Abstract
Proteins in bacteria often deploy to particular places within the cell, but the cues for localization are frequently mysterious. We found that the peripheral membrane protein SpoVM recognizes a geometric cue in localizing to a particular membrane during sporulation in Bacillus subtilis. Sporulation involves an inner cell maturing into a spore and an outer cell nurturing the developing spore. SpoVM is produced in the outer cell where it embeds in the membrane that surrounds the inner cell but not in the cytoplasmic membrane of the outer cell. We found that SpoVM localized by discriminating between the positive curvature of the membrane surrounding the inner cell and the negative curvature of the cytoplasmic membrane. Membrane curvature could be a general cue for protein localization in bacteria.
Proteins often localize to particular positions within bacteria, sometimes in a dynamic manner. A striking but mysterious example of subcellular localization occurs during spore formation in Bacillus subtilis when SpoVM (VM) localizes to a particular patch of membrane (1). How VM discriminates between different membrane surfaces in the same cell is unknown.
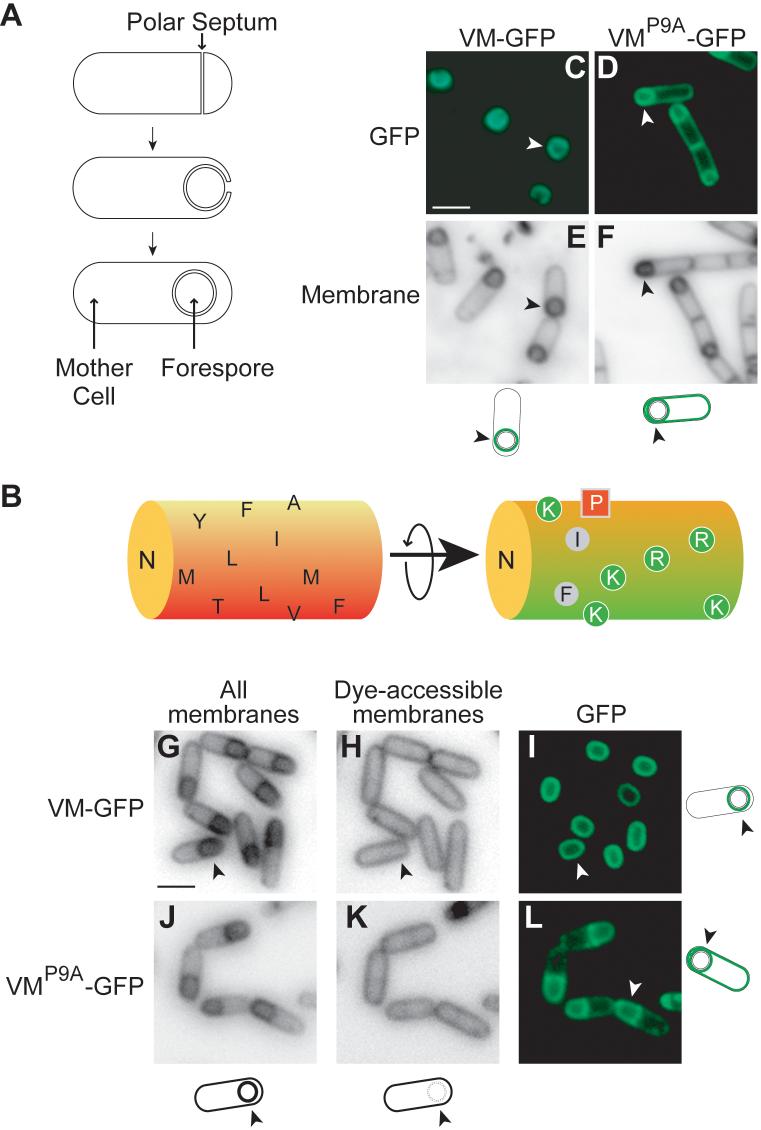
During sporulation, the cell divides asymmetrically to create mother cell and forespore compartments. Next, the mother cell engulfs the forespore, enveloping it with inner and outer membranes (Fig. 1A). Following engulfment a protein coat is deposited around the outer forespore membrane (2). Coat assembly depends on VM, a 26-residue peptide that is produced in the mother cell (3). VM is an amphipathic α-helix (4) that inserts into the membrane with its long axis parallel to the membrane and its hydrophobic face buried in the lipid bilayer (5). During engulfment, VM localizes to the membrane that tracks around the forespore, eventually decorating the entire surface of the forespore, as visualized using a fusion to the Green Fluorescent Protein (VM-GFP; Fig 1C) (1). Proline 9 (P9; Fig. 1B) is critical for this localization (1), as substitution of P9 with alanine (VMP9A-GFP) resulted in localization to both the cytoplasmic and the outer forespore membranes (Fig. 1D).
Figure 1. VM-GFP localizes selectively to the surface of the forespore.
Procedures (11). (A) Stages of sporulation. Top, division creates a mother cell (left) and a forespore (right). Middle, the mother cell engulfs the forespore. Bottom, the forespore is pinched off as a protoplast. (B) α-helical model of VM. Shown on the left is the hydrophobic, membrane-embedded face and on the right a 180° rotation of the helix along its long axis with positively charged residues labeled green. (C) VM-GFP localizes to the surface of the forespore, whereas VMP9A-GFP localizes to all membranes (D). (E-F) Membrane stained cells in (C-D). Arrowheads identify the cell depicted in the cartoon. (G-L) Localization of VM-GFP or VMP9A-GFP produced after topological isolation. Membrane surrounding the forespore was stained by a membrane-permeable (G, J) but not by a membrane impermeable dye (H, K). VM-GFP (I), but not VMP9A-GFP (L), localized selectively to the surface of the forespore. Scale bars are 2 μm.
Following engulfment, the outer forespore membrane becomes topologically isolated from the cytoplasmic membrane. We wondered if VM would adhere to the outer forespore membrane after isolation. We engineered cells to produce VM-GFP in response to an inducer and triggered synthesis of the fusion protein after engulfment. To monitor topological isolation, we stained the membranes with a membrane permeable dye, which stains all membranes, and a membrane impermeable dye, which can only access the engulfment membrane before membrane fusion (6). VM-GFP localized almost exclusively to the outer forespore membrane even when the forespore was topologically isolated (Fig. 1G-I). As a control, VMP9A-GFP synthesized after engulfment localized promiscuously (Fig. 1J-L). Thus, VM-GFP partitions between both membranes but the wild type fusion protein is retained in the outer forespore membrane.
What does VM recognize? We wondered whether the localization cue for VM was geometric, because the outer forespore membrane is the only membrane in the mother cell with positive (convex) curvature. We examined the localization of VM-GFP in cells of a triple mutant (D/M/P) arrested with a straight polar septum (7). VM-GFP behaved indiscriminately in such mutant cells, localizing to the cytoplasmic and the engulfing membranes (Fig. 2B).
Figure 2. Positive curvature is necessary and sufficient for localization.

Localization of VM-GFP in wild type (A) and in mutant cells (ΔD/M/P) with a straight septum (B). (C) Localization of VM-GFP in cells of the ΔD/M/P mutant with bulges. (D-F) Membrane stained cells in (A-C). (G-J) Localization of VM-GFP (G and I) and VMP9A-GFP (H and J) in mutant E. coli cells (ΔmreBCD) that produce internal vesicles. (I and J) Vesicles were visualized by phase contrast microscopy. (K-N) Localization of VM-GFP (K and M) and VMP9A-GFP (L and N) in mutant yeast cells (ΔVPH1) that produced fragmented vacuoles. (I and J) Vacuoles were stained with FM4-64. Scale bars are 2 μm.
As a further test of the idea that VM recognizes curvature, we took advantage of the fact that at low frequency (∼ 1%) the D/M/P triple mutant develops a fissure in the septum, allowing membrane to bulge into the mother cell (8). VM-GFP should localize to this convex membrane protrusion. Indeed, VM-GFP localized with high selectivity to such bulges (Fig. 2C).
To test the curvature hypothesis still further, we examined the localization of VM-GFP in a heterologous host that also exhibits convex membranes, an E. coli mutant lacking the cytoskeletal protein MreB (9) that forms internal vesicles similar in size to the forespore. In mutant cells, VM-GFP localized almost exclusively to the surface of the vesicles (Fig. 2G). As a control, VMP9A-GFP not only localized to the surface of vesicles, but also to the cytoplasmic membrane (Fig. 2H). We also examined the localization of VM-GFP in a mutant of Saccharomyces cerevisiae that produces fragmented vacuoles whose sizes are again similar to that of the forespore (10). In these cells, VM-GFP localized primarily to the surface of vacuoles, whereas VMP9A-GFP localized to the concave periphery of the cells as well (Fig. 2K-L). VM-GFP and VMP9A-GFP were stable and produced at similar levels in B. subtilis, E. coli, or S. cerevisiae (Fig. S1). Taken together, VM appears to respond to a geometric cue, rather than a B. subtilis-specific feature of the membrane.
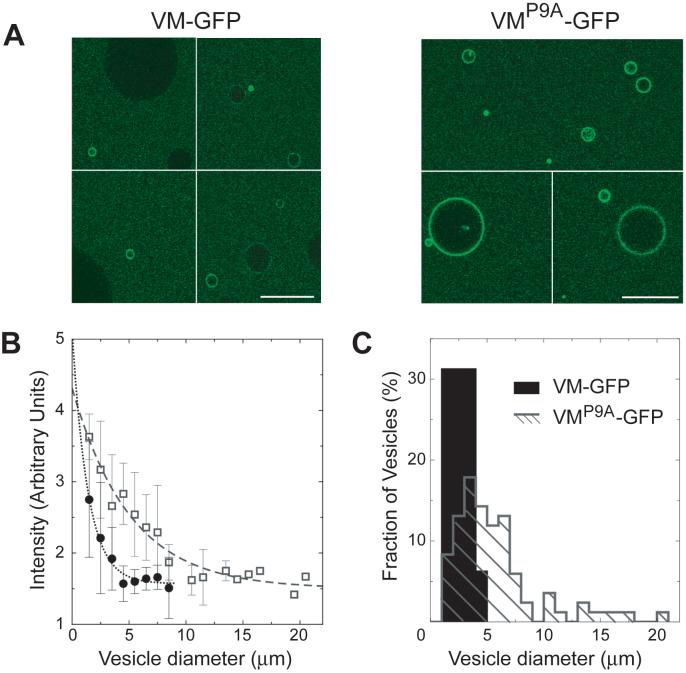
If so, then VM should bind preferentially to any phospholipid bilayer with a curvature similar to that of the outer forespore membrane. We asked whether purified VM-GFP would adsorb to the surface of phosphoplipid vesicles, and whether it would favor vesicles similar in size to the forespore. We prepared by electroformation a heterogeneously sized population of unilamellar phospholipid vesicles, ranging in diameter from approximately 1 to 30 μm, with a distribution that peaked at 8 μm (11). We incubated the vesicles with purified VM-GFP harboring a C-terminal polyhistidine tag and examined the distribution of fluorescence by confocal laser microscopy. Remarkably, VM-GFP preferentially adsorbed to the smallest observable vesicles in the population (Fig. 3A). In contrast, VMP9A-GFP displayed a diminished preference for smaller vesicles and adsorbed to the surfaces of even large vesicles (Fig. 3A). The fluorescent signal resulted from surface-localized VM-GFP-His6 and a label introduced into the lipids resulted in uniform, equally fluorescent vesicles (11).
Figure 3. VM-GFP detects curvature in vitro.
(A) Confocal fluorescence micrographs of purified VM-GFP (left) or VMP9A-GFP (right) incubated with phospholipid vesicles. Scale bars are 20 μm. (B) Membrane fluorescence intensity as a function of vesicle diameter. Vesicles were incubated with similar concentrations of VM-GFP (closed circles) or VMP9A-GFP (open squares) The data points are mean +/- SD measured on N vesicles (N=3-18) and were fit with an exponential decay (11). (C): Size distribution of fluorescent vesicles whose intensity is at least 2.5-fold above background.
The concentration of surface-adsorbed VM-GFP decreased as the diameter of the vesicles increased (Fig. 3B). Thus, smaller vesicles harbored more VM-GFP per surface area than did larger vesicles. When vesicles were incubated with VMP9A-GFP, however, the decrease of fluorescence intensity with increasing vesicle size was much less pronounced, suggesting that VMP9A-GFP was recruited more readily to larger vesicles (Fig. 3B). Next, we measured the number of vesicles whose fluorescence was at least 2.5 times higher than background fluorescence (Fig. 3C). Strong adsorption of VM-GFP occurred for vesicles less than 5 microns in diameter and peaked near the smallest vesicles in the population at around 2.5 microns (by comparison, a forespore is approximately 1 micron in diameter). In our analysis, vesicles less than 1 micron in diameter were grossly underrepresented. Thus, we likely overestimated the peak size of vesicles to which VM-GFP was recruited and VM-GFP may preferentially adsorb onto even smaller vesicles (extrapolation of the data of Fig. 3B; see also Fig. 4A). In contrast to VM-GFP, VMP9A-GFP adsorbed onto vesicles up to 20 microns in diameter, resulting in a broad distribution with a peak at about 5 microns. Substitution of proline 9 does not simply increase the affinity of VM for membranes. Thus, the adsorption of VM-GFP to membranes is sensitive to curvature and dependent on proline 9.
Figure 4. Preferential adsorption of VM-GFP onto smaller vesicles is concentration dependent.
(A) Membrane fluorescence intensity as a function of vesicle diameter for increasing bulk concentrations of VM-GFP. At higher concentrations, vesicles with a diameter below a critical value (Dc) of ∼ 4 μm were preferentially labeled. (B) Membrane fluorescence intensity as a function of VM-GFP concentration for different vesicle diameters (indicated diameters are +/- 0.5 μm). Above Dc, all data points were similar and well approximated by a linear fit (dotted line), whereas below Dc, the isotherms were steeper and deviated from linear. (C) Typical behaviors for small (1.5 +/- 0.5 μm) and large (6.5 +/- 0.5 μm) vesicles, and theoretical curves obtained using a model for cooperativity (11). Data points are mean +/- SD measured on N vesicles (N=3-26), resulting from 3 independent experiments.
How can VM, which is 40 Å in length or less (12), be sensitive to the curvature of micron-sized spheres? A precedent is ArfGAP1, a Golgi-associated protein that preferentially associates with yeast vesicles via a stretch of α-helix (13, 14) and is sensitive to curvature. But the vesicles recognized by ArfGAP1 are far smaller (∼50 nm) and more highly curved than those recognized by VM. For a 40 Å rod lying flat on the surface of a 1-micron-diameter sphere the maximum distance between one end of the rod and the surface of the sphere is less than 0.2 Å. It therefore seems unlikely that partitioning of individual VM molecules between different sized spheres could be influenced by such a small degree of curvature. Several molecules of VM may thus be required to display a collective sensitivity for slightly curved surfaces. We tested if the preferential adsorption of VM-GFP to smaller vesicles was dependent on the concentration of VM-GFP by measuring membrane fluorescence intensity as a function of vesicle size, for different concentrations of VM-GFP. The preferential adsorption of VM-GFP onto smaller vesicles increased with increasing concentrations of protein (Fig. 4A). Moreover, our data suggest a critical value of the vesicle diameter Dc of about 4 μm. For all concentrations, above Dc the amount of VM-GFP adsorbed per unit area of membrane did not vary significantly. However, below Dc, and at higher concentrations, the adsorbed amount strongly increased with decreasing vesicle diameter.
We then constructed an “adsorption isotherm”, which is a signature of the adsorption mechanism. For a given vesicle diameter, we plotted membrane fluorescence intensity as a function of VM-GFP bulk concentration (Fig. 4B). When the vesicle diameter was larger than Dc, all curves were similar and approximately linear, indicating that the adsorption mechanism was the same and not dependent on membrane curvature. However, below Dc the curves deviated from linear and became progressively steeper as vesicle diameter decreased, suggesting an adsorption mechanism that involves cooperative interactions (clustering) among VM-GFP. Preliminary theoretical analysis points to clusters consisting of just a few VM molecules (Fig. S2, (11)). VM-GFP molecules do not appear to interact with each other directly (Fig. S3). An alternative possibility, however, and one that we favor, is that the insertion of a VM molecule into the membrane indirectly recruits other VM molecules to its vicinity analogous to the clustering of certain phospholipids (15) and membrane proteins (16, 17): the energetic cost due to bilayer deformation induced by the insertion of VM could be minimized by clustering of VM molecules, resulting in an apparent cooperativity that does not involve contact between protein molecules.
Might geometric cues represent a strategy by which proteins localize to particular patches of membrane in bacteria? Convex surfaces, such as that of the forespore, are not typical in bacteria. Nonetheless, some bacteria produce organelles with positively curved membranes, such as photosynthetic vesicles and magnetosomes (18, 19). Perhaps amphipathic α-helices are used to identify the membranes of these organelles for protein localization. A major challenge in bacterial cell biology is to identify cues that recruit proteins to the poles of cells. Conceivably, membrane curvature, in this case extreme negative curvature, is used to identify the inside surface at the pole.
Supplementary Material
Acknowledgments
We thank R. Gaudet, D. Rudner, C. Schmidt, L. Shapiro, R. Schekman, D. Weitz, and N. Wingreen for discussions, and S. Lacefield, B. Scheid, and P. de Boer for advice. This work was supported by the Harvard Nanoscale Science and Engineering Center, a Fulbright grant to S.L. and NIH grant GM18568 to R.L.
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without prior, written permission of AAAS.
References and Notes
- 1.van Ooij C, Losick R. J Bacteriol. 2003;185:1391. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henriques AO, Moran CP., Jr Annu Rev Microbiol. 2007 doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 3.Levin PA, et al. Mol Microbiol. 1993;9:761. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- 4.Prajapati RS, Ogura T, Cutting SM. Biochim Biophys Acta. 2000;1475:353. doi: 10.1016/s0304-4165(00)00089-1. [DOI] [PubMed] [Google Scholar]
- 5.Ramamurthi KS, Clapham KR, Losick R. Mol Microbiol. 2006;62:1547. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- 6.Rudner DZ, Pan Q, Losick RM. Proc Natl Acad Sci U S A. 2002;99:8701. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichenberger P, Fawcett P, Losick R. Mol Microbiol. 2001;42:1147. doi: 10.1046/j.1365-2958.2001.02660.x. [DOI] [PubMed] [Google Scholar]
- 8.Blaylock B, Jiang X, Rubio A, Moran CP, Jr., Pogliano K. Genes Dev. 2004;18:2916. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendezu FO, de Boer PA. J Bacteriol. 2008;190:1792. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seeley ES, Kato M, Margolis N, Wickner W, Eitzen G. Mol Biol Cell. 2002;13:782. doi: 10.1091/mbc.01-10-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supporting material on Science Online.
- 12.Barlow DJ, Thornton JM. J Mol Biol. 1988;201:601. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 13.Parthasarathy R, Groves JT. Soft Matter. 2007;3:24. doi: 10.1039/b608631d. [DOI] [PubMed] [Google Scholar]
- 14.Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. Embo J. 2005;24:2244. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang KC, Mukhopadhyay R, Wingreen NS. PLoS Comput Biol. 2006;2:e151. doi: 10.1371/journal.pcbi.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goulian M, Bruinsma R, Pincus P. Europhysics Lett. 1993;22:145. [Google Scholar]
- 17.Kim KS, Neu J, Oster G. Biophys J. 1998;75:2274. doi: 10.1016/S0006-3495(98)77672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackenzie C, et al. Annu Rev Microbiol. 2007;61:283. doi: 10.1146/annurev.micro.61.080706.093402. [DOI] [PubMed] [Google Scholar]
- 19.Komeili A, Li Z, Newman DK, Jensen GJ. Science. 2006;311:242. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.