Abstract
Objective
The aim of this study was to determine if there is a significant difference in the risk of developing Wilms tumour between patients with submicroscopic and those with visible deletions of the WT1 tumour suppressor gene.
Methods
To determine which subjects had WT1 deletions, high‐resolution chromosomal deletion analysis of the 11p13 region was carried out in 193 people with aniridia. The rationale for this was that aniridia is caused by loss of function of one copy of the PAX6 gene, and although most patients with aniridia have intragenic mutations, a proportion has deletions that also include the nearby WT1 gene. Fluorescence in situ hybridisation (FISH) analysis of patients with aniridia identifies people with WT1 deletions regardless of whether they have Wilms tumour, allowing the deletion size to be correlated with clinical outcome.
Results
Wilms tumour was not observed in any case without a WT1 deletion. Of subjects in whom WT1 was deleted, 77% with submicroscopic deletions (detectable only by high‐resolution FISH analysis) presented with Wilms tumour compared with 42.5% with visible deletions (detectable by microscopy). This difference was significant.
Conclusions
High‐resolution deletion analysis is a useful tool for assessing the risk of Wilms tumour in neonates with aniridia. People with submicroscopic WT1 deletions have a significantly increased risk of Wilms tumour, and a high level of vigilance should be maintained in such cases.
Keywords: aniridia, Wilms tumour, WAGR, deletion, FISH
Aniridia (OMIM 106200) is a haploinsufficiency disorder caused by inactivation of one copy of the PAX6 gene at 11p13. The predominant mutational mechanism is intragenic point mutations, but numerous chromosomal rearrangements have also been described.1,2 Aniridia can be familial with dominant inheritance, or sporadic, where the mutation arises de novo and is dominantly inherited in subsequent generations. The highly penetrant phenotype is consistent with the PAX6 expression pattern and typically includes severe iris hypoplasia, foveal hypoplasia, optic nerve defects, cataracts, and a variety of brain and olfactory anomalies.3,4,5
Most cases of aniridia are isolated, but about 5% of infants born with sporadic aniridia go on to develop Wilms tumour (WT; OMIM 194070), a paediatric nephroblastoma.6,7 These individuals have WAGR syndrome (WT, aniridia, genitourinary anomalies and mental retardation; OMIM 194072), caused by 11p13 deletions that encompass both the aniridia gene (PAX6) and the Wilms tumour suppressor gene (WT1), which are ∼700 kb apart.2
A variety of loci and mechanisms are implicated in the onset of WT, but in WAGR syndrome, the most likely cause is loss of function of both copies of WT1 through a classic “two‐hit” mechanism, the first hit being constitutional deletion of one allele.8,9 People with WAGR deletions who develop WT have a relatively poor long‐term prognosis compared with patients with WT without WAGR deletions, and are at significantly higher risk of bilateral disease and end‐stage renal failure.7,10 Several longitudinal studies have documented the phenotype and clinical outcome in patients with WAGR who have presented with WT but much less is known about cases of WAGR without WT or about the likelihood that a person with WAGR deletion will go on to develop WT.
Previously we reported chromosomal aberrations, including deletions of PAX6 and WT1, in a large panel of patients with aniridia.2,11,12,13 We have now extended this work, examining more patients and collating clinical outcomes. We present evidence that deletion size influences the risk of WT, with submicroscopic deletions significantly more likely to result in tumours.
Methods
Patients
In total, 193 aniridia cases, some with associated anomalies, were collected over several years in three centres through clinical genetics and ophthalmology departments around Europe, either with ethics committee approval or for diagnostic testing, and used for deletion analysis by FISH.
Deletion analysis
Chromosomes were prepared for deletion analysis using conventional methods. FISH was performed as previously described13 with three nick‐translated cosmid probes covering a 700 kb interval: FAT5 for PAX6, P60 for D11S324 and B2.1 for WT1.2,12 For patients with visible deletions, cytogenetic breakpoints were determined by the referring laboratories using conventional microscopy. Some of the cases were studied in additional detail as described in the cited references in tables 1 and 2.
Table 1 WT1 deletion cases with Wilms tumour.
| Case No/ID* | Genitourinary anomalies | Mental retardation | Age at WT diagnosis (months) | Sex | Deletion | Confirmed by FISH | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01/SGL325 | + | + | 9 | Male | V | Yes | ||||||
| 02/WT8.1 | + | + | 9 | Male | V | Yes | ||||||
| 03/BAN14,15 | + | + | 30 | Male | V | Yes | ||||||
| 04/SOA14,15 | + | + | 48 | Male | V | Yes | ||||||
| 05/SERV116 | + | + | 28 | Male | V | Yes | ||||||
| 06/A10.111 | + | + | 12 | Female | V | Yes | ||||||
| 07/SGL243 | + | + | 12 | Female | V | Yes | ||||||
| 08/GIFRA12 | + | ? | 18 | Male | V | Yes | ||||||
| 09/GOTY14,15 | – | + | 27 | Female | V | Yes | ||||||
| 10/MARGA14,15 | – | – | 9 | Male | V | Yes | ||||||
| 11/WRGL36 | + | + | 36 | Male | V | Yes | ||||||
| 12/WRGL42 | – | ? | 26 | Female | V | Yes | ||||||
| 13/CRAWI | ? | + | 11 | Male | V | No | ||||||
| 14/CRADB | ? | + | 10 | Male | V | No | ||||||
| 15/CRAEI | ? | ? | 12 | Female | V | No | ||||||
| 16/CRAPD | ? | ? | 12 | Female | V | No | ||||||
| 17/CRAKJ | ? | + | 44 | Male | V | No | ||||||
| 18/SFG037 | + | + | 18 | Male | S | Yes | ||||||
| 19/NAHAS14,17 | + | + | 30 | Male | S | Yes | ||||||
| 20/ALSTA14,17 | + | – | 15 | Male | S | Yes | ||||||
| 21/VIMA | ? | ? | 12 | Male | S | Yes | ||||||
| 22/ANS118 | + | – | 23 | Male | S | Yes | ||||||
| 23/ANS218 | – | – | 21 | Male | S | Yes | ||||||
| 24/SIOP3677 | – | – | 12 | Male | S | Yes | ||||||
| 25/ANS1118 | – | – | 11 | Female | S | Yes | ||||||
| 26/SIOP3824 | – | – | 14 | Female | S | Yes | ||||||
| 27/WRGL16 | + | – | 17 | Male | S | Yes |
+, present; –, absent; ?, unknown; FISH, fluorescence in situ hybridisation; S, submicroscopic; V, visible.
*All patients have aniridia and Wilms' tumour. Superscript numbers are references, and patients without a cited reference number are new to this study.
Table 2 WT1 deletion cases without Wilms tumour.
| Case No/ID* | Genitourinary anomalies | Mental retardation | Tumour‐ free age (years)† | Sex | Deletion | Confirmed by FISH | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 28/A2.111 | + | + | 8 | Male | V | Yes | ||||||
| 29/RIWAR14,15 | + | + | 9 | Male | V | Yes | ||||||
| 30/TRAKE14,15 | + | + | 12 | Male | V | Yes | ||||||
| 31/TAK11 | – | + | 38 | Female | V | Yes | ||||||
| 32/A7.111 | – | + | 22 | Male | V | Yes | ||||||
| 33/ANNA14,15 | + | ? | 30 | Male | V | Yes | ||||||
| 34/NYMI14,15 | – | + | 12 | Female | V | Yes | ||||||
| 35/LEV714,15 | + | + | 7 | Male | V | Yes | ||||||
| 36/SERV216 | ? | ? | 10 | Female | V | Yes | ||||||
| 37/ANS918 | ? | ? | 5 | Female | V | Yes | ||||||
| 38/LIROB | – | + | 21 | Female | V | Yes | ||||||
| 39/AMJA | – | – | 10 | Female | V | Yes | ||||||
| 40/MAFRA12 | ? | ? | 30 | Female | V | Yes | ||||||
| 41/WRGL1413 | + | ? | 10 | Male | V | Yes | ||||||
| 42/WRGL1513 | – | ? | 7 | Female | V | Yes | ||||||
| 43/CAMEL | – | – | 6 | Male | V | No | ||||||
| 44/A9.111 | – | + | 11 | Female | V | Yes | ||||||
| 45/WRGL212 | – | – | 7 | Female | V | Yes | ||||||
| 46/WRGL41 | – | + | 9 | Female | V | Yes | ||||||
| 47/WRGL47 | + | + | 38 | Male | V | Yes | ||||||
| 48/WRGL33 | ? | + | 4.5 | Male | V | Yes | ||||||
| 49/WRGL34 | – | + | 9 | Male | V | Yes | ||||||
| 50/WRGL51 | ? | ? | 5 | Female | V | Yes | ||||||
| 51/WRGL26 | – | – | 6 | Male | S | Yes | ||||||
| 52/KAZHM14,17 | – | + | 10 | Female | S | Yes | ||||||
| 53/PAZO14,15 | + | + | 16 | Male | S | Yes |
+, present; –, absent; ?, unknown; FISH, fluorescence in situ hybridisation; S, submicroscopic; V, visible.
*All patients have aniridia but not Wilms' tumour. Superscript numbers are references, and patients without a cited reference number are new to this study.
†Tumour‐free age in years at last follow‐up.
Statistical analysis
Kaplan–Meier analysis was used to plot the incidence of tumour‐free survival for visible deletions and submicroscopic deletions and the log‐rank test was used to analyse the difference between the two groups. To assess the effect of different risk factors, multivariate analysis was performed using the Cox proportional hazards regression model. Significance was set at p<0.05.
Results
High‐resolution chromosome analysis of 11p13 was performed on 193 aniridia cases. All, except for six early cases with obvious visible deletions (tables 1 and 2), were also analysed by FISH, using cosmids FAT5 for PAX6, P60 for D11S324 (between PAX6 and WT1) and B2.1 for WT1 (700 kb proximal to PAX6).2,12 Of the 193 cases, 140 (72.5%) did not have B2.1 (WT1) deletions. There was no detectable deletion in 136 cases and 4 had only FAT5 (PAX6) deletions. None of the 140 cases with no WT1 deletion presented with Wilms tumour.
The remaining 53 patients (27.5%) had deletions of all three cosmids including the PAX6 and WT1 loci. In all, 40 of the 53 deletions (75.5%) were visible and 13 (24.5%) were submicroscopic. At the time of enquiry, 27 of the 53 deletion cases (51%) had presented with WT. Of these affected individuals, 17 carried visible and 10 submicroscopic deletions. The remaining 26 individuals had not developed a tumour; 23 had a visible deletion and 3 had a submicroscopic deletion (tables 1 and 2).
Although some patients had the classic features of WAGR syndrome, others had no evident genitourinary anomalies or mental retardation and at birth would be indistinguishable from non‐deletion aniridia cases. Our cohort was biased towards those with systemic anomalies and may therefore be enriched for individuals with larger deletions. To ascertain the true incidence of visible and submicroscopic deletions among patients with aniridia, a long‐term prospective study of newborns would be required.
For all subjects with WT, the mean age of tumour presentation was 19.5 months. The mean age of presentation for visible and submicroscopic deletions was 20.8 and 17.3 months, respectively, but this difference was not significant (p = 0.88, Mann–Whitney test). The oldest child to present with WT was 48 months. The youngest tumour‐free child was 4.5 years old and the mean age of tumour‐free subjects with a deletion was 14.25 years. In the study cohort, 17 of 40 patients with visible deletions (42.5%) and 10 of 13 patients with submicroscopic deletions (77%) developed WT.
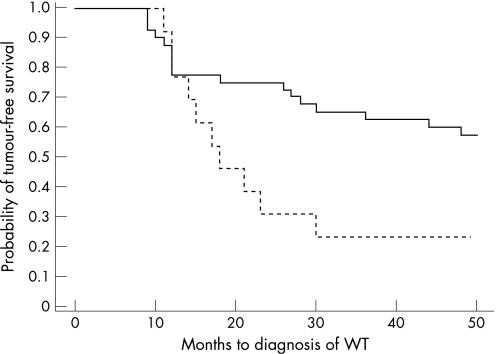
To investigate whether the increased incidence of WT in patients with submicroscopic deletion was significant, the probability of tumour‐free survival for submicroscopic and visible deletions in the first 50 months of life was determined by Kaplan–Meier analysis (fig 1). The analysis was also performed for a time‐frame of 500 months with the same outcome (data not shown). At the age of 4 years, by which time the vast majority of tumours have presented, children with submicroscopic deletions were more than twice as likely to have WT than those with visible deletions. By Cox proportional‐hazards regression analysis, visibility of deletion was a significant predictor of tumour‐free survival (p = 0.03) but sex was not (p = 0.13).
Figure 1 Kaplan–Meier plot comparing tumour‐free survival with age of presentation of Wilms tumour for visible (solid line) and submicroscopic (dashed line) WT1 deletions.
Discussion
In people with WAGR, the second WT1 hit can occur via a variety of mechanisms.19,20 Many of these, such as mitotic recombination and loss of the normal chromosome followed by duplication of the mutant homologue, result in homozygosity for the original deletion.19 A priori, it would be expected that people with large deletions are less likely to develop a tumour, because these deletions are more likely to encompass genes that are essential for cell survival. Such deletions would therefore be cell‐lethal when homozygous. In contrast, homozygosity for a submicroscopic deletion would confer a growth advantage on a kidney cell by removing both copies of WT1 while sparing essential genes, thus increasing the risk of tumour development. The same line of reasoning predicts that the risk of a second or a bilateral tumour will be reduced for large deletions and increased for small deletions, although there is no evidence to support this in the present study.
In an attempt to ascertain where cell‐essential gene(s) might lie, we plotted the end‐points of visible deletions where these had been characterised in detail (see supplementary material; available at http://jmg.bmj.com/supplemental). This analysis was hampered by the fact that detailed breakpoint data were only available for nine cases: four with and five without WT. In addition, it is not known whether the tumours, where present, were homozygous for the germline deletion (in which case they would be informative about the location of cell‐essential loci) or whether they had an independent second hit such as a WT1 point mutation (in which case they would not be informative). Therefore it is not possible at present to identify the location of genes essential for cell viability. Molecular analysis of tumours and detailed breakpoint mapping of many more visible deletion cases would be required to address this point.
The aniridia phenotype, which is recognisable at birth, draws immediate attention to the possibility of a WT1 deletion. This provides a unique opportunity to ascertain patients with a tumour suppressor gene deletion independent of tumour status, and allows correlation of deletion size with clinical outcome. In the case of retinoblastoma, which is caused by loss of function of both copies of the tumour suppressor gene RB1, numerous deletions have been described, but almost all are in patients who have already presented with a tumour. Therefore it is not possible to compare deletion sizes in large numbers of cases with and without tumours. However, if deletion size is compared with the number of tumour foci per individual, visible deletions are associated with fewer tumours than other mutation types, including cryptic deletions.21 Consistent with the idea that large deletions are less likely to become homozygous, one tumour‐free person in the study of Thienpont et al had a 25 Mb deletion encompassing RB1.22 For each tumour‐suppressor locus, the size of deletion that is associated with a decreased risk of tumour development will depend on the proximity of cell‐essential genes.
Patients with WAGR who develop WT are at increased risk of end‐stage renal disease (ESRD).7,10,23 The high frequency of ESRD may be a direct consequence of constitutional absence of one copy of WT1, which is expressed at multiple stages in the developing kidney.24 Indeed, mice with reduced Wt1 expression have a higher incidence of glomerulosclerosis.25 If ESRD is due solely to lack of one copy of WT1, people with deletions without tumour presentation will also be at risk and may need to be monitored for onset of aberrant kidney function. Long‐term follow‐up of tumour‐free deletion cases will help to clarify this issue.
Key points
To investigate the effect of 11p13 deletion size on the risk of developing Wilms tumour, we used the aniridia phenotype to ascertain people who might have deletions of the WT1 tumour suppressor gene.
Chromosomal deletion analysis, including high‐resolution FISH, was used to identify 53 patients with visible or submicroscopic WT1 deletions.
Individuals with submicroscopic WT1 deletions have a significantly higher risk of developing Wilms tumour than those with visible deletions.
All neonates with aniridia should be screened by FISH for deletion of WT1 and those with deletions need to be monitored regularly for tumour development; those with submicroscopic deletions require extra vigilance.
To assess the risk of developing WT, deletion analysis should be used as the primary screening method in the neonatal period in babies with aniridia, particularly de novo sporadic cases. In this study, FISH analysis was used, but deletion studies using dosage‐sensitive genomic PCR methods or array approaches may supersede this. If WT1 and its regulatory regions are not deleted, then a priori it would be expected that the risk of WT should be the same as in the general population. This is supported by empirical observations that patients with aniridia who do not have WT1 deletions do not have an increased risk of WT (Crolla and van Heyningen2, Gronskov et al6, this study).
If WT1 is deleted, the risk of WT is high regardless of deletion size, and regular routine abdominal examinations should be performed. In our study, 77% of individuals with submicroscopic deletions (detectable only by FISH) presented with WT, compared with 42.5% with visible deletions. Individuals with submicroscopic deletions are at particularly high risk of developing Wilms tumour and consequently high levels of vigilance should be maintained in these cases.
Supplementary material is available on the JMG website at http://jmg.bmj.com/supplemental
Acknowledgements
We thank Andrew Carothers and Paul Strike for statistics help, Matthias Drechsler for providing additional information about WT1 deletion cases, and Isabel Hanson for preparing the manuscript.
Abbreviations
ESRD - end‐stage renal disease
FISH - fluorescence in situ hybridisation
OMIM - Online Mendelian Inheritance in Man
WAGR - Wilms tumour, aniridia, genitourinary anomalies and mental retardation
WT - Wilms tumour
Footnotes
Competing interests: none declared.
Supplementary material is available on the JMG website at http://jmg.bmj.com/supplemental
References
- 1.Prosser J, van Heyningen V. PAX6 mutations reviewed. Hum Mutat 19981193–108. [DOI] [PubMed] [Google Scholar]
- 2.Crolla J A, van Heyningen V. Frequent chromosome aberrations revealed by molecular cytogenetic studies in patients with aniridia. Am J Hum Genet 2002711138–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson L B, Spaeth G L, Nowinski T S, Margo C E, Jackson L. Aniridia. A review. Surv Ophthalmol 198428621–642. [DOI] [PubMed] [Google Scholar]
- 4.Sisodiya S M, Free S L, Williamson K A, Mitchell T N, Willis C, Stevens J M, Kendall B E, Shorvon S D, Hanson I M, Moore A T, van Heyningen V. PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet 200128214–216. [DOI] [PubMed] [Google Scholar]
- 5.van Heyningen V, Williamson K A. PAX6 in sensory development. Hum Mol Genet 2002111161–1167. [DOI] [PubMed] [Google Scholar]
- 6.Gronskov K, Olsen J H, Sand A, Pedersen W, Carlsen N, Bak Jylling A M, Lyngbye T, Brondum‐Nielsen K, Rosenberg T. Population‐based risk estimates of Wilms tumor in sporadic aniridia. A comprehensive mutation screening procedure of PAX6 identifies 80% of mutations in aniridia. Hum Genet 200110911–18. [DOI] [PubMed] [Google Scholar]
- 7.Breslow N E, Norris R, Norkool P A, Kang T, Beckwith J B, Perlman E J, Ritchey M L, Green D M, Nichols K E. Characteristics and outcomes of children with the Wilms tumor‐Aniridia syndrome: a report from the National Wilms Tumor Study Group. J Clin Oncol 2003214579–4585. [DOI] [PubMed] [Google Scholar]
- 8.Dome J S, Coppes M J. Recent advances in Wilms tumor genetics. Curr Opin Pediatr 2002145–11. [DOI] [PubMed] [Google Scholar]
- 9.Royer‐Pokora B, Beier M, Henzler M, Alam R, Schumacher V, Weirich A, Huff V. Twenty‐four new cases of WT1 germline mutations and review of the literature: genotype/phenotype correlations for Wilms tumor development. Am J Med Genet A 2004127249–257. [DOI] [PubMed] [Google Scholar]
- 10.Breslow N E, Collins A J, Ritchey M L, Grigoriev Y A, Peterson S M, Green D M. End stage renal disease in patients with Wilms tumor: results from the National Wilms Tumor Study Group and the United States Renal Data System. J Urol 20051741972–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mannens M, Hoovers J M, Bleeker‐Wagemakers E M, Redeker E, Bliek J, Overbeeke‐Melkert M, Saunders G, Williams B, van Heyningen V, Junien C, Haber D, Speleman F, Heyting C, Slater R M, Leschott N J, Westerveld A. The distal region of 11p13 and associated genetic diseases. Genomics 199111284–293. [DOI] [PubMed] [Google Scholar]
- 12.Fantes J A, Bickmore W A, Fletcher J M, Ballesta F, Hanson I M, van Heyningen V. Submicroscopic deletions at the WAGR locus, revealed by nonradioactive in situ hybridization. Am J Hum Genet 1992511286–1294. [PMC free article] [PubMed] [Google Scholar]
- 13.Crolla J A, Cawdery J E, Oley C A, Young I D, Gray J, Fantes J, van Heyningen V. A FISH approach to defining the extent and possible clinical significance of deletions at the WAGR locus. J Med Genet 199734207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fantes J A, Oghene K, Boyle S, Danes S, Fletcher J M, Bruford E A, Williamson K, Seawright A, Schedl A, Hanson I, Zehetner G, Bhogal R, Lehrach H, Gregory S, Williams J, Little P F R, Sellar G C, Hoovers J, Mannens M, Weissenbach J, Junien C, van Heyningen V. A high‐resolution integrated physical, cytogenetic, and genetic map of human chromosome 11: distal p13 to proximal p15.1. Genomics 199525447–461. [DOI] [PubMed] [Google Scholar]
- 15.van Heyningen V, Little P F. Report of the fourth international workshop on human chromosome 11 mapping 1994. Cytogenet Cell Genet 199569127–158. [DOI] [PubMed] [Google Scholar]
- 16.Henry I, Hoovers J, Barichard F, Bertheas M F, Puech A, Prieur F, Gessler M, Bruns G, Mannens M, Junien C. Pericentric intrachromosomal insertion responsible for recurrence of del(11)(p13p14) in a family. Genes Chromosomes Cancer 1993757–62. [DOI] [PubMed] [Google Scholar]
- 17.Kent J, Lee M, Schedl A, Boyle S, Fantes J, Powell M, Rushmere N, Abbott C, van Heyningen V, Bickmore W A. The reticulocalbin gene maps to the WAGR region in human and to the Small eye Harwell deletion in mouse. Genomics 199742260–267. [DOI] [PubMed] [Google Scholar]
- 18.Drechsler M, Meijers‐Heijboer E J, Schneider S, Schurich B, Grond‐Ginsbach C, Tariverdian G, Kantner G, Blankenagel A, Kaps D, Schroeder‐Kurth T, Royer‐Pokora B. Molecular analysis of aniridia patients for deletions involving the Wilms' tumor gene. Hum Genet 199494331–338. [DOI] [PubMed] [Google Scholar]
- 19.Dao D D, Schroeder W T, Chao L Y, Kikuchi H, Strong L C, Riccardi V M, Pathak S, Nichols W W, Lewis W H, Saunders G F. Genetic mechanisms of tumor‐specific loss of 11p DNA sequences in Wilms tumor. Am J Hum Genet 198741202–217. [PMC free article] [PubMed] [Google Scholar]
- 20.Gessler M, Konig A, Moore J, Qualman S, Arden K, Cavenee W, Bruns G. Homozygous inactivation of WT1 in a Wilms' tumor associated with the WAGR syndrome. Genes Chromosomes Cancer 19937131–136. [DOI] [PubMed] [Google Scholar]
- 21.Albrecht P, Ansperger‐Rescher B, Schuler A, Zeschnigk M, Gallie B, Lohmann D R. Spectrum of gross deletions and insertions in the RB1 gene in patients with retinoblastoma and association with phenotypic expression. Hum Mutat 200526437–445. [DOI] [PubMed] [Google Scholar]
- 22.Thienpont B, Vermeesch J R, Fryns J P. 25 Mb deletion of 13q13.3→q21.31 in a patient without retinoblastoma. Eur J Med Genet 200548363–366. [DOI] [PubMed] [Google Scholar]
- 23.Niaudet P, Gubler M C. WT1 and glomerular diseases. Pediatr Nephrol 2006211653–1660. [DOI] [PubMed] [Google Scholar]
- 24.Moore A W, McInnes L, Kreidberg J, Hastie N D, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 19991261845–1857. [DOI] [PubMed] [Google Scholar]
- 25.Guo J K, Menke A L, Gubler M C, Clarke A R, Harrison D, Hammes A, Hastie N D, Schedl A. WT1 is a key regulator of podocyte function: reduced expression levels cause crescentic glomerulonephritis and mesangial sclerosis. Hum Mol Genet 200211651–659. [DOI] [PubMed] [Google Scholar]