Abstract
Aims
MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression at the post-transcriptional level by either degradation or translational repression of a target mRNA. Encoded in the genome of most eukaryotes, miRNAs have been proposed to regulate specifically up to 90% of human genes through a process known as miRNA-guided RNA silencing. For the first time, we sought to test how myocardial ischaemia–reperfusion (IR) changes miR expression.
Methods and results
Following 2 and 7 h of IR or sham operation, myocardial tissue was collected and subjected to miRNA expression profiling and quantification using a Bioarray system that screens for human-, mice-, rat-, and Ambi-miR. Data mining and differential analyses resulted in 13 miRs that were up-regulated on day 2, 9 miRs that were up-regulated on day 7, and 6 miRs that were down-regulated on day 7 post-IR. Results randomly selected from expression profiling were validated using real-time PCR. Tissue elements laser-captured from the infarct site showed marked induction of miR-21. In situ hybridization studies using locked nucleic acid miR-21-specific probe identified that IR-inducible miR-21 was specifically localized in the infarct region of the IR heart. Immunohistochemistry data show that cardiac fibroblasts (CFs) are the major cell type in the infarct zone. Studies with isolated CFs demonstrated that phosphatase and tensin homologue (PTEN) is a direct target of miR-21. Modulation of miR-21 regulated expression of matrix metalloprotease-2 (MMP-2) via a PTEN pathway. Finally, we noted a marked decrease in PTEN expression in the infarct zone. This decrease was associated with increased MMP-2 expression in the infarct area.
Conclusion
This work constitutes the first report describing changes in miR expression in response to IR in the mouse heart, showing that miR-21 regulates MMP-2 expression in CFs of the infarct zone via a PTEN pathway.
KEYWORDS: Reactive oxygen, Non-coding gene, Gene expression, Genomics, Redox
1. Introduction
MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression at the post-transcriptional level by either degradation or translational repression of a target mRNA. Encoded in the genome of most eukaryotes, miRNAs have been proposed to regulate specifically up to 90% of human genes through a process known as miRNA-guided RNA silencing.1 Although miRNA studies predominate in the cancer field, little is known about their expression patterns or role in other diseases, especially cardiovascular disorders.
Cardiac ischaemia leading to post-infarction heart failure particularly in patients with large myocardial infarction is associated with a high mortality. The mouse model of acute myocardial infarction is commonly used to gain mechanistic insight.2 At present, there is no report on how the miR genome responds to myocardial infarction. Recently, we have demonstrated that miRNAs regulate the angiogenic potential of endothelial cells by regulating redox signalling.3 This work constitutes the first report describing changes in global miR expression in response to ischaemia–reperfusion (IR) in the mouse heart. Once the list of IR-sensitive miRs in the myocardium was identified, our goal was to utilize results from expression profiling to develop a new paradigm in IR biology of the murine heart. This is a particularly challenging task because although the IR-sensitive miRs have hundreds of target genes for each miR that have been predicted computationally, very few have targets that have been biologically validated. Our focus on miR-21 was based on the fact that a few target genes for this miR have been biologically validated.
2. Methods
2.1. Survival model for coronary artery occlusion and reperfusion
C57BL/6 (males, 10–12 weeks old) mice were subjected to IR of the heart as described previously.4–6 The studies were approved by the Institutional Laboratory Animal Care and Use Committee of The Ohio State University. The investigation conforms with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. High-resolution (11.7 T) cardiac magnetic resonance imaging was used to determine loss in cardiac function following the IR insult7 (see Supplementary material online, Figure S1). The mice were killed 2 or 7 days after reperfusion. Hearts were either collected frozen in OCT compound for laser capture or in formalin for histological analyses. For microarray analysis and real-time PCR, the hearts were snap frozen in liquid nitrogen. For control, heart tissues were collected from sham-operated mice. These mice were exposed to all surgical procedures except IR.
2.2. MicroRNA expression profiling and quantification
MiRNA expression profiling and quantification was performed using mirVana™ miRNA Bioarray system (miRNA Bioarray V2, 1566V2, Ambion, Austin, TX, USA). Details of the procedure and data analysis are submitted as supplemental text (see Supplementary material online, Table S4).
2.3. Quantification of microRNA expression
Total RNA including the miRNA fraction was isolated using mirVana™ miRNA isolation kit, according to the manufacturer’s protocol (Ambion). miRNA levels were quantified using specific Taqman assays for miRNA (Applied Biosystems, Foster City, CA, USA) and Taqman Universal Master Mix (Applied Biosystems). miRNA levels were quantified with the 2(−ΔΔct) relative quantification method8 using miR-16 as the house-keeping miRNA.
2.4. miR/mRNA expression assay from laser-captured microdissected heart samples
Laser microdissection and pressure catapulting (LMPC) was performed using the Microlaser System from PALM Microlaser Technologies AG (Bernreid, Germany) as described.5,9,10 Briefly, murine hearts subjected to IR were harvested, frozen in OCT compound, and then cut into 10 µm sections using a cryo-microtome. The sections were placed on polyethylene napthalate membrane glass slides (P.A.L.M. Microlaser Technologies AG; Bernreid), which had been RNAZap (Ambion) and UV treated, for cutting and catapulting as described by our group recently.9,11 Sections were stained using a modified haematoxylin QS procedure and the infarct site identified.9,12 Typical settings used for laser cutting were UV-Energy of 75–85 and UV-Focus of 70. Matched area (2 × 106 µm2) of control or non-infarct (NI) and infarct (I) areas were captured into 25 µL of RNA extraction buffer (RNAqueous®-Micro Kit, Ambion). The total RNA including the miRNA fractions was isolated using QIAcube (Qiagen, Valencia, CA, USA) using RNeasy Mini Plus Protocol with the gDNA eliminator columns and RNeasy Mini Spin column. The quantification of miRNA-21 was performed using Taqman assays for miRNA as described in the previous section. Quantification of matrix metalloprotease-2 (MMP-2) was performed using real-time PCR as described.11
2.5. In situ hybridization
miR-21 was localized in mouse heart and subjected to IR using in situ hybridization described previously.13 In brief, the tissue was deparaffinized, treated with protease (2 mg/mL of pepsin for 30 min in RNase free water), washed in sterile water, then treated with 100% ethanol, and air-dried. LNA-modified cDNA probes for miR-21 was used. The probes were labelled with the 3′ oligonucleotide tailing kit (Enzo Diagnostics, Farmingdale, NY, USA) using biotin as the reporter nucleotide. Hybridization was performed at 37°C overnight and followed by a wash in 0.2× SSC and 2% bovine serum albumin. The probe-target complex was seen due to the action of alkaline phosphatase (as part of the streptavidin complex—Enzo Diagnostics) on the chromogen nitroblue tetrazolium and bromochloroindolyl phosphate. Nuclear fast red was used as the counterstain. The negative controls included: omission of the probe, use of a scrambled LNA probe (the same sequence as the miR cDNA but where the nucleotides have been ‘scrambled’ at random so that is very low homology with the target sequence), and the internal negative controls provided by each tissue section.
2.6. Cardiac fibroblast isolation and culture
Experiments were performed using primary cardiac fibroblasts (CFs) isolated from adult (5–6 week old) mouse ventricle using procedures described previously.4,6 Experiments were performed under 5% O2 (in vivo normoxia for cardiac cells) conditions as described before.4–6,14
2.7. miRIDIAN miRNA inhibitor/mimic and siRNA delivery
The cells were seeded (0.2 × 106 cells/plate in 35 mm plate) in antibiotic-free medium for 24 h prior to transfection. DharmaFECT™ 1 transfection reagent (Dharmacon RNA Technologies, Lafayette, CO, USA) was used to transfect cells with miRIDIAN mmu-miR-21 inhibitor or miRIDIAN mimic-miR-21 (Dharmacon RNA Technologies) for 72 h as per the manufacturer’s instructions. miRIDIAN miRNA inhibitor/mimic negative controls (Dharmacon RNA Technologies) were used for control transfections. Samples were collected after 72 h of miRNA inhibitor/mimic transfections for quantification of miRNA and protein expression. Phosphatase and tensin homologue (PTEN) siRNA transfection was performed as described.5
2.8. Western blot
Western blot was performed using primary antibody against PTEN (anti-PTEN, 26H9, Cell Signalling Technology, Inc., Danvers, MA, USA) and p-AKT (anti-phospho AKT, Ser 473, Cell Signalling Technology, Inc.) as described previously.4,5,10
2.9. pGL3-PTEN-3′-UTR luciferase reporter assay
mmu-miR-21 inhibitor or miRIDIAN mimic-miR-21 was transfected to the cells followed by transfection with pGL3-PTEN-3′-UTR firefly luciferase expression construct together with Renilla luciferase pRL-cmv expression construct using Lipofectamine™ LTX PLUS™ reagent (Invitrogen). Luciferase (Renilla and firefly) assays were performed using the dual-luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase expression for each sample.
2.10. Histology
Formalin-fixed paraffin-embedded or OCT-embedded frozen specimens were sectioned. Immunohistochemical staining of sections was performed as described earlier10 using the following primary antibodies: anti-vimentin (dil 1:200, clone V2009, Biomeda), anti-muscle actin (dil. 1:50, clone HHF35, Dako Cytonation), anti-alpha-sarcomeric actin (dil. 1:200, Abcam), and anti-PTEN (dil.1:100, Cell Signalling). Secondary antibody detection and counterstaining were performed as described previously.10
2.11. Statistics
In vitro data are reported as mean ± SD of at least three experiments. Comparisons among multiple groups were made by analysis of variance. P < 0.05 was considered statistically significant. For in vivo studies, data are reported as mean ± SD of at least four to six animals.
3. Results
This study consists of two distinct components. The first part focuses on identifying, for the first time, IR-sensitive miRNA in the myocardium in vivo. This expression profiling component represented a hypothesis-generating mission that would provide results from unbiased interrogation of the miR genome. The second part of this study is based on findings of the first part where the functional significance of miR-21 in the ischaemia-reperfused myocardium was addressed in a hypothesis testing scenario.
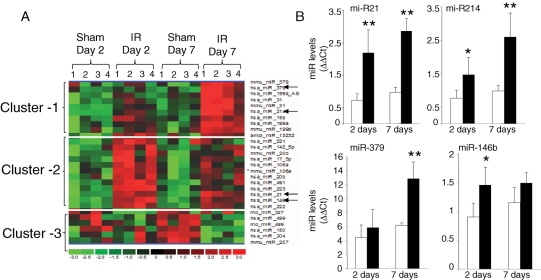
The experimental design and data analysis approach (see Supplementary material online, Figure S2) of this work are based on our previous studies on the analysis of gene expression profiling.10,11 Here, a total of 661 miRs were screened using a mirVana Bioarray V2 platform (http://www.ambion.com/jp/techlib/resources/miRNA_array/Ambion_bioarray_1566V2_Jul06.xls). These bioarrays contain probes targeting a comprehensive selection of mouse miRNA from miRBase (Sanger miRNA Registry: http://microrna.sanger.ac.uk) and a set of novel, proprietary miRNA called Ambi-miR. The data analysis approach identified a total of 4 miRs that were significantly and uniquely up-regulated at day 7 post-IR, 13 miRs that were up-regulated at day 2 post-IR, and 6 miRs that were uniquely down-regulated at day 7 post-IR (see Supplementary material online, Figure S2A). With the objective to validate the normalization approach used in this study, side-by-side box plots of globally normalized log2 ratio of the expression values for each sample were plotted.15 Box plot is typically represented by a box including a central line and tail. The median is represented by a red line in the box (see Supplementary material online, Figure S2B). The ends of the box represent the first and third quartiles of the data, with the box containing 50% of the data. A point beyond the ends of the box on either side is considered a mild outlier and red crosses represent extreme outliers. The box plot illustrated as in Supplementary material online, Figure S2 enables visualization of spatial variation among all samples following scaling and normalization. The distribution of data among all arrays following normalization was centred, thus validating the normalization approach (see Supplementary material online, Figure S2). Hierarchical clustering involves unsupervised sample clustering of candidate genes, obtained by appropriate analysis and filtering, to identify novel clusters that changed in a similar direction in response to an experimental intervention.16 Hierarchical cluster image illustrating IR-sensitive miR induced or down-regulated in the myocardial tissue is presented as in Figure 1A. miRs that were found to be significantly up- or down-regulated in response to IR, compared with the results from sham-operated mice, were subjected to hierarchical clustering. The following three major clusters were thus identified: (i) miR up-regulated 7 days post-IR; (ii) miR up-regulated 2 days post-IR; and (iii) miR down-regulated 7 days post-IR. Annotation of miR in each of the cluster is presented in Table 1.
Figure 1.
miR expression profiling in hearts subjected to ischaemia–reperfusion. (A) Specific clusters of miR showing an increase or decrease in expression following ischaemia (30 min) and reperfusion (2 or 7 days). miRs that were found significantly up- or down-regulated in all replicates following ischaemia–reperfusion (IR) compared with sham-operated samples were selected. These select candidates were subjected to hierarchical clustering to identify clusters of miRs that were induced or down-regulated by ischaemia–reperfusion following 2 or 7 days post-ischaemia–reperfusion. (i) cluster of miR up-regulated specifically 7 days post-ischaemia–reperfusion; (ii) cluster of miR up-regulated 2 days post-ischaemia–reperfusion; (iii) cluster of miR down-regulated specifically 7 days post-ischaemia–reperfusion. Data shown are from four (1–4) individual animals. As is the standard, red to green gradation in colour represents higher to lower expression signals. Annotations of miR in each of the cluster are presented in Table 1. (B) Expression levels of select miR identified using the profiling assay were independently determined using real-time PCR. Real-time PCR data were normalized to miR-16. We chose miR-16 as the normalizing control because the expression of this miR remained unchanged between sham and ischaemia–reperfusion groups. Those marked with a black arrow were verified using quantitative assays. Data represent mean ± SD (n = 4). *P < 0.05 and **P < 0.01 compared with the corresponding sham samples.
Table 1.
miRNA significantly (P < 0.05) changed in hearts subjected to ischaemia–reperfusion compared with sham-treated hearts on day 2 or day 7 post-ischaemia–reperfusion
| Post-IR day 2 (sham vs. IR) |
Post-IR day 7 (sham vs. IR) |
|||||
|---|---|---|---|---|---|---|
| Probe set | Mean | SD | P | Mean | SD | P |
| Cluster 1 | ||||||
| hsa_miR_214a | 1.849 | 0.306 | 0.001 | 2.730 | 0.989 | 0.013 |
| hsa_miR_199a | 1.502 | 0.077 | 0.048 | 2.205 | 0.301 | 0.001 |
| hsa_miR_379 | 1.059 | 0.120 | 0.533 | 1.845 | 0.443 | 0.011 |
| hsa_miR_199a_AS | 1.340 | 0.265 | 0.071 | 1.690 | 0.436 | 0.023 |
| hsa_miR_31 | 1.327 | 0.199 | 0.045 | 1.683 | 0.260 | 0.003 |
| mmu_miR_379a | 1.032 | 0.181 | 0.847 | 1.633 | 0.166 | 0.003 |
| mmu_miR_199b | 1.305 | 0.184 | 0.171 | 1.616 | 0.094 | 0.007 |
| mmu_miR_31 | 1.296 | 0.056 | 0.000 | 1.599 | 0.269 | 0.006 |
| hsa_miR_15b | 1.554 | 0.237 | 0.022 | 1.365 | 0.117 | 0.004 |
| Cluster 2 | ||||||
| hsa_miR_223 | 3.664 | 0.253 | 0.000 | 1.329 | 0.214 | 0.143 |
| hsa_miR_21a | 3.290 | 0.520 | 0.000 | 3.078 | 0.937 | 0.005 |
| hsa_miR_146ba | 2.042 | 0.237 | 0.000 | 1.504 | 0.177 | 0.006 |
| ambi_miR_13232 | 1.986 | 0.234 | 0.001 | 1.433 | 0.132 | 0.036 |
| hsa_miR_491 | 1.978 | 0.170 | 0.000 | 1.558 | 0.255 | 0.007 |
| hsa_miR_142_5p | 1.902 | 0.459 | 0.016 | 1.378 | 0.242 | 0.025 |
| hsa_miR_222 | 1.639 | 0.244 | 0.003 | 1.308 | 0.129 | 0.007 |
| mmu_miR_106a | 1.604 | 0.138 | 0.003 | 1.327 | 0.145 | 0.052 |
| mmu_miR_20b | 1.530 | 0.292 | 0.015 | 1.199 | 0.174 | 0.200 |
| hsa_miR_17_5p | 1.522 | 0.259 | 0.008 | 1.238 | 0.115 | 0.053 |
| hsa_miR_20b | 1.508 | 0.164 | 0.009 | 1.249 | 0.178 | 0.113 |
| hsa_miR_106a | 1.407 | 0.144 | 0.005 | 1.210 | 0.146 | 0.114 |
| hsa_miR_221 | 1.370 | 0.086 | 0.001 | 0.990 | 0.113 | 0.922 |
| Cluster 3 | ||||||
| hsa_miR_499 | −1.602 | 0.607 | 0.146 | −1.639 | 0.191 | 0.003 |
| rno_miR_499 | −1.217 | 0.211 | 0.273 | −1.638 | 0.459 | 0.022 |
| hsa_miR_204 | −0.923 | 0.146 | 0.386 | −1.436 | 0.190 | 0.001 |
| mmu_miR_207 | −0.969 | 0.126 | 0.626 | −1.412 | 0.154 | 0.011 |
| hsa_miR_150 | −1.059 | 0.105 | 0.688 | −1.325 | 0.070 | 0.000 |
| rno_miR_327 | −1.354 | 0.042 | 0.005 | −1.015 | 0.175 | 0.948 |
The corresponding cluster image is shown in Figure 1. Data presented indicate fold change (mean ± SD) in miR expression.
aVerified by quantitative assays.
Mining of the expression profiling data generated three clusters of genes: up-regulated day 2 post-IR, up-regulated day 7 post-IR, and down-regulated day 7 post-IR. In the cluster that was up-regulated 2 days post-IR, roughly half (6 out of 13) of all the miRs were also found to be significantly higher on day 7 post-IR. Thus, this cluster had a subset that was induced transiently, whereas the second subset was found induced at both time-points examined. For the cluster of miRs that were induced on day 7 post-IR, roughly half (five out of nine) were already induced on day 2 post-IR. Therefore, this cluster demonstrated the same pattern of subsets as noted for the 2-day post-IR cluster discussed above. For the miR down-regulated in response to IR, all but one showed significant lowering on day 7 post-IR. The remaining miR, rno_miR_327, was down-regulated on day 2 post-IR (Table 1). To verify the accuracy of miR expression profiling analyses performed, Taqman-based real-time PCR analysis was performed on four candidate miRs (miR-21, miR-214, miR-379, and miR-146b) randomly selected from Table 1. Quantitative real-time PCR analyses of the individual miR provided results that were consistent with microarray findings (Figure 1B).
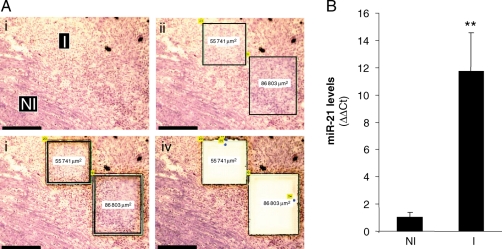
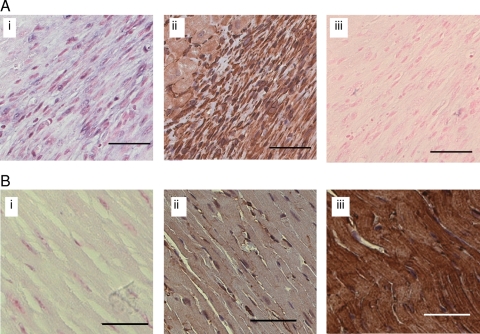
Next, our goal was to utilize results from expression profiling to develop a new paradigm in IR biology of the murine heart. Recently, we have developed novel technical capabilities in LMPC, which enable the execution of high-resolution laser capture and subsequent analyses from the myocardium and vascular tissues.9–12 Our results demonstrated that adoption of the LMPC approach, as reported by our laboratory, substantially improves the confidence in detected changes in gene expression in IR-affected tissue especially in mice where the affected area is so small. Standard surgical techniques do not allow exercise of a high level of stringency in selectively collected IR-affected tissue while rejecting unaffected tissue, resulting in dilution of the observation. Thus, only major changes can be detected, whereas subtle changes in gene expression stand the risk of getting masked while using traditional surgical techniques for tissue harvest.10 LMPC is best suited to address this limitation.9–12 Results of this study represent first report of miRNA quantitation from laser-captured tissue elements. We observed that miR-21 is potently and specifically induced in the infarct region, but not in the non-infarct tissue harvested beyond 500 µm away from the infarct site (Figure 2). To follow-up on the functional significance of this observation, it was important to know whether the noted miR-21 induction was specific to any given cell type. As a result, in situ hybridization to detect miR-21 was performed using an LNA-based approach. Cell-specific localization of miR-21 in heart sections was achieved with an LNA miR-21-specific probe. Interestingly, we observed that miR-21 was indeed induced, but only specifically in the infarct zone of the hearts subjected to IR (Figure 3). Vimentin immunostaining of a serial section shows that CFs represent the major cell type of this zone (Figure 3). The signal was not evident in the serial section when the scrambled LNA miRNA probe was utilized (Figure 3). miR-21-specific signal was not observed in non-IR regions of the heart (Figure 3).
Figure 2.
Induction of miR-21 in the infarct region of ischaemia-reperfused murine heart: a laser-capture microdissection based approach. C57BL/6 mice were subjected to ischaemia (30 min) and reperfusion (IR). Heart samples were collected 7 days post-IR. (A) Frozen sections (10 µm) from collected heart samples were stained with haematoxylin QS to histologically define the infarct (I) and control or non-infarct (NI) regions. These regions were subsequently cut and captured using laser-capture microdissection for miRNA expression analysis. (i) Representative images of heart tissue showing infarct (I) and control or non-infarct (NI) areas following laser-capture microdissection compatible staining with haematoxylin QS; (ii) the identified area is marked; and (iii) laser-assisted cutting and separation of the identified area for catapulting; (iv) the section after the marked area has been catapulted; Scale bar = 150 µm. (B) The laser-captured tissue elements from I and NI regions were used for quantification of miR-21 expression using real-time PCR. Data represent mean ± SD (n = 4). **P < 0.01 compared with control (NI) tissue.
Figure 3.
Localization of miR-21 in ischaemia-reperfused mouse heart regions. Representative image showing localization of miR-21 in a section of a mouse heart subjected to ischaemia for 30 min and reperfusion 7 days. Localization of miR-21 in heart sections was achieved using in situ hybridization analysis with an LNA miR-21-specific probe. (A) miR-21 signal (blue) is evident in the infarct region (i). Fibroblast represented the major cell type in this region as identified using immunohistochemical analysis with an anti-vimentin antibody (ii, brown) The miR-21 signal [as in (i)] was not evident in the serial section when the scrambled LNA miRNA probe was utilized (iii). (B) Representative images from the non-infract region showing no or weak signal (blue) for miR-21 (i). The distribution of fibroblasts and myocytes in this non-infarct (control) region was shown using anti-vimentin (ii, brown, fibroblasts), or anti-muscle actin (iii brown, myocytes) antibody immunohistochemical staining. Nuclear red fast (ISH) and haematoxylin (nuclear, blue, immunohistochemical) were used as counterstains. Scale bar=50 µm. Additional higher magnification images of the infarct region for the ISH and immunohistochemical stains are also presented in Supplementary material online, Figure S3.
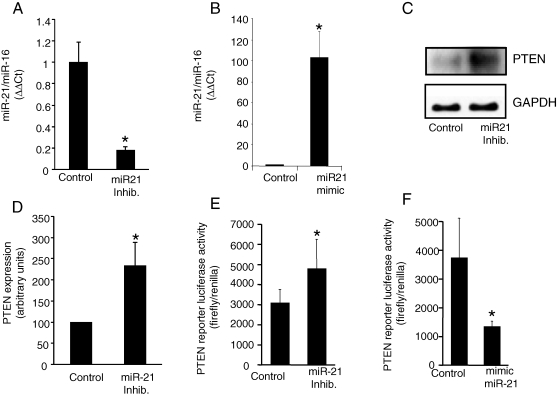
Although computational algorithms have predicted target genes for specific miR, most such predictions remain to be experimentally validated. PTEN has been recently identified as a target for miR-21.17 In addition, PTEN is the major phosphatase that has been recently linked to the biology of myocardial IR injury as well as ischaemic preconditioning.18 Furthermore, PTEN has been suggested to play a significant role in the regulation of cardiac fibrosis.19 Thus, we sought to examine the significance of miR-21 in regulating PTEN in the CFs. Experiments were performed using primary CFs isolated from adult (5–6 week old) mouse ventricle.4,6 miR-21 levels in CFs were modulated by transfecting the cells with miRIDIAN mmu-miR-21 inhibitor or mimic. These miR inhibitors/mimics are single- or double-stranded RNA oligonucleotides designed to inhibit or increase the function of specific miRNAs. Oligonucleotides are chemically modified to improve efficiency and biological stability. A negative control miR inhibitor/mimic was used as control. Transfection of CFs with miR-21 inhibitor significantly inhibited miR-21 expression in these cells (Figure 4A). Likewise, miR-21 mimic increased the expression of miR-21 in CFs (Figure 4B). Inhibition of miR-21 in CFs resulted in an increase in PTEN protein abundance (Figure 4C and D). To assess whether PTEN is a direct target of miR-21 in CFs, a fragment of the 3′-UTR of PTEN mRNA containing the putative miR-21-binding sequence cloned into a firefly luciferase reporter construct was used. This construct was co-transfected with a control Renilla luciferase reporter construct into CFs along with control or miR-21-specific inhibitor/mimic.17 miR-21 inhibitor significantly increased the luciferase activity, whereas increasing miR-21 expression in CFs using mimic resulted in significantly decreased luciferase activity, demonstrating a direct and inverse control of miR-21 on PTEN levels in CFs (Figure 4E and F). These observations, for the first time, identify a direct role of miR-21 in regulating PTEN levels in CFs.
Figure 4.
Direct regulation of phosphatase and tensin homologue in cardiac fibroblasts by miR-21. After isolation, cardiac fibroblasts were cultured for 5 days. The cells were transfected with miRIDIAN mmu-miR-21 inhibitor or miRIDIAN mmu-miR-21 mimic to down-regulate or increase miR-21 levels. (A and B) miR-21 expression levels in cardiac fibroblasts 72 h post-transfection were determined using real-time PCR. miRIDIAN miRNA inhibitor/mimic negative controls were used for control transfections. Mean ± SD (n = 4), *P < 0.01 compared with control-transfected cardiac fibroblasts. (C) Phosphatase and tensin homologue protein levels in cardiac fibroblasts following knockdown of miR-21 in cardiac fibroblasts. (D) Quantification of phosphatase and tensin homologue level was performed using densitometry. Data were normalized to GAPDH. Data shown represent mean ± SD (n = 3). *P < 0.01 compared with control-transfected cells. (E and F) To demonstrate that phosphatase and tensin homologue is a direct target for miR-21 in cardiac fibroblasts, the cells were transfected with a pGL3-PTEN-3′-UTR firefly luciferase expression construct and co-transfected with pRL-TK Renilla luciferase expression construct along with either miR-21 inhibitor or mimic. An increase in relative firefly luciferase activity in the presence of miR-21 inhibitor or a decrease in the presence of miR-21 mimic indicates that the 3′-UTR of phosphatase and tensin homologue contains a target that is modulated by miR-21. Data represent mean ± SD (n = 3). *P < 0.05 compared with control-transfected cells.
Work directed at understanding the significance of PTEN regulation by miR-21 in CFs led to the observation that MMP-2 expression in CFs is regulated by miR-21 via a PTEN-AKT-phosphorylation-dependent pathway. PTEN knockdown in CFs resulted in marked phosphorylation of Akt (Figure 5A–C). Transfection of CFs with miR-21 mimic significantly increased MMP-2 expression (Figure 5D). This effect was significantly enhanced under conditions of PTEN knockdown (Figure 5D). Taken together, these results demonstrate that PTEN, an miR-21 target in CFs, negatively regulates Akt phosphorylation. miR-21-dependent lowering of PTEN results in Akt phosphorylation and induction of MMP-2. These results, combined with our observation that miR-21 is expressed in the infarct zone of the IR heart (Figure 3A), were in indirect agreement with our finding that PTEN expression is lowered and MMP-2 expression is elevated in the infarct region of the heart (Figure 6).
Figure 5.
miR-21 regulates MMP-2 expression in cardiac fibroblasts via a PTEN-AKT-phosphorylation-dependent pathway. After isolation, cardiac fibroblasts were cultured for 5 days. The cells were transfected with phosphatase and tensin homologue siRNA. (A) Western blots of AKT phosphorylation and phosphatase and tensin homologue expression in cardiac fibroblasts following transfection of the cells with phosphatase and tensin homologue or control siRNA. β-actin was used for data normalization. (B and C) Quantification of p-AKT and phosphatase and tensin homologue levels were performed using densitometry. Data were normalized to β-actin. Data represent mean ± SD (n = 3). *P < 0.05 compared with control siRNA-transfected cells. (D) MMP-2 expression in cardiac fibroblasts transfected with miR-21 mimic to increase miR-21 levels. MMP-2 expression in cardiac fibroblasts was measured using real-time PCR. Data represent mean ± SD (n = 3). *P < 0.05 compared with mimic control-transfected cells. (E) MMP-2 expression in cardiac fibroblasts transfected either with miR-21 mimic or co-transfected with phosphatase and tensin homologue siRNA. Data represent mean ± SD (n = 3). *P < 0.05 compared with mimic control-transfected cells. #P < 0.05 compared with miR-21 mimic + control siRNA-transfected cells.
Figure 6.
Decreased phosphatase and tensin homologue and increased MMP-2 expression in the infarct region of heart. Mice were subjected to left anterior descending coronary artery ligation for 30 min followed by reperfusion (IR). (A) Representative images showing phosphatase and tensin homologue expression in a section of mouse heart subjected to 7 days post-IR. Localization of phosphatase and tensin homologue (green) in heart sections was achieved using anti-phosphatase and tensin homologue antibody. The sections were counterstained with DAPI (nuclear, blue). Myocytes were stained using anti-alpha-sarcomeric actin antibody (red). (i) Representative image of phosphatase and tensin homologue (green) and DAPI (blue), infarct (I) or non-infarct (NI) regions are indicated; (ii) alpha-sarcomeric actin (red) and DAPI (blue) image. Scale bar = 50 µm. (B) Quantification of phosphatase and tensin homologue signal in infarct (I) vs. non-infarct (NI) regions. Data represent mean ± SD (n = 3). *P < 0.05 compared with NI region. (C) Histologically defined infarct (I) and non-infarct (NI) regions were captured using laser-capture microdissection as described in Figure 2. The laser-captured tissue elements from infarct and non-infarct regions were used for quantification of MMP-2 expression using real-time PCR. Data represent mean ± SD (n = 4). **P < 0.01 compared with control (NI) tissue.
4. Discussion
Expression profiling of miR in a given disease paradigm represents a powerful approach for unbiased comprehensive interrogation of the miR genome.20,21 The first expression profiling of miR in the heart was reported only recently.21 These studies aimed at examining heart failure and cardiac hypertrophy. This report provides first evidence temporally characterizing global changes in murine myocardial miRNA expression following IR.
The functional significance of miR-21 largely depends on the target genes that it regulates. Recently, PTEN has been validated as a target gene for miR-21.17 PTEN is a dual protein–lipid phosphatase which dephosphorylates the secondary messenger produced by PI3K and interrupts the downstream activation of Akt.22 Of note in the context of this study is the observation that PTEN is implicated in ischaemic heart diseases.23 Therefore, the blocking of PTEN has been proposed to be an important strategy to improve myocardial survival following an ischaemic episode.19 MMP-2 has emerged as a key protease in various pathologies associated with oxidative stress, including myocardial IR, heart failure, or inflammation.24 In the IR heart, MMP-2 directly contributes to cardiac mechanical dysfunction.25 Heart-specific transgenic MMP-2 directly impairs contractility26 and causes mitochondrial dysfunction.27 MMP-2 degrades the cytoskeletal protein alpha-actinin in peroxynitrite-mediated myocardial injury.28 Findings of this study present first evidence demonstrating that in CFs, miR-21 negatively regulates PTEN expression up-regulating phospho-Akt as well as MMP-2 expression. miR-21-induced MMP-2 expression was significantly increased in response to PTEN knockdown, directly implicating PTEN in the inverse regulation of MMP-2 expression. This observation is consistent with previous reports characterizing PTEN as a suppressor of MMP-2 expression.29–31 The hypothesis that Akt induces MMP-2 expression is also supported by evidence from other experimental systems.32,33 While this work was in review, two other publications have recognized the significance of miR-21 in the heart.34,35 Changes in miR-21, specifically in CFs, can regulate heart failure.34 Another short report noted the significance of miR-21 in influencing myocardial infarction outcomes.35
Taken together, the current study presents first evidence documenting changes in miR expression following myocardial IR in vivo. Comprehensive expression profiling was conducted to identify clusters of miRs that significantly changed in expression on days 2 and 7 post-IR. IR rapidly induced miR-21 in the heart, and elevated miR-21 levels were noted on day 7 post-IR as well. In the heart, miR-21 induction in response to IR was limited to CFs, where PTEN was identified as a target gene for miR-21. The current findings indicate that IR-induced miR-21 limits PTEN function and therefore causes activation of the Akt pathway and increased MMP-2 expression in CFs of the infarct region of the IR heart.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Funding
This work was supported by National Institutes of Health grant RO1-HL073087 to C.K.S.
Supplementary Material
References
- 1.Perron MP, Provost P. Protein interactions and complexes in human microRNA biogenesis and function. Front Biosci. 2008;13:2537–2547. doi: 10.2741/2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajasingh J, Bord E, Hamada H, Lambers E, Qin G, Losordo DW, et al. STAT3-dependent mouse embryonic stem cell differentiation into cardiomyocytes: analysis of molecular signaling and therapeutic efficacy of cardiomyocyte precommitted mES transplantation in a mouse model of myocardial infarction. Circ Res. 2007;101:910–918. doi: 10.1161/CIRCRESAHA.107.156786. [DOI] [PubMed] [Google Scholar]
- 3.Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:471–477. doi: 10.1161/ATVBAHA.107.160655. [DOI] [PubMed] [Google Scholar]
- 4.Roy S, Khanna S, Bickerstaff AA, Subramanian SV, Atalay M, Bierl M, et al. Oxygen sensing by primary cardiac fibroblasts: a key role of p21(Waf1/Cip1/Sdi1) Circ Res. 2003;92:264–271. doi: 10.1161/01.res.0000056770.30922.e6. [DOI] [PubMed] [Google Scholar]
- 5.Roy S, Khanna S, Rink T, Radtke J, Williams WT, Biswas S, et al. p21waf1/cip1/sdi1 as a central regulator of inducible smooth muscle actin expression and differentiation of cardiac fibroblasts to myofibroblasts. Mol Biol Cell. 2007;18:4837–4846. doi: 10.1091/mbc.E07-03-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy S, Khanna S, Wallace WA, Lappalainen J, Rink C, Cardounel AJ, et al. Characterization of perceived hyperoxia in isolated primary cardiac fibroblasts and in the reoxygenated heart. J Biol Chem. 2003;278:47129–47135. doi: 10.1074/jbc.M308703200. [DOI] [PubMed] [Google Scholar]
- 7.Ojha N, Roy S, Radtke J, Simonetti O, Gnyawali S, Zweier JL, et al. Characterization of the structural and functional changes in the myocardium following focal ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2008;294:H2435–H2443. doi: 10.1152/ajpheart.01190.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn DE, Roy S, Radtke J, Gupta S, Sen CK. Laser microdissection and pressure-catapulting technique to study gene expression in the reoxygenated myocardium. Am J Physiol Heart Circ Physiol. 2006;290:H2625–H2632. doi: 10.1152/ajpheart.01346.2005. [DOI] [PubMed] [Google Scholar]
- 10.Roy S, Khanna S, Kuhn DE, Rink C, Williams WT, Zweier JL, et al. Transcriptome analysis of the ischemia-reperfused remodeling myocardium: temporal changes in inflammation and extracellular matrix. Physiol Genomics. 2006;25:364–374. doi: 10.1152/physiolgenomics.00013.2006. [DOI] [PubMed] [Google Scholar]
- 11.Roy S, Patel D, Khanna S, Gordillo GM, Biswas S, Friedman A, et al. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci USA. 2007;104:14472–14477. doi: 10.1073/pnas.0706793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn D, Roy S, Radtke J, Khanna S, Sen CK. Laser microdissection and capture of pure cardiomyocytes and fibroblasts from infarcted heart regions: perceived hyperoxia induces P21 in peri-infarct myocytes. Am J Physiol Heart Circ Physiol. 2007;292:H1245–H1253. doi: 10.1152/ajpheart.01069.2006. [DOI] [PubMed] [Google Scholar]
- 13.Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Sen CK, Khanna S, Roy S. Perceived hyperoxia: oxygen-induced remodeling of the reoxygenated heart. Cardiovasc Res. 2006;71:280–288. doi: 10.1016/j.cardiores.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Davison TS, Johnson CD, Andruss BF. Analyzing micro-RNA expression using microarrays. Methods Enzymol. 2006;411:14–34. doi: 10.1016/S0076-6879(06)11002-2. [DOI] [PubMed] [Google Scholar]
- 16.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mocanu MM, Yellon DM. PTEN, the Achilles’ heel of myocardial ischaemia/reperfusion injury? Br J Pharmacol. 2007;150:833–838. doi: 10.1038/sj.bjp.0707155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oudit GY, Sun H, Kerfant BG, Crackower MA, Penninger JM, Back PH. The role of phosphoinositide-3 kinase and PTEN in cardiovascular physiology and disease. J Mol Cell Cardiol. 2004;37:449–471. doi: 10.1016/j.yjmcc.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 21.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosivatz E. Inhibiting PTEN. Biochem Soc Trans. 2007;35:257–259. doi: 10.1042/BST0350257. [DOI] [PubMed] [Google Scholar]
- 23.Sasaoka T, Wada T, Tsuneki H. Lipid phosphatases as a possible therapeutic target in cases of type 2 diabetes and obesity. Pharmacol Ther. 2006;112:799–809. doi: 10.1016/j.pharmthera.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Viappiani S, Nicolescu AC, Holt A, Sawicki G, Crawford BD, Leon H, et al. Activation and modulation of 72 kDa matrix metalloproteinase-2 by peroxynitrite and glutathione. Biochem Pharmacol. 2008 doi: 10.1016/j.bcp.2008.11.004. Epub ahead of print (PMID: 19046943) [DOI] [PubMed] [Google Scholar]
- 25.Cheung PY, Sawicki G, Wozniak M, Wang W, Radomski MW, Schulz R. Matrix metalloproteinase-2 contributes to ischemia-reperfusion injury in the heart. Circulation. 2000;101:1833–1839. doi: 10.1161/01.cir.101.15.1833. [DOI] [PubMed] [Google Scholar]
- 26.Wang GY, Bergman MR, Nguyen AP, Turcato S, Swigart PM, Rodrigo MC, et al. Cardiac transgenic matrix metalloproteinase-2 expression directly induces impaired contractility. Cardiovasc Res. 2006;69:688–696. doi: 10.1016/j.cardiores.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 27.Zhou HZ, Ma X, Gray MO, Zhu BQ, Nguyen AP, Baker AJ, et al. Transgenic MMP-2 expression induces latent cardiac mitochondrial dysfunction. Biochem Biophys Res Commun. 2007;358:189–195. doi: 10.1016/j.bbrc.2007.04.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung MM, Schulz CG, Wang W, Sawicki G, Bautista-Lopez NL, Schulz R. Matrix metalloproteinase-2 degrades the cytoskeletal protein alpha-actinin in peroxynitrite mediated myocardial injury. J Mol Cell Cardiol. 2007;43:429–436. doi: 10.1016/j.yjmcc.2007.07.055. [DOI] [PubMed] [Google Scholar]
- 29.Park MJ, Kim MS, Park IC, Kang HS, Yoo H, Park SH, et al. PTEN suppresses hyaluronic acid-induced matrix metalloproteinase-9 expression in U87MG glioblastoma cells through focal adhesion kinase dephosphorylation. Cancer Res. 2002;62:6318–6822. [PubMed] [Google Scholar]
- 30.Zheng H, Takahashi H, Murai Y, Cui Z, Nomoto K, Niwa H, et al. Expressions of MMP-2, MMP-9 and VEGF are closely linked to growth, invasion, metastasis and angiogenesis of gastric carcinoma. Anticancer Res. 2006;26:3579–3583. [PubMed] [Google Scholar]
- 31.Furukawa K, Kumon Y, Harada H, Kohno S, Nagato S, Teraoka M, et al. PTEN gene transfer suppresses the invasive potential of human malignant gliomas by regulating cell invasion-related molecules. Int J Oncol. 2006;29:73–81. [PubMed] [Google Scholar]
- 32.Park MJ, Kwak HJ, Lee HC, Yoo DH, Park IC, Kim MS, et al. Nerve growth factor induces endothelial cell invasion and cord formation by promoting matrix metalloproteinase-2 expression through the phosphatidylinositol 3-kinase/Akt signaling pathway and AP-2 transcription factor. J Biol Chem. 2007;282:30485–30496. doi: 10.1074/jbc.M701081200. [DOI] [PubMed] [Google Scholar]
- 33.Park CM, Park MJ, Kwak HJ, Lee HC, Kim MS, Lee SH, et al. Ionizing radiation enhances matrix metalloproteinase-2 secretion and invasion of glioma cells through Src/epidermal growth factor receptor-mediated p38/Akt and phosphatidylinositol 3-kinase/Akt signaling pathways. Cancer Res. 2006;66:8511–8519. doi: 10.1158/0008-5472.CAN-05-4340. [DOI] [PubMed] [Google Scholar]
- 34.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 35.Yin C, Wang X, Kukreja RC. Endogenous microRNAs induced by heat-shock reduce myocardial infarction following ischemia–reperfusion in mice. FEBS Lett. 2008;582:4137–4142. doi: 10.1016/j.febslet.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.