1997). Moreover, hFXR mediates bile acid activation of phospholipid transfer protein gene expression (Urizar et al., 2000), which suggests that bile acids and hFXR may play a role in high-density lipoprotein metabolism via the regulation of this gene. Several more bile acid transporters, including organic anion transport protein 2 and the bile salt export pump, which promotes bile acid efflux from the liver (Bjorkhem, 1985), have been recognized as hFXR target genes activated by bile acids (Ananthanarayanan et al., 2001).
It has been shown that hFXR also controls the metabolism of triglycerides. Bile acid-activated hFXR suppresses apolipoprotein A-I and apolipoprotein C-III transcription (Claudel et al., 2002, 2003). It was shown that the suppression of apolipoprotein A-I occurs via a novel negative hFXR response element (NFRE) (Claudel et al., 2002). Also, enzymes involved in bile acid detoxification, especially of lithocholic acid, have been identified as being under the control of bile acid-mediated regulation. Very recently, direct involvement of hFXR in the regulation of human UDP-glucuronosyltransferase (UGT) 2B4 has been demonstrated in human primary hepatocyte cultures (Barbier et al., 2003).
Recent findings indicate that three additional nuclear receptors, PXR, PPARα, and hVDR, accept bile acids as ligands and regulate bile acid synthesis and transport. It has been demonstrated that PPARα may modulate metabolic pathways common to hFXR. The first molecular evidence for cross talk between hFXR and PPARα pathways in humans has been found (Torra et al., 2003). Bile acid-mediated activation of PXR has been postulated in the regulation of multidrug resistance-associated protein 3 (Staudinger et al., 2003) and has been shown to regulate cytochrome P450 3A4 expression (Takeshita et al., 2002). Bile acids seem to be involved in the regulation of several biologically important metabolic pathways by the induction and/or suppression of hFXR, PPARα, and PXR target genes. hVDR is also activated by bile acids (Makishima et al., 2002). It has been demonstrated that lithocholic acid is a higher affinity ligand for hVDR than for either hFXR or PXR. The identification of hVDR as an intestinal bile acid sensor may indicate an additional mechanism for the protective effects of vitamin D and its receptor against toxic lithocholic acid. It has been demonstrated that activation of hVDR by lithocholic acid or vitamin D induces expression in vivo of cytochrome P450 3A enzymes that detoxify lithocholic acid in the liver and intestine.
Bile acids have been identified as excellent substrates for human UGTs (Gall et al., 1999). UGTs are phase II biotransformation enzymes, localized to the endoplasmic reticulum (Chowdhury et al., 1985), that catalyze the conjugation of glucuronic acid to a broad spectrum of endobiotic and xenobiotic substrates with diverse chemical structures. Endogenous substrates for UGTs include bilirubin, bile acids, fatty acids, prostaglandins, serotonin, thyroid hormones, biogenic amines, and steroid hormones (Radominska-Pandya et al., 1999). Additionally, UGTs have a large number of xenobiotic substrates, including fat-soluble vitamins, carcinogens, plant metabolites, environmental pollutants, and drugs such as acetaminophen, chloramphenicol, diethylstilbestrol, morphine, and salicylic acid (Burchell et al., 1998; Radominska-Pandya et al., 1999; Tukey and Strassburg, 2000). UGT2B7 is one of two major human isoforms involved in the glucuronidation of both the hydroxyl and carboxyl functions of bile acids.
In this work, we have identified the gene encoding human UGT2B7 as a new target of lithocholic acid-mediated suppression. The transcription of UGT2B7 is down-regulated by lithocholic acid, and we have collected reliable evidence that hFXR is involved in this process. This down-regulation could result in a dramatic accumulation of toxic lithocholic acid in cells and the development of severe cholestasis. It is anticipated that identification of the mechanism of lithocholic acid toxicity will result in the successful development of drugs that will prevent cholestasis and other lithocholic acid-mediated toxicities.
Materials and Methods
Cell Culture and Transient Transfections
The human hepatocellular carcinoma cell line, HepG2 (ATCC HB-8065), and the human colorectal adenocarcinoma cell line, Caco-2 (ATCC HTB-37), were obtained from the American Type Culture Collection (Manassas, VA). Both cell lines were maintained at 37°C, 5% CO2 in high glucose Dulbecco’s modified Eagle’s medium (DMEM) with Earle’s salts and L-glutamine (Invitrogen, Carlsbad, CA), supplemented with 1% nonessential amino acids, 1 mM sodium pyruvate, and 10% fetal bovine serum (Invitrogen). The culture medium was changed twice weekly during maintenance, and untransfected cells used for RNA isolation were harvested when they neared confluence. In experiments with transcription inhibitor, 1 μg/ml actinomycin-D was added to the culture medium together with vehicle or lithocholic acid, and mRNA was collected at various time points.
For transient transfection, HepG2 or Caco-2 cells were seeded in six-well plates at 1 × 106 and 2.5 × 105 cells per well, respectively. Transfections were performed in triplicate using the indicated amount of expression plasmid or empty parent vector and 10 μl of Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Five hours after transfection, the cells were washed in DMEM and replacement medium containing bile acids (Sigma-Aldrich, St. Louis, MO) at the concentration given in the figures or vehicle (0.1% DMSO) was added. The sodium salts of lithocholic acid, chenodeoxycholic acid, hyodeoxycholic acid, and ursodeoxycholic acid, were solubilized in 100% DMSO, and during experiments, control cells were exposed to the same DMSO concentration as the cells treated with bile acids. At 24 h post-transfection, the cells were treated again with bile acids or solvent in fresh DMEM. After a total of 48 h, all cells were washed in phosphate-buffered saline and harvested for RNA extraction.
Cell Viability Assay
Cell viability was determined by trypan blue exclusion. Cells were plated at a density of 5 × 105 cells per well in six-well plates and cultured for 24 h. Lithocholic acid, at the concentrations indicated in the figures, was then added. After an additional 48-h incubation, the cells were released from attachment to the plates with trypsin/EDTA (Invitrogen), incubated with 0.4% trypan blue (Mediatech, Herndon, VA) for 5 min, and counted using a hemocytometer. Since the monolayers were not washed prior to trypsin/EDTA treatment or scraping of adherent cells, cells that detached during incubation with lithocholic acid were detected.
RNA Isolation
Total RNA was isolated from cell cultures using a phenol and guanidine isothiocyanate RNA extraction method (TRIzol; Invitrogen), following the instructions of the supplier. To avoid any contamination of the RNA by genomic DNA, DNase treatment was performed using RQ1 RNase-Free DNase (Promega, Madison, WI).
cDNA was synthesized by mixing 1 μg of total RNA from each sample with 100-pmol random hexamer primers in 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol containing 100 U of Moloney murine leukemia virus reverse transcriptase, 20 U of RNase inhibitor, and a 1 mM concentration of each dNTP (Promega) in a total volume of 20 μl. The samples were incubated at 37°C for 60 min and then heated at 95°C for 5 min to inactivate the reverse transcriptase. The reaction mixture was diluted to 100 μl with sterile diethylpyrocarbonate-treated H2O.
Semiquantitative RT-PCR of hFXR, UGT2B7, PXR, and I-BABP
Primers for UGT2B7, hFXR, PXR, I-BABP, and GAPDH are described in Table 1. hFXR, PXR, UGT2B7, and I-BABP primers were designed in this study. PCRs were performed as follows. A 10-μl aliquot of cDNA was added to a reaction mixture containing 10 mM Tris-HCl buffer (pH 8), 20 mM KCl, 0.1% Triton X-100, 1.5 mM MgCl2, a 0.2 mM concentration of each dNTP, 50 pmol of each primer, and 2 units of TaqDNA polymerase (Promega), in a total volume of 50 μl. Amplification of the ubiquitously expressed GAPDH cDNA was performed under the same conditions in separate experiments. Table 1 details the annealing temperature and cycle number required for each template. Primer pairs were designed to specifically amplify across exon boundaries in mRNA from GAPDH, I-BABP, and UGT2B7 (Table 1). The specificity of all primer pairs was confirmed through sequencing of the PCR products. The PCR products were resolved by 2% agarose electrophoresis and detected by ethidium bromide. The bands were visualized under UV light and photographed by a computer-assisted camera. Quantification of each band was performed by densitometric analysis by using NIH Image Software (National Institutes of Health, Bethesda, MD).
TABLE 1.
Primers and conditions for semi-quantitative PCR analysis. Nucleotide sequences are reported for primer pairs used in RT-PCR analysis experiments in HepG2 and Caco-2 cells.
| Target Transcript | Primer Set | Annealing Temperature | Amplicon Size | No. of Cycles on HepG2 cDNA | No. of Cycles on Caco-2 cDNA |
|---|---|---|---|---|---|
| °C | bp | ||||
| FXR | F: ggaaccatactcgcaatacagc | 56 | 402 | 31 | 32 |
| R: tcgcatgtacatatccatcacac | |||||
| PXR | F: caagccaagtgttcacagtgag | 60 | 818 | 35 | 35 |
| R: caaagagcacagatcttccg | |||||
| UGT2B7 | F: agttggagaatttcatcatgcaacaga | 58 | 232 | 26 | 30 |
| R: tcagcccagcagctcaccacaggg | |||||
| I-BABP | F: gacttaggggctgagcctcagca | 60 | 491 | 34 | |
| R: ttgctcacgcgctcataggtcac | |||||
| GAPDH | F: acccactcctccacctttg | 64 | 178 | 25 | 25 |
| R: ctcttgtgctcttgctggg |
F, forward primer 5′-3′; R, reverse primer 5′-3′.
For each primer pair, we performed PCR with different cycle numbers and plotted these data to form a standard curve. We chose the cycle that was found to be within the nonsaturable range of amplification for use in further experiments. All other conditions were kept consistent unless significant changes in mRNA level were observed.
Real-Time PCR Assay
Real-time PCR analyses were performed with lithocholic acid-treated Caco-2 cells for comparison with the data obtained from semiquantitative PCR. Real-time PCR primer sequences for UGT2B7 and GAPDH were obtained from Congiu et al. (2002) and Knerr et al. (2002), respectively.
Inhibition of UGT2B7 transcription in the presence of varying concentrations of lithocholic acid was evaluated by real-time RT-PCR and used to find the IC50 value, which was determined using Prism 4 software (GraphPad Software Inc., San Diego, CA). The real-time PCR reaction mixture contained 5 μl of the sample and 20 μl of a master mix containing forward and reverse primers (300 nM each), IQ SYBR Green Supermix (2×; Bio-Rad, Hercules, CA) and the appropriate volume of sterile water. PCR conditions were 3 min at 95°C, followed by 40 cycles of 95°C for 10 s and 60°C for 45 s. Optical data were collected during the 60°C step. PCRs were carried out in 96-well thin-wall PCR plates covered with optically clear sealing film (Bio-Rad). Amplification, detection, and data analysis were performed with an iCycler IQ real-time detection system (Bio-Rad).
A standard curve was generated from a cDNA dilution series (1 ng, 5 ng, 25 ng) constructed as reference samples from the DMSO control sample. Real-time PCRs of UGT2B7 and GAPDH were performed in duplicate on both the experimental cDNA samples (10 ng) and reference standards. Values for target abundance in each experimental sample were extrapolated from the standard curve generated with the reference standards. The final data were expressed as the ratio of the values for the genes of interest and the corresponding values for GAPDH.
Isolation of UGT2B7 Promoter Fragments
Clones containing UGT2B7 promoters were isolated from a human placenta lambda genomic library (BD Biosciences Clontech, Palo Alto, CA) after several rounds of screening. Briefly, DNA fragments from the 5′-ends of cDNAs encoding the UGTs of interest were labeled with [32P]dATP by random priming using the DECA-prime II DNA labeling kit (Ambion, Austin, TX). Hybond-N membranes (Amersham Biosciences Inc., Piscataway, NJ) from plaque lifts were denatured, neutralized, and UV cross-linked according to the manufacturer’s protocol (PT1010-1). Prehybridization was carried out in a solution of 5× SSC (0.375 M NaCl, 0.0375 M sodium citrate), 5× Denhardt’s (Ficoll, polyvinylpyrrolidone, and bovine serum albumin, 1 mg/ml each), and 0.5% SDS for 4 h at 42°C before hybridization with 1 × 106 cpm/μl of probe overnight. Membranes were washed twice in 2× SSC, 0.1% SDS for 5 min at room temperature, and twice in 0.1× SSC, 0.1% SDS for 30 min at 65°C followed by exposure to autoradiography film with intensification screens at −70°C overnight. Positive plaques were picked and screened by restriction digestion and sequencing for appropriate UGT promoter inserts. A UGT2B7 1.3-kb proximal promoter was amplified from these clones by PCR, using primers designed with information from the sequencing of the genomic clones and/or from the human genome databases. The primers contained KpnI and Mlu1 sites for directional cloning into the KpnI and MluI sites of the pGL3-Basic luciferase reporter vector (Promega).
Generation of the UGT2B7 Promoter-Luciferase Reporter Constructs
A series of 5′ deletion constructs containing different promoter fragments were generated from the 1.3-kb proximal UGT2B7 promoter construct [2B7(p1328)] using the Erase-a-Base system (Promega). The plasmid containing the insertion was double-digested by MluI and KpnI. The linear fragment was further digested by Exonuclease III and aliquots were removed at different times to obtain a series of 5′ deletions. S1 nuclease was added to remove the remaining 5′ overhang, and the resulting blunt ends were linked by adding Klenow fragment, dNTPs, and T4 DNA ligase. Plasmids were transformed into the E. coli DH10B strain and incubated at 37°C overnight. Positive clones containing the UGT2B7 promoter were verified by direct DNA sequencing.
Deletions and Site-Directed Mutagenesis
For deletion of the NFRE sequence in 2B7(p1328)-Luc to create 2B7(p1315)NFREΔ-Luc, two oligonucleotides were designed by Oligo Lite software (Molecular Biology Insights, Inc., Cascade, CO) and synthesized by Integrated DNA Technologies (Coralville, IA): sense, 5′-GGCATGTCCATACAATAGCTGAAGGATAGCAC-3′; antisense, 5′-GTGCTATCCTTCAGCTATTGTATGGACATGCC-3′. The NFRE was in the middle of the oligonucleotides with 16 bases of correct sequence on either side. Constructs with deletions within the NFRE, 2B7(p1328)mut-Luc and 2B7(p428)mut-Luc, were prepared using the following primers: sense, 5′-CCATACAAGATCCTTATAGTGTGCTGAAGGATAGC-3′; antisense, 5′-GCTATCCTTCAGCACACTATAAGGATCTTGTATGG-3′ (bases within the NFRE are underlined and the mutated bases are shown in bold). The deletions were done using the Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) and verified by direct DNA sequencing.
Dual-Luciferase Assay
Caco-2 cells were seeded in 12-well plates at 0.5 × 105 cells per well the day before transfection. Transfections were performed in triplicate using 0.5 μg of UGT2B7 promoter-driven luciferase plasmid in the absence or presence of 100 ng of the hFXR and/or hRXR expression vectors, pCMX-hFXR and pCMX-hRXR. The total amount of DNA was supplemented by pCMX-null vector to 1.5 μg per well. Five hours after transfection, the cells were washed in DMEM and DMEM containing 1% fetal bovine serum, and either bile acid at the concentrations shown in the figures or vehicle (0.1% DMSO) was added. After a total of 48 h, all cells were washed in phosphate-buffered saline, lysed with Passive Lysis buffer (Promega), and centrifuged at 16,000g for 5 min at 4°C, and 20 μl of each supernatant was used to determine firefly luciferase activity. To normalize transfection efficiency, cells were cotransfected with 0.5 μg of the Renilla luciferase reporter plasmid pRL-TK (Promega), and dual luciferase activity was recorded using a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). Results are expressed as relative luciferase units after normalization to the Renilla luciferase activity.
Electrophoretic Mobility Shift Assay (EMSA)
For EMSAs, nuclear extracts were prepared from Caco-2 cells using a Nuclear Extract Kit (Active Motif Inc., Carlsbad, CA). Protein concentrations were determined by a Bio-Rad Protein Assay (Bio-Rad). The following single-stranded oligonucleotides and their antisense complements were synthesized by Integrated DNA Technologies: inverted repeat-1 (IR-1), (5′-GATCTCAAGAGGTCATTGACCTTTTTG-3′); apolipoprotein A-I, (5′-CAGAGCTGATCCTTGAACTCTTAAGTT-3′); NFRE, (5′-CATACAAGATCCTTGATATTAGCTGAA-3′); mutant IR-1, (5′-CAAATGAGCAAATTAACAATTCCAGT-G-3′); cyclic AMP response element-binding protein (CREB), (5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′).
Sense and antisense strands of the same element were annealed by heating at 65°C for 10 min and cooling to room temperature. Double-strand probes were labeled by γ- [32P]ATP by T4 polynucleotide kinase (Promega). Binding reactions contained 5 μg of crude nuclear extract and 2 μg of poly(dI-dC) · (dI-dC) in a buffer containing 10 mM HEPES, pH 7.9, 1 mM MgCl2, 50 mM KCl, 1 mM DTT, 0.1 mM EDTA, 10% glycerol, and, when appropriate, various molar excesses of unlabeled double-stranded oligonucleotides. Reactions were carried out at room temperature for 20 min and protein-DNA complexes were separated by electrophoresis on 6% polyacrylamide gels in 0.25× Tris/borate/EDTA buffer at 4°C. The gel was dried and exposed to blue sensitive film (Midwest Scientific, Valley Park, MO) and analyzed by NIH Image Software. Unlabeled double-stranded oligonucleotides corresponding to IR-1, apolipoprotein A-I, and NFRE mutant IR-1 and CREB were used as competitor DNAs.
Results
Lithocholic Acid Down-Regulates the Expression of UGT2B7
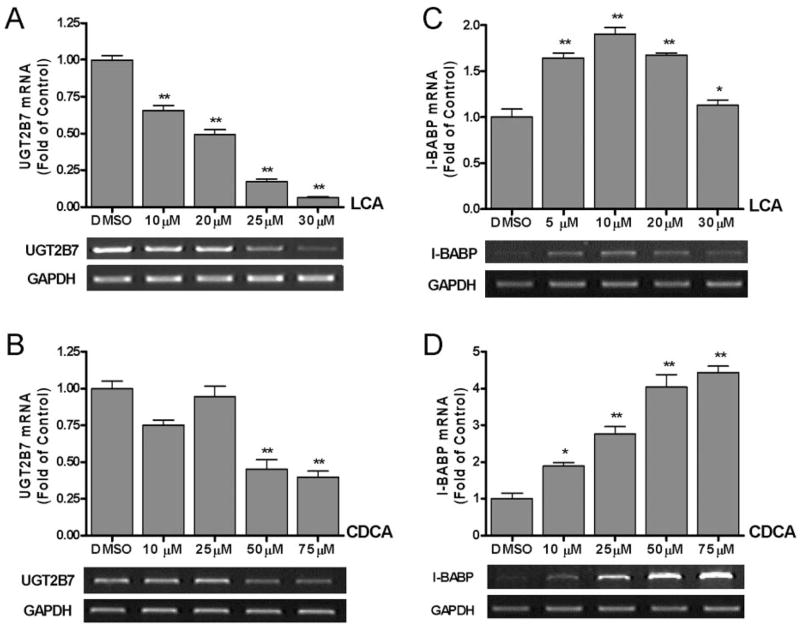
To determine whether UGT2B7 is a target of hFXR, Caco-2 and HepG2 cells were treated with various concentrations of lithocholic acid and chenodeoxycholic acid for 48 h. UGT2B7 mRNA levels at each concentration were analyzed by semiquantitative RT-PCR, and the final data were obtained with real-time RT-PCR. Lithocholic acid treatment of Caco-2 cells, which contain significant endogenous levels of both human hFXR and UGT2B7 mRNA (data not shown), resulted in a significant dose-dependent decrease in UGT2B7 mRNA levels as compared with vehicle-treated cells (Fig. 1A). Incubation of Caco-2 cells with increasing concentrations of chenodeoxycholic acid, a known hFXR activator, for 48 h had no significant effect on UGT2B7 mRNA levels until the concentration reached 50 μM (Fig. 1B). To confirm these results quantitatively, real-time RT-PCR experiments were performed (data not shown). An IC50 value of 13.7 μM for lithocholic acid-mediated inhibition of UGT2B7 mRNA expression was calculated from these real-time RT-PCR experiments.
Fig. 1.
The effect of lithocholic acid and chenodeoxycholic acid on UGT2B7 mRNA expression. Caco-2 cells seeded in six-well plates at a density of 0.5 × 106 cells per well were treated with various concentrations of lithocholic acid (10, 20, 25, 30 μM) (A and C) and chenodeoxycholic acid (10, 25, 50, 75 μM) (B and D) for 48 h. Total RNA was prepared, and UGT2B7 (A and B) and I-BABP (C and D) mRNA expression was analyzed by semiquantitative RT-PCR (as described under Materials and Methods). Results are expressed as the ratio of treated cells to control cells treated with vehicle (fold change from control), and are the mean ± S.E.M. of three experiments. The results were analyzed by one-way ANOVA followed by Dunnett’s multiple comparison test. Measurements that vary significantly from the DMSO control are indicated (★, p < 0.05; ★★, p < 0.01; ★★★, p < 0.001).
To test the response of UGT2B7 gene expression to other, nontoxic bile acids, hyodeoxycholic acid and ursodeoxycholic acid were tested in Caco-2 cells, and RT-PCR showed that neither hyodeoxycholic acid nor ursodeoxycholic acid induced or suppressed UGT2B7 mRNA levels (data not shown). Experiments carried out with HepG2 cells showed that neither lithocholic acid nor chenodeoxycholic acid, up to a concentration of 75 μM, affected UGT2B7 mRNA levels (data not shown). The lack of effect of chenodeoxycholic acid on UGT2B7 mRNA in HepG2 cells was in an agreement with recently published data by Barbier et al. (2003).
In the next series of experiments, we investigated and compared the effect of lithocholic acid and chenodeoxycholic acid on I-BABP expression. We selected I-BABP because it was the first gene identified as a target of chenodeoxycholic acid-activated hFXR, representing hFXR-mediated induction (Grober et al., 1999). As shown in Fig. 1C, lithocholic acid acted as an activator of I-BABP expression up to a concentration of 10 μM, but at 20 μM, mRNA levels began to decrease, and by 30 μM, mRNA expression had returned to basal levels. In contrast, chenodeoxycholic acid showed a strong activating effect on I-BABP up to 75 μM (Fig. 1D), in agreement with previous reports (Grober et al., 1999). These experiments with I-BABP confirmed that both lithocholic acid and chenodeoxycholic acid up-regulate the expression of I-BABP genes. Strong negative regulation by bile acids, especially lithocholic acid, was observed exclusively with UGT2B7. Moreover, UGT2B7 was also more sensitive to the higher concentrations of chenodeoxycholic acid. The direct comparison of the effect of chenodeoxycholic acid on UGT2B7 and I-BABP can be seen in Fig. 1, B and D. The experiments with I-BABP serve as important positive controls for our studies with lithocholic acid and chenodeoxycholic acid, proving that the effects of these two ligands are, in fact, gene-dependent.
To exclude the possibility that toxic lithocholic acid affects the integrity of Caco-2 cells, we checked their viability by trypan blue staining. All concentrations of lithocholic acid, including those that significantly decreased UGT2B7 mRNA expression, were found to be well tolerated by the cells, and no obvious cytotoxicity was observed (data not shown).
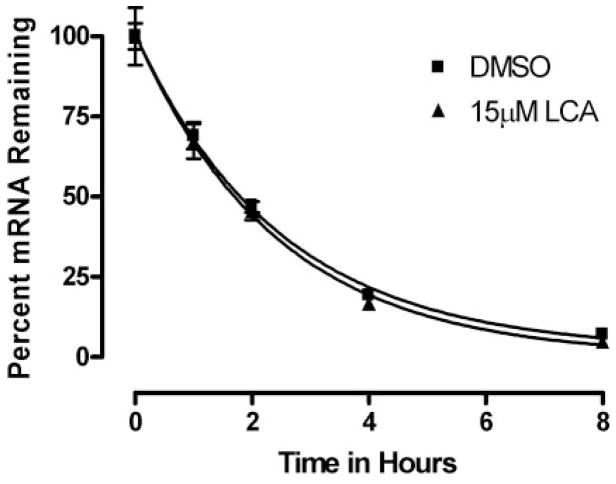
Finally, to assure that UGT2B7 mRNA suppression was not due to increased instability of mRNA in the presence of lithocholic acid, we used actinomycin-D, an inhibitor of transcription, to evaluate the stability of UGT2B7 mRNA in the presence of lithocholic acid. As shown in Fig. 2, the half-life of UGT2B7 mRNA was calculated by plotting the percentage of RNA remaining in actinomycin-D-treated cells versus time and fitting that plot using GraphPad Prism 4 software. The half-lives of mRNA decay in the presence and absence of lithocholic acid were 1.68 and 1.72 h, respectively. Thus, treatment with lithocholic acid did not affect the stability of UGT2B7 mRNA, indicating that the suppression of UGT2B7 mRNA is at the transcriptional level.
Fig. 2.
Analysis of stability of UGT2B7 mRNA in Caco-2 cells. Stability of UGT2B7 mRNA was analyzed by addition of actinomycin-D (1 μg/ml). Total RNA was extracted at various time points, and RT-PCR of UGT2B7 mRNA was performed. Plotted data are expressed as percentage of mRNA remaining and are representative of three independent experiments. The half-life of UGT2B7 mRNA in actinomycin-D-treated cells was calculated by fitting the percentage of mRNA remaining versus time data with a one-phase exponential decay equation using GraphPad Prism 4. (Y = span · e−K*X + plateau, where X = time, and Y = % mRNA remaining. Y = span + plateau at X = 0 and decreases to plateau with a rate constant, K. The half-life of the decay is 0.6932/K.)
Lithocholic Acid Down-Regulation of UGT2B7 Expression Does Not Involve PXR
In our previously published experiments in Caco-2 cells (Gardner-Stephen et al., 2004), three UGT1A isoforms, which have been proven to be PXR target genes, were induced by rifampicin (Rif) treatment. Since lithocholic acid is also a ligand for PXR (Xie et al., 2001), we investigated whether UGT2B7 is also a target gene of PXR. To eliminate the possibility that negative regulation of UGT2B7 by lithocholic acid could be mediated via PXR, Caco-2 cells were treated with Rif, a PXR-specific ligand (Kliewer and Willson, 2002), and it was demonstrated that Rif had no effect on UGT2B7 transcription (data not shown). In HepG2 cells, Rif did not change UGT2B7 mRNA expression. Additional experiments were conducted involving the transfection of Caco-2 cells with activated PXR in the presence and absence of lithocholic acid, and it was demonstrated that PXR transfection had no effect on UGT2B7 transcription (data not shown). The outcome of both sets of experiments suggests that PXR is not involved in UGT2B7 down-regulation.
The Effect of Chenodeoxycholic Acid and Lithocholic Acid on the 1.3-kb UGT2B7 Promoter Fragment
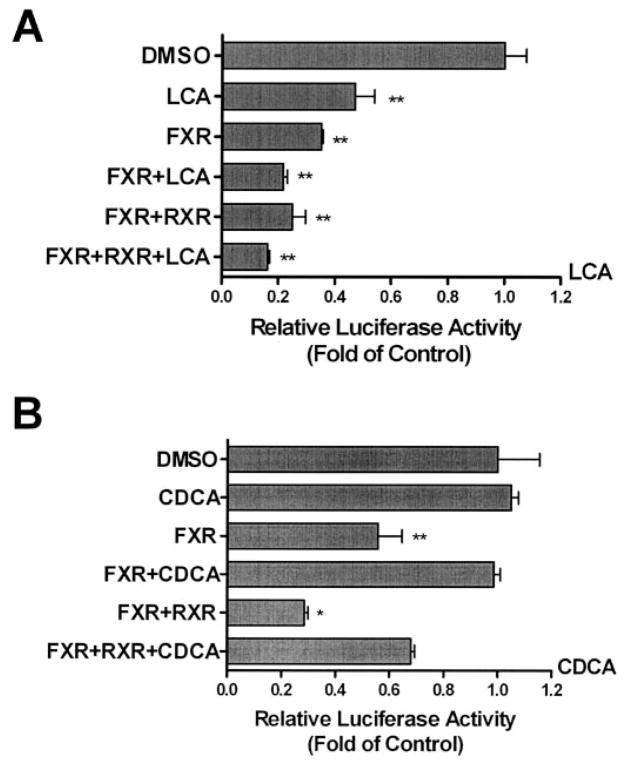
In the first series of experiments, we compared the effects of lithocholic acid and chenodeoxycholic acid on the 1.3-kb UGT2B7 promoter construct in Caco-2 cells cotransfected with hFXR and/or hRXR (Fig. 3). As observed previously, hFXR cotransfected into Caco-2 cells with 2B7(p1328)-Luc caused a decrease in luciferase activity, even in the absence of ligand, and cotransfection with hRXR additionally decreased the luciferase activity. However, the two bile acids had different effects on luciferase activity in these cotransfected cells. In the absence of receptors, 25 μM chenodeoxycholic acid did not suppress the luciferase activity, whereas 15 μM lithocholic acid resulted in a suppression of approximately 50%. In cells cotransfected with hFXR and then exposed to ligand, additional suppression was observed in cells exposed to lithocholic acid, but chenodeoxycholic acid seemingly protected the cells from some or all of the suppressive effects of hFXR and hRXR on luciferase activity.
Fig. 3.
Effect of chenodeoxycholic acid and lithocholic acid on the 1.3-kb UGT2B7 promoter. Caco-2 cells were transfected with 2B7(p1328)-Luc and either pCMX-hFXR, pCMX-hRXR, and/or pRL-TK and treated with vehicle, 15 μM lithocholic acid (A), or 25 μM chenodeoxycholic acid (B). Lysed cells were assayed for luciferase activity 48 h later. The results, normalized to Renilla luciferase activity, are expressed as fold change from control and are the mean ± S.D. of triplicate determinations. The results were analyzed by one-way ANOVA followed by Dunnett’s multiple comparison test. Measurements that vary significantly from the DMSO control are indicated (★, p < 0.05; ★★, p < 0.01; ★★★, p < 0.001).
Mapping of an hFXR Response Element in the Human UGT2B7 Promoter
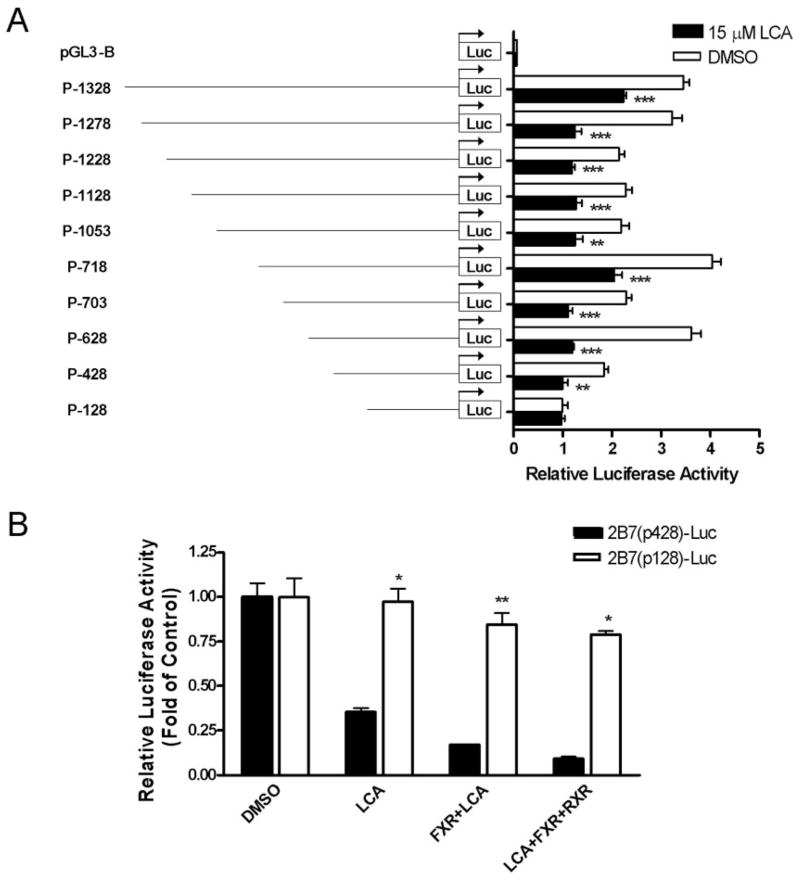
We next investigated whether the observed lithocholic acid-mediated changes in UGT2B7 gene expression were due to the response elements located in the gene promoter. To localize the region within the UGT2B7 promoter that contains transcriptional responsiveness to lithocholic acid, serial deletion constructs of the promoter, from 2B7(p1328)-Luc to 2B7(p428)-Luc, were generated, cloned in the pGL3-luciferase reporter plasmid, and transfected into Caco-2 cells, which were then treated with lithocholic acid (Fig. 4A). A significant decrease in reporter activity in response to lithocholic acid was observed for each deletion construct except p-128, indicating that the region controlling the suppressive effect of lithocholic acid on the UGT2B7 promoter was between −428 bp and −128 bp.
Fig. 4.
Analysis of the proximal promoter region of human UGT2B7 gene and the localization of a NFRE in the 428-kb UGT2B7 promoter fragment. In A, a series of DNA UGT2B7 promoter fragments was cloned into the pGL3-Basic luciferase reporter vector and tested for suppression of UGT2B7 in the presence of 15 μM lithocholic acid. Constructs were transiently transfected into Caco-2 cells together with pRL-TK. Cells were treated with vehicle or 15 μM lithocholic acid, and lysed cells were assayed for luciferase activity 48 h later. The results, normalized to Renilla luciferase activity, are expressed as relative luciferase units and are the mean ± S.D. of triplicate determinations. The results were analyzed by one-way ANOVA followed by Dunnett’s multiple comparison test. Measurements for lithocholic acid treatment that vary significantly from the DMSO control are indicated (★, p < 0.05; ★★, p < 0.01; ★★★, p < 0.001). In B, Caco-2 cells were transfected with 2B7(p428)-Luc or 2B7(p128)-Luc and either pCMX-hFXR, pCMX-hRXR, or pRL-TK. Cells were then treated with vehicle or 15 μM lithocholic acid, and lysed cells were assayed for luciferase activity 48 h later. The results, normalized to Renilla luciferase activity, are expressed as fold change from control, and are the mean ± S.D. of triplicate determinations. The results were analyzed by two-way ANOVA followed by the Bonferroni post hoc test. Measurements for the p-128 construct that vary significantly from the p-428 construct are indicated (★, p < 0.05; ★★, p < 0.01; ★★★, p < 0.001).
To further demonstrate that the −428- to −128-bp region of the UGT2B7 promoter was responsible for the hFXR-mediated lithocholic acid suppression, we measured the effect of lithocholic acid treatment and cotransfection with hFXR and/or hRXR on these promoter constructs. Caco-2 cells were transfected with the luciferase reporter plasmids driven by these constructs and cotransfected with the expression vectors for hFXR and hRXR in the presence and absence of lithocholic acid (Fig. 4B). Whereas the activity of the −428-bp promoter was suppressed by lithocholic acid, hFXR, and hRXR, this effect was completely abolished in the −128-bp promoter fragment. These results demonstrate the importance of the region between −428 and −128 bp in the regulation of the UGT2B7 promoter.
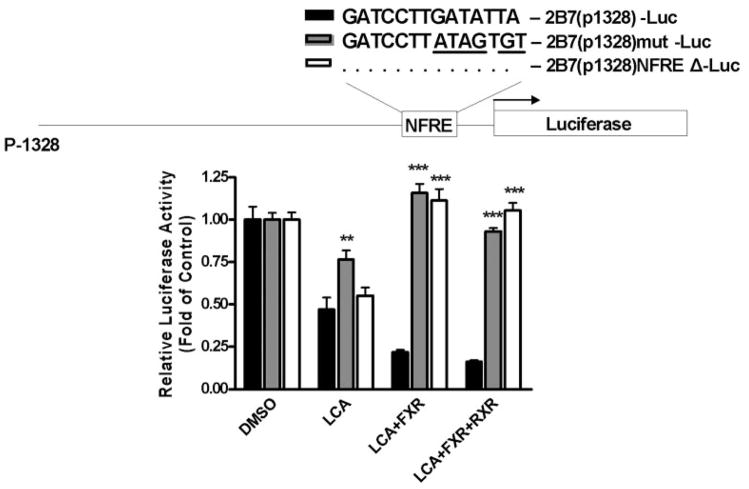
Identification and Mutation of a NFRE in the −428- to −128-bp Promoter Region
Computer analysis of the −428- to −128-bp region of the UGT2B7 promoter revealed the presence of a GATCCTTGATATTA sequence between nucleotides −148 and −134, which resembles the NFRE, 3′-GATCCTTGAACTCT-5′, previously identified in apolipoprotein A-I (Claudel et al., 2002). To determine whether this sequence could be a NFRE, this 14-bp region was deleted from the 1.3-kb promoter luciferase construct, resulting in the 2B7(p1315)NFREΔ-Luc construct. We also mutated this response element in the luciferase construct to create 2B7(p1329)mut-Luc. Both of these new constructs were then analyzed for lithocholic acid suppression and compared to that of 2B7(p1328)-Luc. As shown in Fig. 5, these mutations abolished the suppression of UGT2B7 promoter activity by hFXR and hRXR. However, the effect of lithocholic acid alone was not completely reversed. Two conclusions can be drawn from these data. First, deletion of the NFRE eliminates the suppression of UGT2B7 mRNA by hFXR and hRXR, suggesting that this is a true NFRE. Second, lithocholic acid suppression of UGT2B7 mRNA is only partially related to the presence of this NFRE; thus, a significant portion of the suppression is the result of another signaling pathway.
Fig. 5.
Mutation and deletion of NFRE in the 1.3-kb UGT2B7 promoter. A deletion construct, 2B7(p1315)NFREΔ-Luc, and a mutation construct, 2B7(p1315)mut-Luc, were prepared from 2B7(p1328)-Luc by PCR-based site-directed mutagenesis. Caco-2 cells were transfected with 2B7(p1315)NFREΔ-Luc, 2B7(p1315)mut-Luc, or 2B7(p1328)-Luc, and/or pCMX-hFXR, pCMX-hRXR and pRL-TK. Cells were then treated with vehicle or 15 μM lithocholic acid, and lysed cells were assayed for luciferase activity 48 h later. The results, normalized to Renilla luciferase activity, are expressed as fold of control, and are the mean ± S.D. of triplicate determinations. The results were analyzed by two-way ANOVA followed by a Bonferroni post hoc test. Measurements for the 2B7(p1315)NFREΔ-Luc or 2B7(p1315)mut-Luc constructs that vary significantly from the 2B7(p1328)-Luc construct are indicated (★, p < 0.05; ★★, p < 0.01; ★★★, p < 0.001).
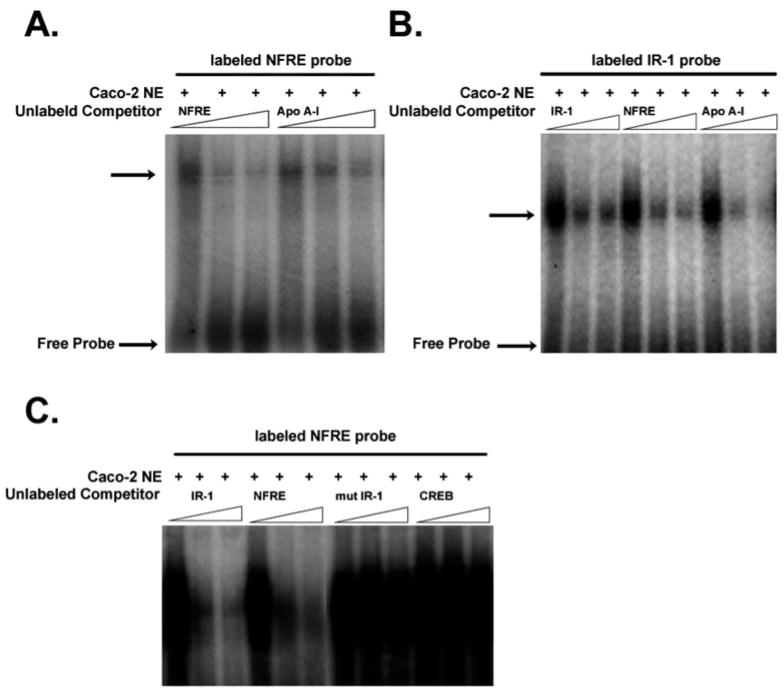
EMSA
Suppressive regulation of UGT2B7 by the NFRE indicated that this motif might be a binding site for hFXR. Binding of the hFXR complex to the NFRE in the UGT2B7 promoter was examined by EMSA (Fig. 6). As a control, a known NFRE from apolipoprotein A-I was also used as a probe. When nuclear extract prepared from Caco-2 cells treated with lithocholic acid was incubated with a 32P-labeled UGT2B7-NFRE probe, induced DNA-protein complexes were detected (Fig. 6A). Binding of hFXR was inhibited in a concentration-dependent manner by increasing concentrations of unlabeled oligonucleotides. A similar experiment was done with a 32P-labeled IR-1 probe (Fig. 6B). Competition for the labeled probe was observed when a 50-fold excess of unlabeled IR-1, UGT2B7-NFRE, or apolipoprotein A-I-NFRE was included in the reaction. To eliminate unspecific binding, experiments were done using mutated IR-1 and CREB oligonucleotides as nonspecific inhibitors (Fig. 6C). In contrast to the IR-1 and UGT2B7-NFRE probes, binding of the mutated IR-1 and CREB oligonucleotides to Caco-2 nuclear extract was not observed under these experimental conditions. These results indicated that hFXR binds specifically to the UGT2B7-NFRE.
Fig. 6.
A NFRE in the UGT2B7 promoter can bind to the hFXR/hRXR heterodimer in a nuclear extract of Caco-2 cells. EMSA analysis was carried out by incubating 32P-labeled NFRE and IR-1 sites with 5 μg of Caco-2 nuclear extract. In addition to specific competitors, mutant IR-1 and CREB oligonucleotides were used as nonspecific competitors. Competition experiments were performed by adding 0-, 50-, and 100-fold molar excesses of the indicated unlabeled probes. The specific hFXR/hRXR complex in nuclear extract is indicated by an arrow.
Discussion
In these studies, we have identified human UGT2B7 as a target gene of lithocholic acid-activated hFXR. However, UGT2B7 transcription is very strongly suppressed, rather than induced, by lithocholic acid. We have collected evidence that this lithocholic acid suppression occurs via hFXR binding to a novel NFRE in a 1.3-kb UGT2B7 promoter fragment. Our results also show that this suppression by hFXR is increased upon cotransfection with hRXR, indicating that hFXR binds to the NFRE as a heterodimer with hRXR. Thus, we have demonstrated that UGT2B7 is an hRXR-dependent, negatively regulated target gene of hFXR. This is in contrast to the bile acid-activated hFXR suppression of apolipoprotein A-I transcription mediated by a similar NFRE, but in a manner independent of hRXR (Claudel et al., 2002). Recently, it has been reported that another human UGT isoform, UGT2B4, is a target gene for hFXR, and induction of this UGT increases bile acid glucuronidation in HepG2 cells (Barbier et al., 2003). In this case, hFXR binds as a monomer to the bile acid response element in the UGT2B4 gene promoter. In addition, activation of hRXR represses the UGT2B4 gene expression induced by chenodeoxycholic acid, and over-expression of UGT2B4 in HepG2 cells results in reduced hFXR-mediated induction of the small heterodimeric partner (SHP).
Our results reveal that, in Caco-2 cells, lithocholic acid dramatically decreased expression of UGT2B7 mRNA levels (Fig. 1) and also decreased the luciferase activity of the UGT2B7 promoter. In contrast, this effect was not observed in HepG2 cells. We carried out a series of experiments to examine the effect of transfection of hFXR on UGT2B7 mRNA expression in Caco-2 cells. Transfection of cells with hFXR resulted in a very significant suppression of UGT2B7 expression in the absence of lithocholic acid, and additional suppression was observed when the cells were treated with lithocholic acid. The observation that hFXR and hFXR/hRXR suppress UGT2B7 transcription in the absence of ligand is not surprising. Many orphan receptors, such as human liver X receptor, are able to stimulate gene transcription without ligand binding (Peet et al., 1998). Another example has been presented for cytochromes P450 where, without bile acid activation, hFXR binds and stimulates rat cytochrome P450 7A1 mRNA expression in HepG2 cells; on the other hand, in the absence of hFXR, bile acids suppress the transcription of the cytochrome P450 7A1 gene (Chiang et al., 2000). Our data demonstrate that hFXR can act as a UGT2B7 transcriptional suppressor when over-expressed and presumably unliganded, as well as in the presence of one of its natural ligands, lithocholic acid.
hFXR may compete with other nuclear receptors for limiting co-activators, such as steroid receptor coactivator-1. For example, hepatic nuclear factor 4 binds to the direct repeat-1 motif in bile acid response element-II and plays a role in the transactivation of cytochrome P450 7A1 transcription (Crestani et al., 1998). Evidence has also been presented showing that the suppression of apolipoprotein A-I by bile acid, mediated by hFXR, is carried out in a manner independent of the SHP regulatory pathway, but via a NFRE (Claudel et al., 2002). It has been documented that PXR is a bile acid receptor that plays a critical role in bile acid detoxification by the liver; specifically, lithocholic acid has been identified as a PXR activator ligand (Xie et al., 2001). In the present studies, we found no evidence that lithocholic acid activation of PXR suppresses the transcription of UGT2B7. Specifically, the PXR-specific ligand, Rif, had no effect on the repression of UGT2B7 transcription in Caco-2 cells, indicating that PXR is not involved in this phenomenon. The exclusion of SHP and PXR involvement suggests that the down-regulation of UGT2B7 by lithocholic acid is mediated though the suppressor activity of hFXR.
To understand the mechanism of this negative regulation, we cloned a 1.3-kb fragment of the UGT2B7 promoter and carried out deletion analysis to identify the region of the promoter involved in the repression of UGT2B7 transcription. Promoter deletion identified a promoter fragment between −428 and −128 bp as being involved in the bile acid-activated hFXR-mediated decrease in UGT2B7 promoter activity. Computer-assisted analysis of this fragment revealed nucleotide sequences relevant to the binding of hFXR and regulation via lithocholic acid. It is recognized that hFXR response elements are inverted repeats in which consensus receptor-binding hexamers are separated by one nucleotide (IR-1). hFXR/hRXR heterodimers bind to these specific DNA motifs. However, hFXR/hRXR can also bind to direct repeats of the AGGTCA hexamer separated by 0, 3, or 4 nucleotides (Laffitte et al., 2000). Moreover, hFXR also binds as a monomer to a negative regulation site in the apolipoprotein A-1 gene promoter (Claudel et al., 2002). In this study, we have demonstrated that hFXR can bind to the slightly modified NFRE in the human UGT2B7 promoter in a manner similar to that described for the apolipoprotein A-1 promoter. Mutations of this element in the UGT2B7 −1328-bp promoter fragment abolished the repression of UGT2B7 promoter activity by hFXR and/or hRXR, indicating an absolute requirement of this fragment for the hFXR-induced repression.
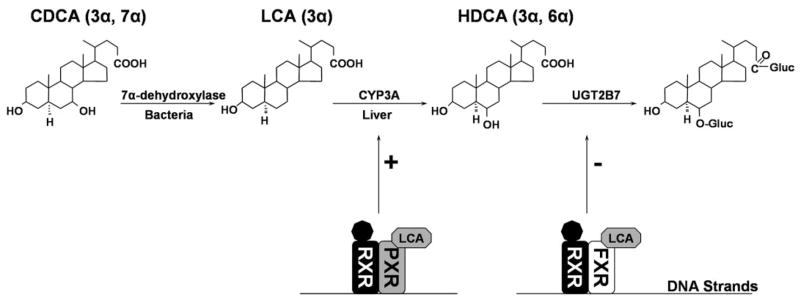
The physiological role of the UGT2B7 suppressive regulation pathway remains to be clearly delineated. We have demonstrated experimentally that cholestatic lithocholic acid is converted to the hydrophilic hyodeoxycholic acid in a reaction catalyzed by cytochrome P450 3A4 (Radominska et al., 1991; Xie et al., 2001). Since, in humans, hyodeoxycholic acid-6-O-glucuronide, rather than hyodeoxycholic acid, itself, is excreted in bile and urine, we postulated that coupling between cytochrome P450 3A4 and UGT2B7 occurs as an indispensable pathway for lithocholic acid detoxification (Radominska et al., 1991; Zimniak et al., 1991). We have also demonstrated that, in rats, lithocholic acid can be directly glucuronidated at the 3-OH position, forming the 3-O-glucuronide of lithocholic acid, one of the most cholestatic compounds identified thus far (Oelberg et al., 1984). The sequential detoxification of lithocholic acid by cytochrome P450 3A4 and UGT2B7 is highly specific to humans. We propose the pathway depicted in Fig. 7 as a putative mechanism for hFXR/PXR-mediated bile acid regulation of the transcription of several genes involved in the synthesis and detoxification of lithocholic acid. In Fig. 7, lithocholic acid is produced from chenodeoxycholic acid by the action of bacterial 7α-dehydroxylase. Subsequently, lithocholic acid is specifically hydroxylated at carbon 6 by cytochrome P450 3A4 and then conjugated at this hydroxyl group to produce a water-soluble glucuronide. We have shown previously that the 6-hydroxyl group of hyodeoxycholic acid and both the carboxyl and hydroxyl groups of lithocholic acid can be glucuronidated by human UGT2B7 (Gall et al., 1999).
Fig. 7.
Putative hFXR/PXR-mediated mechanism of bile acid regulation of gene transcription.
As presented in Fig. 7, there are three enzymes involved in the biosynthesis (7α-dehydroxylase) and detoxification (cytochrome P450 3A and UGT2B7) of lithocholic acid. Detoxification of lithocholic acid by cytochrome P450 3A4 is up-regulated by activation of cytochrome P450 3A4 expression by lithocholic acid-activated PXR. Cytochrome P450 3A4 converts lithocholic acid to hyodeoxycholic acid, which, as a substrate for UGT2B7, is effectively glucuronidated and then excreted in bile and/or urine. Up-regulation of cytochrome P450 3A4 via lithocholic acid is a significant hepatoprotective mechanism. However, we have unexpectedly discovered that UGT2B7, the final enzyme involved in this scheme, is down-regulated in Caco-2 cells by the presence of high levels of lithocholic acid. The down-regulation of this last enzyme involved in the lithocholic acid detoxification process could be highly deleterious both in the intestine and the liver.
We would like to emphasize that suppression of UGT2B7 mRNA levels might occur only in high concentrations of lithocholic acid. It has been demonstrated that low, micromolar concentrations of lithocholic acid are effectively cleared via sulfation (Radominska et al., 1993; Halvorsen et al., 1999), whereas at high concentrations, glucuronidation takes over the lithocholic acid detoxification process. Therefore, we postulate that the suppression of UGT2B7 expression might only occur under pathological conditions, such as liver damage and/or cholestasis, which are known to produce significant levels of lithocholic acid.
All this work is in contrast to work done by Barbier et al. (2003) in human hepatocytes, where hFXR activated via chenodeoxycholic acid induces UGT2B4 and may contribute to a reduction of bile acid toxicity, positively affecting bile acid metabolism and cholesterol homeostasis in human hepatocytes. The effect of lithocholic acid on UGT2B7 expression was not investigated in this hepatocyte model. In our experiments with UGT2B7 in Caco-2 or HepG2 cells, chenodeoxycholic acid did not show any activating effect. Nevertheless, these two human UGTs, both of which are involved in the glucuronidation of bile acid, despite their differing repressive/inductive mechanisms, are significantly regulated by their bile acid substrates.
In this study, we have demonstrated that hFXR negatively regulates UGT2B7 gene expression by binding to a NFRE. This is the first, novel demonstration of suppression of UGT genes in general and, specifically, in response to their bile acid substrates. In addition, we cannot exclude that other, as yet unknown, mechanisms distinct from hFXR may contribute to the observed repression of UGT2B7 transcription by lithocholic acid. Several mechanisms could underlie the distinct behavior of hFXR with differing hFXR response elements. These alternatives are currently under investigation.
Acknowledgments
This work was supported in part by National Institutes of Health Grants DK56226 and DK60109 (A.R.-P.) and 20 RR016460 (T.L.). A.R-P. is also supported by tobacco settlement funds from University of Arkansas for Medical Sciences.
We thank Joanna Little for critical review of the manuscript. Dr. Piotr Czernik is acknowledged for cloning the 1.3-kb UGT2B7 promoter fragment and generating the deletion constructs. We would like to emphasize that it was Dr. Czernik’s wish not to be listed as a coauthor since he did not have any intellectual involvement.
ABBREVIATIONS
- hFXR
human farnesoid X receptor
- UGT
UDP-glucuronosyltransferase
- I-BABP
ileal-bile acid-binding protein
- PPAR
peroxisome proliferator-activated receptor
- HepG2
human hepatocellular carcinoma cell line
- Caco-2
human colorectal adenocarcinoma cell line
- CREB
cyclic AMP response element-binding protein
- hRXR
human retinoid X receptor
- EMSA
electrophoretic mobility shift assay
- PXR
pregnane X receptor
- hPXR
human PXR
- NFRE
negative farnesoid X receptor response element
- hVDR
human vitamin D receptor
- DMEM
Dulbecco’s modified Eagle’s medium
- dNTP
deoxynucleoside-5′-triphosphate
- PCR
polymerase chain reaction
- RT-PCR
reverse transcriptase-PCR
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- SSC
standard saline citrate
- kb
kilobase(s)
- bp
base pair(s)
- IR-1
inverted repeat-1
- Rif
rifampicin
- SHP
small heterodimer partner
- ANOVA
analysis of variance
References
- Ananthanarayanan M, Balasubramanian N, Makishima M, Mangeldorf DJ, Suchy FJ. Human bile salt export pump is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276:28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- Barbier O, Peneda Torra I, Sirvent A, Claudel T, Blanquart C, Duran-Sandoval D, Kuipers F, Kosykh V, Fruchart J-C, Staels B. FXR induces the UGT2B4 enzyme in hepatocytes: a potential mechanism of negative feedback control of FXR activity. Gastroenterology. 2003;124:1926–1940. doi: 10.1016/s0016-5085(03)00388-3. [DOI] [PubMed] [Google Scholar]
- Bjorkhem I. Mechanism of bile acid biosynthesis in mammalian liver. In: Danielsson H, Sjovall J, editors. Sterols and Bile Acids. New Comprehensive Biochemistry. Elsevier; Amsterdam: 1985. pp. 231–278. [Google Scholar]
- Borgstrom B, Barrowman JR, Lindstrom M. Roles of bile acids in intestinal lipid digestion and absorption. In: Danielsson H, Sjovall J, editors. Sterols and Bile Acids. New Comprehensive Biochemistry. Elsevier; Amsterdam: 1985. pp. 405–425. [Google Scholar]
- Burchell B, Brierley CH, Monaghan G, Clarke DJ. The structure and function of the UDP-glucuronosyltransferase gene family. Adv Pharmacol. 1998;42:335–338. doi: 10.1016/s1054-3589(08)60758-9. [DOI] [PubMed] [Google Scholar]
- Chiang JY, Kimmel R, Weinberger C, Stroup D. Farnesoid X receptor responds to bile acids and represses cholesterol 7α-hydroxylase gene (CYP7A1) transcription. J Biol Chem. 2000;275:10918–10924. doi: 10.1074/jbc.275.15.10918. [DOI] [PubMed] [Google Scholar]
- Chiang JYL. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr Rev. 2002;23:443–463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- Chowdhury JR, Novikoff PM, Chowdhury NR, Novikoff AB. Distribution of UDP glucuronosyltransferase in rat tissue. Proc Natl Acad Sci USA. 1985;82:2990–2994. doi: 10.1073/pnas.82.9.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claudel T, Inoue Y, Barbier O, Duran-Sandoval D, Kosykh V, Fruchart J, Fruchart J-C, Gonzalez FJ, Staels B. Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology. 2003;125:544–555. doi: 10.1016/s0016-5085(03)00896-5. [DOI] [PubMed] [Google Scholar]
- Claudel T, Sturm E, Duez H, Torra IP, Sirvent A, Kosykh V, Fruchart J-C, Dallongeville J, Hum DW, Kuipers F, et al. Bile acid-activated nuclear receptor FXR suppresses apolipoprotein A-I transcription via a negative FXR response element. J Clin Investig. 2002;109:961–971. doi: 10.1172/JCI14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congiu M, Mashford ML, Slavin JL, Desmond PV. UDP-glucuronosyltransferase mRNA levels in human liver disease. Drug Metab Dispos. 2002;30:129–134. doi: 10.1124/dmd.30.2.129. [DOI] [PubMed] [Google Scholar]
- Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang JY. Transcriptional activation of the cholesterol 7α-hydroxylase gene (CYP7A) by nuclear hormone receptors. J Lipid Res. 1998;39:2192–2200. [PubMed] [Google Scholar]
- Gall WE, Zawada G, Mojarrabi B, Tephly TR, Green MD, Coffman BL, Mackenzie PI, Radominska-Pandya A. Differential glucuronidation of bile acids, androgens and estrogens by the human recombinant UDP-glucuronosyltransferases UGT1A3 and 2B7. J Steroid Biochem Mol Biol. 1999;70:101–108. doi: 10.1016/s0960-0760(99)00088-6. [DOI] [PubMed] [Google Scholar]
- Gardner-Stephen DA, Heydel J-M, Goyal A, Lu Y, Xie W, Lindblom T, Mackenzie P, Radominska-Pandya A. Human PXR variants and their differential effects on the regulation of human UDP-glucuronosyltransferase gene expression. Drug Metab Dispos. 2004;32:340–347. doi: 10.1124/dmd.32.3.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grober J, Zaghini I, Fujii H, Jones SA, Kliewer SA, Willson TM, Ono T, Besnard P. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J Biol Chem. 1999;274:29749–29754. doi: 10.1074/jbc.274.42.29749. [DOI] [PubMed] [Google Scholar]
- Halvorsen B, Kase BF, Prydz K, Garagozlian S, Andrresen MS, Kolset SO. Sulphation of lithocholic acid in the colon-carcinoma cell line Caco-2. Biochem J. 1999;343:533–539. [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Willson TM. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J Lipid Res. 2002;43:359–364. [PubMed] [Google Scholar]
- Knerr I, Dachert C, Beinder E, Metzler M, Dotsch J, Repp R, Rascher W. Adrenomedullin, calcitonin gene-related peptide and their receptors: evidence for a decreased placental mRNA content in preeclampsia and HELLP syndrome. Eur J Obstet Gynecol Reprod Biol. 2002;101:47–53. doi: 10.1016/s0301-2115(01)00519-x. [DOI] [PubMed] [Google Scholar]
- Laffitte B, Kast H, Nguyen C, Zavacki A, Moore D, Edwards P. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem. 2000;725:10638–10647. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin D receptor as an intestinal bile acid sensor. Science (Wash DC) 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- Oelberg DG, Chari MV, Little JM, Adcock EW, Lester R. Lithocholate glucuronide is a cholestatic agent. J Clin Investig. 1984;73:1507–1514. doi: 10.1172/JCI111356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet DJ, Janowski BA, Mangelsdorf DJ. The LXRs: a new class of oxysterol receptors. Curr Opin Genet Dev. 1998;8:571–575. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- Princen HMG, Post SM, Twisk J. Regulation of bile acid biosynthesis. Curr Pharm Des. 1997;3:59–84. [Google Scholar]
- Radominska A, Treat S, Little J. Bile acid metabolism and the pathophysiology of cholestasis. Semin Liver Dis. 1993;13:219–234. doi: 10.1055/s-2007-1007351. [DOI] [PubMed] [Google Scholar]
- Radominska A, Zimniak P, Lester R. The detoxification of bile acids. In: Paumgartner G, Stiehl A, Gerok W, editors. Bile Acids as Therapeutic Agents: From Basic Science to Clinical Practice. Kluwer Academic Publishers; Dordrecht: 1991. pp. 99–109. [Google Scholar]
- Radominska-Pandya A, Czernik P, Little JM, Battaglia E, Mackenzie PI. Structural and functional studies of UDP-glucuronsyltransferases. Drug Metab Rev. 1999;31:817–900. doi: 10.1081/dmr-100101944. [DOI] [PubMed] [Google Scholar]
- Russell DW, Setchell KDR. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- Staudinger JL, Madan A, Carol KM, Parkinson A. Regulation of drug transporter gene expression by nuclear receptors. Drug Metab Dispos. 2003;31:523–527. doi: 10.1124/dmd.31.5.523. [DOI] [PubMed] [Google Scholar]
- Takeshita A, Taguchi M, Koibuchi N, Ozawa Y. Putative role of the orphan nuclear receptor SXR (steroid and xenobiotic receptor) in the mechanism of CYP3A4 inhibition by xenobiotics. J Biol Chem. 2002;277:32453–32458. doi: 10.1074/jbc.M111245200. [DOI] [PubMed] [Google Scholar]
- Torra IP, Claudel T, Duval C, Kosykh V, Fruchart J-C, Staels B. Bile acids induce the expression of the human peroxisome proliferator-activated receptor α gene via activation of the farnesoid X receptor. Mol Endocrinol. 2003;17:259–272. doi: 10.1210/me.2002-0120. [DOI] [PubMed] [Google Scholar]
- Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- Urizar NL, Dowhan DH, Moore DD. The farnesoid X-activated receptor mediates bile acid activation of phospholipid transfer protein gene expression. J Biol Chem. 2000;275:39313–39317. doi: 10.1074/jbc.M007998200. [DOI] [PubMed] [Google Scholar]
- Vlahcevic ZR, Eggersten G, Bjorkhem I, Hylemon PB, Redford K, Pandak WM. Regulation of sterol 12α-hydroxylase and cholic acid biosynthesis in the rat. Gastroenterology. 2000;118:599–607. doi: 10.1016/s0016-5085(00)70267-8. [DOI] [PubMed] [Google Scholar]
- Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. An essential role for SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci USA. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak P, Radominska A, Lester R. Phase I and II biotransformation of bile acids. In: Bock KW, Gerok W, Matern S, Smith R, editors. Hepatic Metabolism and Disposition of Endo- and Xenobiotics. Kluwer Academic Publishers; Dordrecht: 1991. pp. 183–192. [Google Scholar]