Abstract
Transcription termination is an important event in the transcription cycle that has been exploited in a variety of genetic regulatory mechanisms. Analysis of transcription termination is greatly facilitated by in vitro approaches. We describe a basic protocol for analysis of transcription termination in vitro, and include descriptions of parameters that can be modified for specific types of experimental questions.
Keywords: RNA polymerase, transcription termination, regulation
1. Introduction
Transcription termination in bacteria occurs by two major mechanisms. The more common mechanism, termed intrinsic termination, involves folding of the nascent RNA transcript into a G+C-rich helix followed by a U-rich track. Pausing of RNA polymerase (RNAP) during transcription of the U residues allows the RNA helix to fold and stimulate release of the enzyme and the RNA from the DNA template. Alternatively, a protein factor (e.g., Rho protein) can interact with the nascent RNA and track along the RNA until it encounters a paused transcription elongation complex (TEC); interaction between the protein and RNAP promotes termination and transcript release. In both types of termination mechanisms RNAP must pause to allow folding of the transcript into the terminator helix or to allow access of the protein to the TEC.
A large array of systems in which gene regulation occurs at the level of premature termination of transcription have been uncovered (reviewed in [1,2]). Genes regulated by mechanisms of this type are characterized by the presence of a termination signal in the region between the promoter and the start of the regulated gene(s). In many of these systems, the structure of the nascent RNA is modulated by binding of a regulatory factor (e.g., a protein, a trans-acting RNA, or a small molecule) to determine whether it forms the helix of the intrinsic terminator. In many cases, the nascent RNA has the potential to fold into an alternate structure, the “antiterminator,” formation of which sequesters sequences that would otherwise participate in formation of the intrinsic terminator helix. Placement of a regulatory factor on the nascent RNA can also affect binding and processivity of Rho protein. Furthermore, regulatory factors (like bacteriophage λ N protein) can modulate termination efficiency by interacting with RNAP, often by affecting the processivity of the TEC.
Many of these regulatory systems were first characterized by genetic analyses in vivo. The ability to reproduce the activity of a regulatable terminator in a purified in vitro transcription system provides crucial information about the requirements of the regulatory mechanism, and insight into the role of RNA folding and kinetics of transcription. We will describe the basic parameters for the use of in vitro transcription systems for the analysis of transcription termination and its control, and will also discuss variations on the assay system that facilitates analysis of a variety of regulatory mechanisms.
2. In vitro transcription termination assays
In vitro transcription assays have been developed using RNAP of varying degrees of purity, from many different bacterial species. In this section we will provide protocols for application of in vitro transcription assays to the analysis of transcription termination, with a focus on termination sites that are subject to regulation in vivo.
2.1 Assay design: an overview
We will first list general considerations for the assay design (with the important points emphasized), and then give a detailed protocol that represents a good starting point for this analysis.
2.1.1 DNA template design
Transcription termination assays generally involve use of a DNA template that includes a promoter, a short region preceding the terminator, and an additional short region following the termination site. The template is usually a linear fragment of DNA, generated by PCR, and transcription elongation through the terminator results in a run-off transcription product when RNAP reaches the end of the DNA fragment (Fig. 1). This allows discrimination between terminated (shorter) and readthrough (longer) RNA products after polyacrylamide gel electrophoresis. Supercoiled templates can be designed in which the promoter and test terminator are inserted into a plasmid vector upstream of one or more highly efficient terminators, such as the E. coli rrnB T1 or T7Te terminators [3] or B. subtilis rpsD gene terminator [4]. Transcription through the test terminator stops at the downstream terminator, again allowing discrimination between terminated and readthrough transcripts.
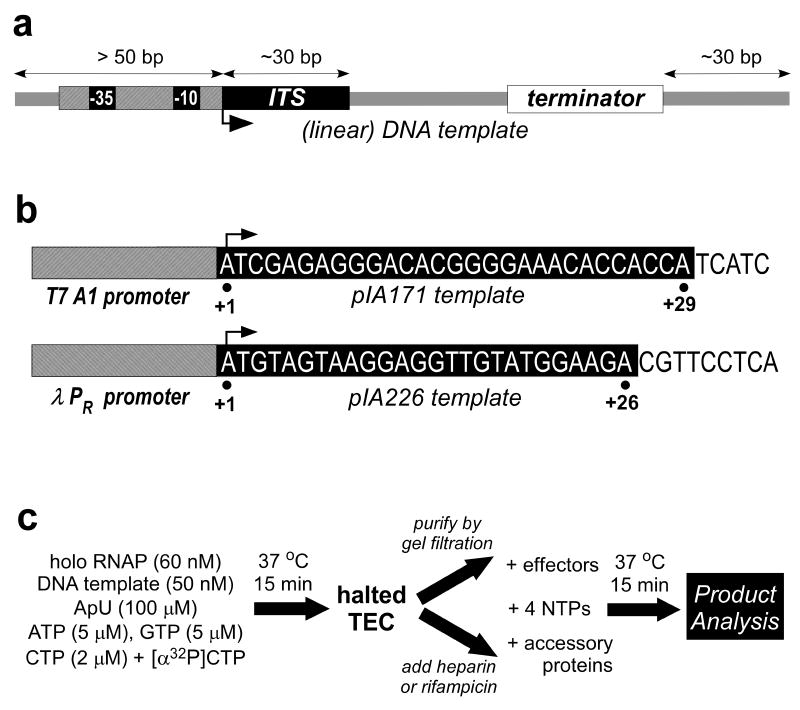
Fig. 1.
Single round termination assay design. (a) General features of the template. The template should encode a strong promoter (with -35 and -10 hexamers elements recognized by primary σ factors), a good initial transcribed sequence (ITS, at least 20 nt long and missing one of the bases), and a region of interest. For linear templates, at least 50 bp of DNA upstream of the transcription start site (a bent arrow, +1) and at least 30 bp downstream from the studied termination site(s) should be included to allow for proper recognition and for differentiation of terminated and readthrough products. (b) Examples of promoter-ITS combinations. The sequence of the non-template strand is shown, the RNA sequence is the same but with Us in place of Ts). pIA171 [32] has a phage T7A1 promoter followed by a 29 nt long T-less region; pIA226 [32] has a λ PR promoter and a 26 nt long C-less region. (c) Assay design (for pIA171). This protocol is suitable for RNAPs from E. coli or other mesophilic species; for RNAPs from thermophilic organisms (e.g., Thermus), the initiation reaction must be carried out at 55°C. Halted complexes will be radiolabeled in the 5′ end at several positions (10 if α-[32P]-GTP is used with pIA226-derived template).
2.1.2 Single-round vs. multi-round
Transcription assays can be run in multi- or single-round format; as the name implies, in the latter case each RNAP molecule transcribes the template only once. Multi-round assays are easier to design, and give a higher yield of RNA product. These assays involve combining all components in a single step, and can be complicated by effects of the added components on transcription initiation. Single-round assays are carried out in two steps, an initiation step and a separate elongation step, allowing addition of components that might otherwise interfere with initiation exclusively during the elongation reaction. The most common approach to single-round transcription reactions is to form a radiolabeled TEC by halting RNAP early within the transcriptional unit by withholding one NTP. This requires designing a template with an initial transcribed sequence (ITS) consisting of at least 15 nt comprised of only three of the four NTPs (Fig. 1). Importantly, even in single-round assays the yield of radiolabeled RNA is sufficient for reliable detection and quantification.
TECs are stable (on ice for hours), and many active, readily extendable variants have been characterized. However, it is important to recognize that when a new ITS is designed, the resulting TEC may turn out to be poorly active, and will resume elongation slowly (and sometimes never); this usually happens because RNAP backtracks and arrests [5]. Furthermore, a sequence that works well for one RNAP does not always behave equally well with enzymes from other species.
A complete set of NTPs is added to the halted TEC, followed by incubation at a target temperature to restart elongation and allow RNAP to reach the termination signal. A fraction of RNAP molecules will release the nascent RNA at the terminator, and the rest will continue past the termination site and run off the end of DNA template (or terminate at a downstream terminator). Many variables can be tested during the elongation reaction. For example, E. coli RNAP elongates and terminates (with varied efficiencies) at 10-60°C, in 0.02-1 M KCl, in 20% ethanol, and in NTP concentrations ranging between 1 μM and 1 mM. Regulatory molecules (e.g., proteins, small RNAs or small molecules) also can be added during the elongation reaction to determine their effect on termination.
To restrict elongation to a single round, rifampicin or heparin are added to block subsequent initiation. However, some caution is needed, as some forms of RNAP (such as Thermus) are rather resistant to rifampicin [6], and heparin is a DNA mimic and thus will trap not only free RNAP but also any DNA-binding regulatory protein, e.g., Rho [7]. Alternatively, halted TECs can be purified by gel filtration using Sephadex G50 or other inert matrix spin columns (e.g., AutSeq columns from GE Health) pre-equilibrated with the transcription buffer, or by immobilization using RNAP with an engineered histidine tag [8] or a biotinylated template generated using a primer containing a biotin moiety [9]; these manipulations will remove the starting dinucleotide used to prime initiation, but will not completely block reinitiation, which can also (albeit less efficiently) be primed with NTPs present during the chase reaction.
RNA release at a terminator is monitored by gel electrophoresis in a denaturing urea-acrylamide gel, followed by imagining and quantification (Fig. 2). The efficiency of termination (i.e., the fraction of RNAP molecules that release the transcript at a given position) can be measured accurately and the point of RNA release can be determined with a single nucleotide precision.
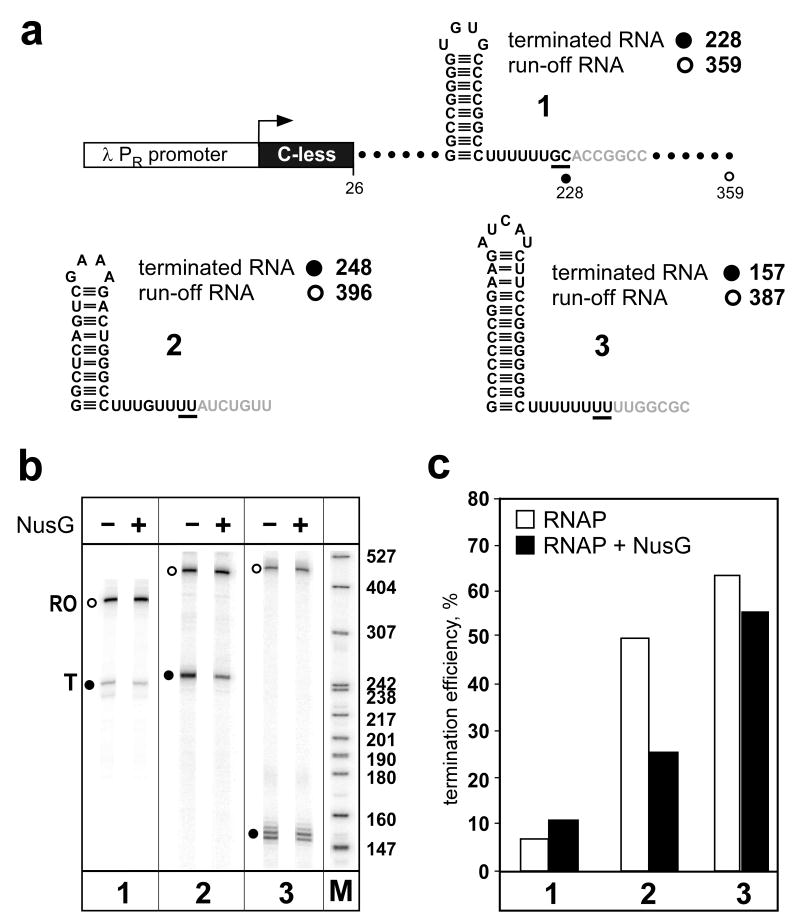
Fig. 2.
Differential effect of NusG protein on termination at three representative intrinsic terminators. (a) Template structure. All three templates share the λ PR promoter and the 26 nt C-less ITS; the downstream regions differ and each encodes an RNA hairpin followed by a U-rich region. The sizes of the terminated and run-off RNA products are shown. (b) Products of the single-round termination reactions. Reactions were carried out as in 2.5) with E. coli RNAP and E. coli NusG (added at 100 nM where indicated). RNA products were separated on a 6% denaturing gel; terminated (T, filled circles) and run-off (RO, open circles) RNAs are indicated for each terminator. The “M” lane was loaded with [32P]-labeled pBR322 MspI digest, with sizes indicated on the right; note that the RNA species migrate more slowly than the DNA fragments of the same size. (c) Quantification of termination efficiency. Termination efficiency is defined as the fraction of RNA at the terminator (T) relative to the total RNA (which equals the sum of T and RO if no other products are present). The three terminators tested differ in their termination efficiencies as well as in their response to NusG.
We prepare all solutions used for in vitro transcription with DEPC-treated H2O (see 2.5) and wear gloves at all times. RNases can be introduced at any point, including RNAP purification and template preparation.
2.2 Source of RNAP
The RNAP preparation is probably the most important factor in your assays! The promoter-containing templates described in these protocols require RNAP holoenzyme that consists of core (α2ββ′ω polypeptides) and a σ factor that matches the promoter sequence in your template (E. coli σ70 holoenzyme in the protocol described here).
Commercial sources are suitable for initial experiments. Currently, two bacterial holoenzyme species are available from Epicentre: E. coli (S90050) and T. thermophilus (T90250). The E. coli enzyme is the best-studied model for biochemical analysis, whereas T. thermophilus RNAP has been captured in the atomic-resolution structures and is therefore a model for structure-function studies.
Affinity purification is the next easiest way to obtain RNAP for analysis. Several strains or plasmids in which one of the RNAP core subunits contains a hexa-histidine tag are available for B. subtilis [10], E. coli [11,12], and Thermus aquaticus [13] RNAPs, and detailed purification protocols have been developed. These protocols are commonly abbreviated to make a partially purified enzyme [14]; in this case, however, the resulting preparation contains a mix of enzyme populations (with associated factors present in unknown ratios to RNAP), complicating data interpretation. A complete purification protocol produces homogeneous highly purified RNAP; the only drawback is that usually both core and holoenzyme are present (and can be separated), and full utilization of the preparation requires addition of extra separately purified σ. Several expression vectors are available for E. coli σ factors [15,16], B. subtilis σA [17,18], T. aquaticus σA [6], etc.
Conventional purification is an alternative when a tagged version of RNAP is not available. In this case, the purification protocol is followed by monitoring RNAP activity. Both core and holoenzyme are active on a non-specific template (e.g., poly dA/dT template); holoenzyme (but not core) is active on a template containing a strong bacterial (or bacteriophage) promoter that is recognized by the corresponding σ factor. A good place to start is to follow a purification protocol developed for the E. coli enzyme [19], with modifications [20].
2.3. Template DNA
To simplify the analysis of termination and its regulation, it is important to design an assay in which initiation of transcription is not sensitive to changes in substrate concentrations, addition of protein factors, or presence of small molecule effectors. The easiest approach is to use a template in which the transcribed region of interest is placed downstream of a strong (commonly bacteriophage) promoter. Good model promoters do not require activators and thus display high activity in vitro, are well-characterized (most importantly, their start-sites are precisely known), are relatively insensitive to DNA supercoiling, and even work well with RNAP from different bacteria. For example, the bacteriophage λ PR promoter is readily recognized by the E. coli, B. subtilis and T. thermophilus enzymes [21,22]. In contrast, cellular promoters are frequently weak, depend on accessory proteins, and their start-sites are not always known. An exception is ribosomal protein gene promoters, which like phage promoters are very strong and constitutively active.
Quality of template DNA is very important in in vitro transcription assays - we find that low quality DNA template is the primary cause of sub-optimal results. Linear, PCR-generated templates (Fig. 1) work well for most assays; if effects of the template topology on termination are studied, circular plasmid templates prepared by standard approaches [23] can also be used. As discussed above, in this case the promoter and region of interest, including the test terminator, are placed upstream of one or more strong, constitutively active terminators within a plasmid vector, and supercoiled plasmid DNA is isolated from the cell and used directly as the template. Traditionally, linear template DNAs were made by subcloning the transcribed region of interest under control of the λ PR or T7A1 promoter, followed by truncation of the DNA at a restriction endonuclease cleavage site downstream of the terminator site, or PCR amplification with suitable flanking primers; PCR amplification is preferable because it decreases the likelihood that the template contains additional promoter sequences that could direct transcription from sites other than the region of interest. The primers should correspond to a region ∼100 bp upstream from the start-site (to include all of the promoter elements) and at least 30 bp downstream from the expected termination site of a test terminator, since both the sequence and the length of the downstream DNA may affect termination efficiency [24]. However, since synthetic oligonucleotides have become very inexpensive, it is also practical to order a longer upstream primer that contains the core promoter elements (50 nt upstream of the start-site is sufficient for both λ PR and T7A1 promoters) and a suitable initial transcribed sequence (ITS, Fig. 1a and b), and is complementary at its 3′ end to the region of interest.
To prepare transcription templates, we use Taq DNA polymerase (Sigma) and either genomic DNA or an existing plasmid as a PCR template. Misincorporation has never been a problem with these rather short (<500 bp) templates, but to eliminate this concern it is always wise to make two template preparations in parallel and use them in duplicate reactions. We purify templates using the QIAquick PCR purification kit (Qiagen); if the product is not homogenous, we purify the desired species from agarose gels using the QIAquick gel extraction kit. We elute the DNA (at 1-15 μM) with DEPC-treated water and store -20°C.
QIAquick (and other commercial) kits are not very efficient in removing >50 nt-long primers (that can be used to add the entire promoter to a region of interest without a cloning step); in this case, we instead use spermine precipitation as follows:
pool PCR reactions (a 1-ml PCR reaction generates template sufficient DNA for multiple experiments, but smaller scale reactions can be used as appropriate), extract once with an equal volume of phenol (equilibrated with Tris·HCl, pH 7.5) and transfer the aqueous layer to a fresh tube;
extract once with CHCl3:isoamyl alcohol (24:1) and transfer the aqueous layer carefully to a fresh tube, avoiding inclusion of any organic phase;
add spermine (Sigma) to 5 mM, incubate for 15 min at room temperature, and centrifuge at 15,000 × g for 15 min to pellet the DNA;
remove all of the liquid with a pipette, add 1 ml of 75% ethanol, 10 mM MgAc to the DNA pellet, and incubate on ice for one h, vortexing occasionally;
recover the pellet by centrifugation (15,000 × g for 15 min), wash with cold 70% EtOH, dry in vacuo, and dissolve in DEPC-treated H2O.
2.4. Substrates
We store all ribonucleotide triphosphates (NTPs) at 10-100 mM in small aliquots at-80°C for long-term storage; stocks in current use are stored at -20 °C for 2-6 months. In our hands, FPLC-purified, 100 mM stock solutions (GE Health) work best when absence of cross-contamination is important (e.g., during preparation of halted complexes). Deamination of CTP to UTP is a typical (and not easily avoidable) issue, particularly for generating U-less halted complexes with labeled CTP. Each batch should be checked before use (e.g., by testing whether a single halted complex species is formed when one NTP is omitted). We obtain α-[32P]NTPs (3000 Ci/mmol) from Perkin Elmer.
RNAP requires high (∼1 mM) concentration of the first two substrates for efficient initiation (cellular concentrations of NTPs are in 0.5-10 mM range), whereas elongation can be carried out even at NTP concentrations as low as 1 μM. During formation of halted complexes, NTPs are used at μM concentrations to enable high efficiency of labeling and prevent readthrough beyond the halt position (due to NTP contamination). To balance these requirements, we use dinucleotides (as specified by the template sequence) that correspond to the +1 and +2 positions of the transcript; eight dinucleotide variants are currently available from Ribomed Products.
At steps following the formation of the halted TEC, NTPs can be used at various concentrations, typically ranging from 50 μM to 1 mM. During this stage of the reaction, cross-contamination is less of an issue and other, less expensive sources of NTPs can be used. Since only cold NTPs are used at this step, the α-[32P]NTP present during initiation is diluted significantly, and the transcript is essentially end-labeled (at several positions within the ITS). For multi-round transcription reactions, NTP concentrations can range from 0.1 to 5 mM; use of a dinucleotide or high concentration (∼1 mM) of the first two NTPs is required for efficient initiation, and a low concentration of the NTP corresponding to the labeled nucleotide is necessary for efficient incorporation. In a multi-round assay the RNA transcript is being labeled continuously (in contrast to the end-labeled RNA generated in the single-round assay); thus the extent of labeling will be proportional to the number of incorporated α- [32P]NMP residues. In practice, this means that the signal has to be corrected for the different number of labeled residues in terminated vs. read-through RNA products.
2.5. Protocol for a single-round termination assay with E. coli RNAP
For all transcription assays, we use nuclease-free pre-siliconized 1.7 ml tubes (AVSC1510, Midwest Scientific) to avoid protein binding to the tube surface. Assays will be described for the pIA226 template (Fig. 1b), which initiates with ApU and has a 26 nt C-less ITS.
Halted complex formation
To prepare 100 μl of a halted TEC, combine the following reagents on ice in the following order:
76.5 μl DEPC-treated water (can be purchased from many suppliers, or home-made: add DEPC to 0.1%, stir for 2-4 hours, autoclave to remove traces of DEPC).
10 μl 10X TGA (200 mM Tris-acetate, pH 8.0, 20 mM Na-acetate, 20 mM Mg-acetate, 40% glycerol, 1 mM DTT, 1 mM EDTA); this buffer works well for many promoters, including T7A1, λ Pr, rrnB, etc., and 20-50 mM final concentration of the buffering agent is a good starting point. Many other buffers can be used, at least for most promoters, including HEPES, MOPS, Tris-HCl, etc.; however, phosphate buffers should be avoided. In general, transcription initiation is much more sensitive to solution conditions than elongation; in particular, high (>200 mM) ionic strength strongly inhibits initiation but not elongation. Glycerol will inhibit elongation if present at >10%, and Mg2+ is required during both initiation and elongation, but transcription initiation activity reaches maximum at ∼ 2 mM Mg2, at least on T7A1 and λ PR templates.
10 μl of 10X initiating NTPs in H2O (10 μM GTP, 50 μM ATP, 50 μM UTP, 1 mM ApU, for a promoter initiating with AU at the +1, +2 positions, and a C-less ITS [final concentrations: 1 μM GTP, 5 μM ATP, 5 μM UTP, 100 μM ApU; note that the labeled NTP (see step 4) is at lower concentration to increase labeling efficiency).
1.5 μl of [α-32P]GTP (3000 Ci/mmol, Perkin Elmer, cat #BLU-006H).
1–2 μl of linear DNA template generated by PCR from pIA226 (5 μM in H2O).
1–3 μl of E. coli σ70 RNAP, 2.5 mg/ml (5.6 μM) in storage buffer (20 mM Tris-HCl, pH 7.9, 100 mM NaCl, 50% glycerol, 1 mM DTT), stored at -20°C for 1-2 years, longer storage at -80°C. See [25] for a purification protocol. The best RNAP preparations reach 90% activity (as measured by DNA binding), and such a highly active enzyme preparation should be added to the template at a slight molar excess (e.g., 11:10). For a less active RNAP preparation, one can use a larger excess. However, adding a large excess of RNAP is not only wasteful but also favors formation of aberrant TECs, in which the second RNAP molecule initiates transcription immediately behind the leading RNAP, which can potentially affect termination efficiency.
Incubate at 37°C for 15 min, then shift to ice. The resulting A26 TEC (last nt added is A at position 26) is stable for at least 2 h but should be used as soon as practically possible. We perform all transcription reactions in digital dry baths filled with sand; AccuBlock incubators (Labnet) maintain constant (within 0.1°C) temperature from 30 to 90°C.
Addition of protein factors
If a regulatory protein or small molecule is tested for effects on termination, it must be added to the halted TEC before NTPs since the nucleotide addition rate is >20 nt/sec. Add the regulatory factor to the halted complex on ice, mix, and transfer the tube to the 37°C bath; incubate for 5 min to equilibrate. To the control reactions, add an equal volume of the storage buffer or compound diluent (specific for each factor added). To increase reproducibility, we make a single preparation of the halted complex and then use aliquots for separate reactions. At this step, it is also possible to add a partially purified protein or even a bacterial cellular extract, for example to compare extracts from a wild-type and mutant strain. E. coli S30 extracts work well [26] if RNasin (Promega) is also added (at 5U/100 μl reaction) to inhibit cellular RNAses.
Chase
Add 2 μl of pre-warmed (37°C) 5X chase solution (1 mM each NTP [200 μM final concentration], 100 μg/ml rifampicin (Sigma) in 1X TGA) to 8 μl of the pre-warmed A26 TEC. Incubate 10 min at 37°C. Stop the reaction by adding an equal volume of STOP solution. To prepare 25 ml of STOP solution, mix 12 g urea, 2 ml 10X TBE, 50 mg xylene cyanol (Sigma), 50 mg bromophenol blue (Sigma), 0.5 ml 0.5 M EDTA, pH 8.0, dissolve by heating to 65 °C, adjust volume to 25 ml, aliquot 1 ml per tube, store at -20°C. Thaw at 55°C before use. Heparin (at 20 μg/ml final concentration) can be used in place of rifampicin to block transcription initiation.
2.6. Mapping the site of termination
We use the standard “chain-termination” protocol to determine the position of termination sites [27]. This procedure is also useful for identifying the position of pause sites during transcription elongation. Set up 4 separate tubes (labeled A, U, G, C), each containing 1 μl of 10X SEQ chase (250 μM NTPs, 500 μg/ml heparin in 1X TGA) and 1 μl of the corresponding 3′ O-methyl NTP stocks (Trilink Biotechnologies; 500 μM in 1X TGA). Add 8 μl aliquots of the A26 TEC to each tube, incubate at 37°C for 5-10 min and stop the reaction by adding 10 μl STOP solution (2.5). Reactions can be allocated and frozen at -20°C for later use if desired.
2.7. Analysis of termination products
Denaturing PAGE
The quality of the gel is very important, particularly if nucleotide-level resolution is necessary for precise mapping of termination sites. For most termination reactions, 6% denaturing (DNA sequencing) gels can be used. A mix for gel casting (without buffer!) can be made as follows and stored up to 2 months at 4°C, with important modifications (steps 4 and 5) that dramatically improve the band separation:
Pour 200 ml 30% Acrylamide:Bis Solution (19:1; Bio-Rad 161-0154) into a 1000 ml glass beaker with a magnetic stir bar.
Add 300 ml MilliQ-grade water, set beaker on hot plate/stirrer preheated to 55°C.
Add 420.42 g of urea (USB, Ultrapure, cat # 57-13-6) incrementally, while stirring. When the urea is fully dissolved (yielding 7 M solution), turn off the heat. Adjust volume to 1000 ml with MilliQ-grade water.
While stirring, add 12 g of mixed bed resin TMD-8 (Sigma, M8157). Continue stirring for 30 min.
Filter the solution using a bottle-top filter into a 1000 ml bottle and degas for ∼30 min in a dessicator. We use a Welch 2545 Vacuum Pump (max. 27″Hg), but any comparable pump can substitute.
Any gel size can be used depending on the complexity and size of analyzed RNA products. We usually run standard size sequencing gels (35×43 cm, 0.4 mm spacers) that require 60 ml of premix to pour. Prepare 75 ml to allow for spills. Mix 75 ml premix with 3.75 ml 10X TBE (0.45 M Tris, 0.45 M H3BO3, 10 mM EDTA, filtered; final concentration is 0.5X TBE for faster electrophoresis time), add 200 μl of 10% ammonium persulfate (APS, stored at −20°C in single-use aliquots) and 30 μl TEMED (Amersham, cat # 17-1312-01; room temperature). Pour gel using a 60-ml syringe. Insert the comb last (to avoid bubbles) so that the entire well is in the gel solution. Wrap the top with plastic wrap. Leave to polymerize ∼1 h). Can store at room temperature for several h. Do not refrigerate!
Running a gel
The set up depends on your particular apparatus (we use OWL sequencing system, Fisher); the most important point is to run gels at a high temperature (to keep RNA well denatured) - pre-run gels until they reach 55°C before loading. We use 0.5X TBE (in place of 1X TBE) for the running buffer for faster electrophoresis times. For a 35×43-cm 6% gel run in 0.5X TBE, it should take ∼35 min at 120 W to reach a constant temperature of ∼55°C (expected current ∼40 mA, voltage ∼3 kV); we use EC4000P power supplies (Thermo EC, Fisher). It is necessary to wash the wells with buffer as soon as the gel is set up in the electrophoresis apparatus (i.e., before prerunning) and again immediately before loading the samples. Aliquot samples into microfuge tubes and preheat for (exactly) 2 min at 90°C in a dry incubator (see 2.5). Spin down for 1 min in a microfuge at top speed. Load 1-3 μl into the bottom of each well (if using a 60 or 20 well comb) with a dedicated pipette (such as Drummond Sequencing Pipet, # 3-000-203) or with a gel-loading tip. Run at 120 W (constant power setting) using dye migration as a guide: in a 6% gel, bromophenol blue migrates at the size of ∼20-mer RNA, and xylene cyanol as ∼100-mer RNA. As a size marker, we use pBR322 DNA digested with MspI (NEB) and end-labeled with γ-[32P]ATP (3000 Ci/mmol, Perkin Elmer, cat #BLU-002A) and T4 polynucleotide kinase (NEB), or the RNA sequencing ladders generated as in 2.6; the former is sufficient for many applications (Fig. 2).
While not necessary, drying the gel is beneficial, as low percentage gels rarely crack and can be stored dry without diffusion of the bands, a definite plus when longer exposures are necessary. Transfer the gel to a drying paper (such as blotting paper from Life Science Products, # LS238-3543), cover with plastic (any food wrap), and use a standard set up (such as BioRad GelDryer 583 and HydroTech vacuum pump); it takes ∼1 hour at 80°C to completely dry a full-size sequencing gel. Expose the dried gel (2 h-overnight) to X-ray film or a phosphor screen (both sold by Kodak); it is not necessary to remove the plastic wrap.
RNA products are most commonly visualized and quantified using a Molecular Dynamics Phosphorimaging System and ImageQuant Software (GE Healthcare) or a comparable scanner/software combination. X-ray film densitometry can also be used but offers a smaller linear range of signal detection. After the terminated and readthrough products are quantified, the termination efficiency is determined as a ratio of the terminated RNA to the sum of the terminated and readthrough species (if no other RNA species are observed).
3. Modification of the basic termination assay for regulatory analyses
As noted above, the general in vitro transcription termination assay can be readily adapted to investigation of specific regulatory mechanisms by small changes in the assay procedure. These changes include addition of putative regulatory factors, including proteins, small RNAs and molecular effectors. In addition, sequence changes in the template, or mutant forms of RNAP, can be tested for their effect on the efficiency of termination. We will illustrate these effects using a few specific examples.
3.1 Effects of template changes and regulatory proteins: E. coli NusG
Variations in the length and stability of the helix of an intrinsic terminator, and in the length of the U run, have a dramatic effect on the efficiency of termination. This is demonstrated in Fig. 2 by the placement of three different terminators downstream of the same promoter and ITS. Terminator 1 is very inefficient, while terminators 2 and 3 show 50-60% termination. The E. coli NusG protein is a transcription factor that affects the processivity of RNAP and its sensitivity to pause and termination signals [28]. Addition of NusG protein to the halted TEC prior to the chase step (triggered by addition of all 4 NTPs) has little effect on terminators 1 and 3, but results in a 2-fold reduction in the activity of terminator 2. These results illustrate not only that subtle changes in terminator architecture can have major effects (note that terminator 1 has a more stable helix than that of terminator 2, and a longer uninterrupted U-run, although terminator 2 has one additional U residue), but also that different terminators may have very different sensitivity to transcription factors. Similar studies can be used to determine the effect of RNA-binding regulatory proteins (like E. coli BglG and B. subtilis TRAP) that bind to specific sequence or structural elements in the nascent transcript to affect RNA structure, therefore affecting formation of the terminator helix (see [2] for review).
3.2. Binding of small molecule effectors to the nascent transcript
Riboswitches are small RNA elements present in the 5′ region of regulated genes or operons that directly recognize physiological signals and exhibit changes in RNA structure that impact expression of the downstream genes (1). The S box riboswitch is a member of this group, and binds SAM to regulate expression of genes involved in methionine metabolism. In most genes in the S box family, binding of SAM results in stabilization of a terminator element, which promotes termination of transcription before RNAP reaches the regulated coding sequences. The S box system was initially characterized in vivo, and genetic experiments demonstrated regulation in response to methionine availability, and the role of the proposed terminator and competing antiterminator elements [29].
Proof that the nascent RNA transcript can directly bind SAM, and that addition of SAM is sufficient to promote transcription termination, was derived from in vitro transcription termination assays [30,31]. In the absence of SAM, the termination site in the 5′ region of an S box gene is inefficient, and most TECs readthrough the terminator (Fig. 3). Addition of SAM results in increased termination, in a dose-dependent, saturable response [30,31], and no cellular factors other than RNAP are required.
Fig. 3.
SAM-dependent transcription termination of the S box riboswitch. (a) In vitro transcription of a B. subtilis yitJ template in the presence or absence of SAM. The template consists of the yitJ 5′ sequence (residues +14 to +289, relative to the yitJ transcription start-site) preceded by the constitutive B. subtilis glyQS promoter. Transcription by B. subtilis RNAP was initiated with ApC (corresponding to the +1/+2 positions of the transcript) in the presence of ATP, CTP and UTP to generate a halted TEC at +17. SAM was added as indicated (0-2.4 μM final concentration), and the products were resolved by 6% denaturing PAGE. Terminated (T, 185 nt, filled circles) and run-off (RO, 235 nt, open circles) RNAs are labeled. (b) Quantification of the SAM response. Termination efficiency is defined as the fraction of RNA at the terminator (T) relative to the total RNA (T + RO).
4. Concluding remarks
In this article, we describe an assay for transcription termination in bacteria along with general considerations for alternative assay designs. In vitro transcription termination assays are relatively easy, work with RNAPs from different bacteria (in our labs, we use them with E. coli, B. subtilis, and T. thermophilus RNAPs), and allow rapid and reproducible testing of the effects of accessory proteins (such as NusG), small molecules (such as metabolites), temperature, solution conditions, etc. The main reason for the robust nature of these assays is the high stability of the transcription elongation complexes, which allows addition of a variety of test reagents. We describe a basic protocol for single-round transcription reactions. This protocol has been streamlined as much as possible, and all the steps that can be omitted (such as RNA precipitation after reaction) have been omitted already. Particular attention should be paid to the quality of RNAP, templates, and substrates, as poor quality of any of these reagents will translate into substandard and poorly reproducible results.
Acknowledgments
We thank Frank Grundy and Vineeta Pradhan for help with the SAM-dependent termination assay. This work was supported by NIH grants GM67153 (IA) and GM47823 and GM63615 (TH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grundy FJ, Henkin TM. Crit Rev Biochem Mol Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 2.Henkin TM, Yanofsky C. Bioessays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds R, Bermudez-Cruz RM, Chamberlin MJ. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 4.Grundy FJ, Moir TR, Haldeman MT, Henkin TM. Nucl Acids Res. 2002;30:1646–1655. doi: 10.1093/nar/30.7.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komissarova N, Kashlev M. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 6.Minakhin L, Nechaev S, Campbell EA, Severinov K. J Bacteriol. 2001;183:71–76. doi: 10.1128/JB.183.1.71-76.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau LF, Roberts JW, Wu R. Proc Natl Acad Sci USA. 1982;79:6171–6175. doi: 10.1073/pnas.79.20.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashlev M, Nudler E, Severinov K, Borukhov S, Komissarova N, Goldfarb A. Methods Enzymol. 1996;274:326–334. doi: 10.1016/s0076-6879(96)74028-4. [DOI] [PubMed] [Google Scholar]
- 9.Park JS, Roberts JW. Proc Natl Acad Sci USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anthony LC, Artsimovitch I, Svetlov V, Landick R, Burgess RR. Protein Expr Purif. 2000;19:350–354. doi: 10.1006/prep.2000.1272. [DOI] [PubMed] [Google Scholar]
- 11.Artsimovitch I, Svetlov V, Murakami KS, Landick R. J Biol Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 12.Vassylyev DG, Svetlov V, Vassylyeva MN, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Artsimovitch I. Nat Struct Mol Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuznedelov K, Lamour V, Patikoglou G, Chlenov M, Darst SA, Severinov K. J Mol Biol. 2006;359:110–121. doi: 10.1016/j.jmb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Tang H, Severinov K, Goldfarb A, Ebright RH. Proc Natl Acad Sci USA. 1995;92:4902–4906. doi: 10.1073/pnas.92.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeda H, Fujita N, Ishihama A. Nucl Acids Res. 2000;28:3497–3503. doi: 10.1093/nar/28.18.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMahan SA, Burgess RR. Biochemistry. 1999;38:12424–12431. doi: 10.1021/bi990824x. [DOI] [PubMed] [Google Scholar]
- 17.Chang BY, Doi RH. J Bacteriol. 1990;172:3257–3263. doi: 10.1128/jb.172.6.3257-3263.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juang YL, Helmann JD. J Mol Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- 19.Burgess RR, Jendrisak JJ. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 20.Hager DA, Jin DJ, Burgess RR. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- 21.Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 22.Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R. J Bacteriol. 2000;182:6027–6035. doi: 10.1128/jb.182.21.6027-6035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albert AC, Roman AM, Bouche G, Leng M, Rahmouni AR. J Biol Chem. 1994;269:19238–19244. [PubMed] [Google Scholar]
- 24.Reynolds R, Chamberlin MJ. J Mol Biol. 1992;224:53–63. doi: 10.1016/0022-2836(92)90575-5. [DOI] [PubMed] [Google Scholar]
- 25.Artsimovitch lab website; http://www.osumicrobiology.org/faculty/iartsimovitch.htm
- 26.Artsimovitch I, Landick R. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 27.Landick R, Wang D, Chan CL. Methods Enzymol. 1996;274:334–353. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- 28.Burova E, Hung SC, Sagitov V, Stitt BL, Gottesman ME. J Bacteriol. 1995;177:1388–1392. doi: 10.1128/jb.177.5.1388-1392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grundy FJ, Henkin TM. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 30.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Proc Natl Acad Sci USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. J Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ederth J, Artsimovitch I, Isaksson LA, Landick R. J Biol Chem. 2002;277:37456–37463. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]