Abstract
We report the first genome-wide association study (GWAS) whose sample size (1,053 Swedish subjects) is sufficiently powered to detect genome-wide significance (p<1.5×10−7) for polymorphisms that modestly alter therapeutic warfarin dose. The anticoagulant drug warfarin is widely prescribed for reducing the risk of stroke, thrombosis, pulmonary embolism, and coronary malfunction. However, Caucasians vary widely (20-fold) in the dose needed for therapeutic anticoagulation, and hence prescribed doses may be too low (risking serious illness) or too high (risking severe bleeding). Prior work established that ∼30% of the dose variance is explained by single nucleotide polymorphisms (SNPs) in the warfarin drug target VKORC1 and another ∼12% by two non-synonymous SNPs (*2, *3) in the cytochrome P450 warfarin-metabolizing gene CYP2C9. We initially tested each of 325,997 GWAS SNPs for association with warfarin dose by univariate regression and found the strongest statistical signals (p<10−78) at SNPs clustering near VKORC1 and the second lowest p-values (p<10−31) emanating from CYP2C9. No other SNPs approached genome-wide significance. To enhance detection of weaker effects, we conducted multiple regression adjusting for known influences on warfarin dose (VKORC1, CYP2C9, age, gender) and identified a single SNP (rs2108622) with genome-wide significance (p = 8.3×10−10) that alters protein coding of the CYP4F2 gene. We confirmed this result in 588 additional Swedish patients (p<0.0029) and, during our investigation, a second group provided independent confirmation from a scan of warfarin-metabolizing genes. We also thoroughly investigated copy number variations, haplotypes, and imputed SNPs, but found no additional highly significant warfarin associations. We present power analysis of our GWAS that is generalizable to other studies, and conclude we had 80% power to detect genome-wide significance for common causative variants or markers explaining at least 1.5% of dose variance. These GWAS results provide further impetus for conducting large-scale trials assessing patient benefit from genotype-based forecasting of warfarin dose.
Author Summary
Recently, geneticists have begun assaying hundreds of thousands of genetic markers covering the entire human genome to systematically search for and identify genes that cause disease. We have extended this “genome-wide association study” (GWAS) method by assaying ∼326,000 markers in 1,053 Swedish patients in order to identify genes that alter response to the anticoagulant drug warfarin. Warfarin is widely prescribed to reduce blood clotting in order to protect high-risk patients from stroke, thrombosis, and heart attack. But patients vary widely (20-fold) in the warfarin dose needed for proper blood thinning, which means that initial doses in some patients are too high (risking severe bleeding) or too low (risking serious illness). Our GWAS detected two genes (VKORC1, CYP2C9) already known to cause ∼40% of the variability in warfarin dose and discovered a new gene (CYP4F2) contributing 1%–2% of the variability. Since our GWAS searched the entire genome, additional genes having a major influence on warfarin dose might not exist or be found in the near-term. Hence, clinical trials assessing patient benefit from individualized dose forecasting based on a patient's genetic makeup at VKORC1, CYP2C9 and possibly CYP4F2 could provide state-of-the-art clinical benchmarks for warfarin use during the foreseeable future.
Introduction
Warfarin is the most widely prescribed anticoagulant for reducing thromboembolic events that often give rise to stroke, deep vein thrombosis, pulmonary embolism or serious coronary malfunctions [1]. A combination of genetic and non-genetic factors cause Caucasians to exhibit 20-fold interindividual variation in required warfarin dose needed to achieve the usual therapeutic level of anticoagulation as measured by the prothrombin international normalized ratio or INR [2]–[4]. Thus, in the absence of information (genotypic, clinical, etc.) for predicting each patient's required warfarin dose, initial prescribed doses may be too low (risking thrombosis) or too high (risking over-anticoagulation and severe bleeding). Warfarin's risk of serious side effects, narrow therapeutic range, and wide interindividual variation in warfarin dose have focused attention on the need to better predict dose in the initial stage(s) of treatment.
We and others have shown that the warfarin drug target VKORC1 (vitamin K epoxide reductase complex, subunit 1) contains common polymorphisms that account for a major portion (∼30%) of the variance in required warfarin dose [5],[6], and we have recently evaluated ∼1500 Swedish patients of the Warfarin Genetics (WARG) cohort in the largest study to date showing likely patient benefit from genetic forecasting of dose [3]. The study confirmed that SNPs in VKORC1 and in the warfarin-metabolizing gene CYP2C9 (cytochrome P450, family 2, subfamily C, polypeptide 9) predict ∼40% of dose variance while non-genetic factors (age, sex, etc.) jointly account for another ∼15%. The robust and now widely replicated associations of warfarin dose with VKORC1 and CYP2C9 have provided one of the most successful applications of pharmacogenetics to date [7] and offer promise for genetic predication of required dose in a clinical setting [3].
Knowledge of major predictors of warfarin dose also impacts the methodology for finding further dose-related genes. In early candidate gene work with a small sample of 201 patients [8], we noted that univariate regression (with tested SNP as the only dose predictor) could statistically detect warfarin association with VKORC1 and with one of two non-synonymous CYP2C9 SNPs (*3) known to influence warfarin dose (Table 1 in [8]). However, a second non-synonymous CYP2C9 SNP (*2) with known but weaker influence on warfarin dose was not detected by univariate regression, but *2 was statistically significant in multivariate regression adjusted for the other known genetic and non-genetic predictors of dose (Table 3 in [8]). These empirical results in a small warfarin sample provided a signpost underscoring the potential importance of multivariate regression for detecting weak effects in studies now searching for additional warfarin genes across the genome.
Table 1. Association (p-value) of SNPs tested by univariate regression or multiple regression with progressive addition of known dose predictorsa.
| Predictors in regression analysis | Tested SNP | ||||
| VKORC1 | CYP2C9*3 | CYP2C9*2 | CYP4F2 | Distribution of all SNPs | |
| rs9923231 | rs1057910 | rs1799853 | rs2108622 | ||
| None | 5.4E-78 | 4.5E-17 | 8.8E-13 | 1.6E-05 | Figure 1A |
| Age, sex | 7.3E-97 | 1.2E-24 | 2.4E-14 | 4.8E-06 | – |
| Age, sex, VKORC1 | – | 3.8E-43 | 1.0E-15 | 4.6E-07 | – |
| Age, sex, VKORC1, CYP2C9*3 | – | – | 1.4E-26 | 8.3E-08 | – |
| Age, Sex, VKORC1, CYP2C9*3 and *2 | – | – | – | 8.3E-10 | Figure 1B |
Linear regression on warfarin dose was calculated for the 1,053 GWAS subjects.
Table 3. Power to detect a dose-altering SNP as a function of its contribution to dose variance (R 2) and adjustment by other predictors in the multiple regression modela.
| Predictors adjusted in multiple regression | Tested SNP | ||||||
| VKORC1 | CYP2C9*3 | CYP2C9*2 | CYP4F2 | Unknown | Unknown | ||
| description | total | rs9923231 | rs1057910 | rs1799853 | rs2108622 | SNP of | SNP of |
| R 2 | (R 2 = 0.283) | (R 2 = 0.080) | (R 2 = 0.038) | (R 2 = 0.011) | R 2 = 0.015 | R 2 = 0.010 | |
| None | 0.000 | 1.00 | 1.00 | 0.88 | 0.03 | 0.10 | 0.02 |
| Age, sex | 0.168 | 1.00 | 1.00 | 0.96 | 0.06 | 0.19 | 0.04 |
| Age, sex, VKORC1 | 0.452 | – | 1.00 | 1.00 | 0.26 | 0.56 | 0.19 |
| Age, sex, VKORC1, CYP2C9*3 | 0.532 | – | – | 1.00 | 0.40 | 0.73 | 0.31 |
| Age, Sex, VKORC1, CYP2C9*3 and *2 | 0.570 | – | – | – | 0.48 | 0.81 | 0.39 |
| Age, Sex, VKORC1, CYP2C9*3 and *2, CYP4F2 | 0.580 | – | – | – | – | 0.82 | 0.41 |
Power calculations assumed a sample size of 1,053 subjects and significance level of 1.5E-7 as employed in our GWAS.
A genome-wide association study (GWAS) enables a systematic search of the entire genome for genetic factors that cause any inherited trait. This method has successfully identified susceptibility loci for common diseases [9], and is beginning to be applied to pharmacogenomics. A recent warfarin GWAS in 181 patients did not detect other genetic factors with major effects on warfarin dose beyond VKORC1 [10] but was underpowered for identifying loci with a moderate contribution. We have now genotyped 325,997 SNPs in 1053 patients of the WARG cohort and here report the first GWAS that is sufficiently powered to detect additional genetic factors that may only modestly influence warfarin dose.
Results
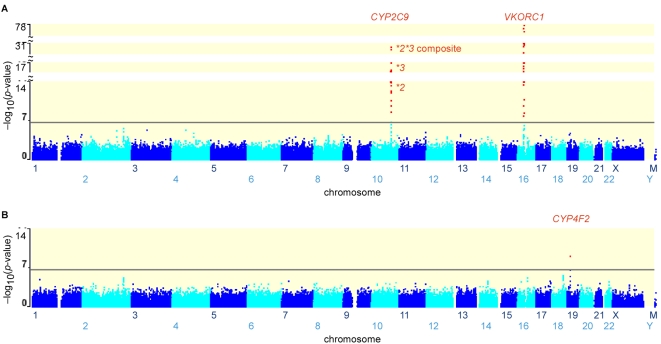
Figure 1A and the first line of Table 1 summarize results of testing 325,997 GWAS SNPs for association with warfarin dose by univariate regression. The strongest associations were at multiple SNPs in and near VKORC1 (Figure 1A) with the lowest p-value given by rs9923231 (P = 5.4×10−78). In prior fine-mapping of the VKORC1 locus [8], we identified rs9923231 as one of three SNPs located in introns or immediately flanking VKORC1 that exhibit almost perfectly concordant genotypes yielding pairwise linkage disequilibrium (LD) r 2≈1 and which define the warfarin-sensitive A-T-T haplotype at rs9923231-rs9934438-rs2359612 (see also [11]). These highly concordant SNPs were the best predictors of warfarin dose in our previous study and in this GWAS analysis (p<5.4×10−78) and completely accounted for the dose variance explained by all other fine-mapping SNPs near VKORC1 [8]. The group of SNPs with the second lowest univariate p-values clustered around CYP2C9 which contains two non-synonymous exonic SNPs whose minor alleles (*2, *3) impair warfarin metabolism and are well known to be associated with warfarin dose. In our previous work [8], we discovered an unusual SNP (rs4917639) whose minor allele is almost perfectly associated with the “composite” CYP2C9 allele formed by combining *2 and *3 into a single allele. Indeed, the GWAS results (1053 subjects) confirmed that LD is nearly perfect (pairwise r 2≈1.0) between rs4917639 and the composite of *2 and *3. Thus, the highly significant univariate result for rs4917639 (R 2 = 0.121, p<3.1×10−31) reflects the combined effect of CYP2C9*2 rs1799853 (R 2 = 0.038, p<8.8×10−13) and CYP2C9*3 rs1057910 (R 2 = 0.080, p<4.5×10−17). Figure 1A therefore indicates p-values for this composite SNP as well as for *2 and *3.
Figure 1. P-values for each GWAS SNP tested for association with warfarin dose.
Horizontal axis shows SNP location and vertical axis is −log10(p-value) for each SNP tested by univariate regression (A) or multivariate regression (B). Red dots and red lettering show SNPs and implicated genes with p-values beyond the genome-wide significance threshold (1.5×10−7) which is denoted by a horizontal line. (A) Univariate regression shows genome-wide significant association to SNPs clustering near the warfarin drug target VKORC1 (e.g., P = 5.4×10−78, rs9923231) and near the warfarin-metabolizing gene CYP2C9 (P = 4.5×10−17 for non-synonymous *3 SNP rs1057910, P = 8.8×10−13 for non-synonymous *2 SNP rs1799853, P = 3.1×10−31 for *2*3 “composite” SNP rs4917639). (B) Multivariate regression adjusting for the contributions of VKORC1 and CYP2C9 had greater power than univariate regression and detected genome-wide significant association to the CYP4F2 gene (P = 8.3×10−10, non-synonymous SNP rs2108622).
Figure 1B and Table 1 (lines 2 to 5) show the results of multivariate regression analysis in which individual SNPs were tested for association with warfarin dose after adjustment for established genetic and non-genetic predictors of dose. The only SNP reaching genome-wide significance (p<1.5×10−7) was a non-synonymous SNP (rs2108622) in exon 2 of CYP4F2 (cytochrome P450, family 4, subfamily F, polypeptide 2) introducing a Val to Met amino acid change at position 433 (V433M). SNP rs2108622 predicts additional dose variance (∼1.1%) that is independent of the variance already explained by VKORC1 and CYP2C9. As noted in the Introduction, our early studies with a small sample of 201 Swedish patients failed to detect the weak CYP2C9*2 effect on dose by univariate regression but *2 was significant in multiple regression [8]. The results in Table 1 with rs2108622 of CYP4F2 show the same phenomenon with a p-value of 1.6×10−5 in univariate regression (line 1) but progressively lower p-values as known predictors are added to the multivariate model so that for the full model a p-value of 8.3×10−10 is achieved which is far below genome-wide significance (p<1.5×10−7). The CYP4F2 association was further confirmed by testing an independent replication panel of 588 Swedish warfarin patients who gave a multivariate p-value of 0.0029 and a total overall p-value of 3.3×10−10 when combined with the GWAS subjects (Table 2). During preparation of this paper, a candidate gene study of drug-metabolizing and transporter genes independently discovered the association of rs2108622 and CYP4F2 with warfarin dose, providing further confirmation [12].
Table 2. Multiple regression analysis of warfarin dose in the GWAS, replication and combined panels.
| Predictor | WARG GWAS (1053) | Replication (588) | Combined (1641) | ||||||
| Effect on doseb | R 2 | P-value | Effect on doseb | R 2 | P-value | Effect on doseb | R 2 | P-value | |
| Estimate (95% CI) | Estimate (95% CI) | Estimate (95% CI) | |||||||
| VKORC1 rs9923231 (C->T, 0.402)a | −0.96 (−1.03, −0.89) | 0.283 | 1.6E-122 | −0.99 (−1.09, −0.88) | 0.284 | 5.0E-62 | −0.97 (−1.02, −0.91) | 0.283 | 2.7E-181 |
| CYP2C9*3 rs1057910 (Ile359Leu, 0.070)a | −1.13 (−1.26, −1.00) | 0.075 | 2.6E-55 | −1.08 (−1.27, −0.89) | 0.089 | 2.3E-26 | −1.11 (−1.22, −1.00) | 0.080 | 2.6E-79 |
| CYP2C9*2 rs1799853 (Arg144Cys, 0.109)a | −0.63 (−0.74, −0.52) | 0.048 | 1.7E-28 | −0.40 (−0.55, −0.24) | 0.023 | 5.5E-07 | −0.54 (−0.63, −0.45) | 0.038 | 1.1E-31 |
| CYP4F2 rs2108622 (Val433Met, 0.240)a | 0.25 (0.17, 0.33) | 0.016 | 8.3E-10 | 0.16 (0.05, 0.27) | 0.005 | c0.0029 | 0.21 (0.14, 0.27) | 0.011 | 3.3E-10 |
| Age | −0.04 (−0.04, −0.03) | 0.170 | 1.9E-63 | −0.03 (−0.04, −0.03) | 0.129 | 1.7E-31 | −0.04 (−0.04, −0.03) | 0.155 | 1.2E-111 |
| Sex (male) | 0.35 (0.25, 0.45) | 0.017 | 7.6E-12 | 0.25 (0.10, 0.40) | 0.009 | 0.001 | 0.30 (0.22, 0.38) | 0.013 | 1.6E-12 |
In parenthesis are major/minor allele, and minor allele frequency.
Effect of individual predictor on dose is indicated by regression coefficient and 95% confidence interval, proportion of explained variance (R 2) and P-value.
Association in same direction as GWAS was assessed by a one-tailed test.
To increase the power of our multivariate regression model and possibly detect additional weak effects, we added CYP4F2 (rs2108622) to the model as a predictor and conducted further analyses. First, we retested the GWAS SNPs, but no new SNPs reached genome-wide significance and there was also no apparent excess of SNPs at lower significance thresholds (Figure S1). We also tested warfarin association with haplotypes and with ungenotyped SNPs imputed at 2.2 million HapMap SNPs, but no haplotype or imputed SNP approached genome-wide significance in a genomic region not containing VKORC1, CYP2C9 or CYP4F2. To explore whether copy number variations (CNVs) detectable by the HumanCNV370 array might influence warfarin dose, we used rigorous quality control and retained 879 samples calling 2530 CNVs (see Materials and Methods). None of the CNV loci were significantly associated with dose after correction for multiple testing (lowest CNV p-value was 1.1×10−4 which exceeds 0.05/2530≈2.0×10−5). We note that probe density in many of the detected CNVs is not optimal for conducting association analyses and these results should therefore be viewed as preliminary.
Finally, after excluding SNPs near VKORC1, CYP2C9 and CYP4F2, we identified 40 other loci containing one or more GWAS SNPs with p-values below 2.0×10−4 and we genotyped 40 SNPs representing these loci in a follow-up sample of 588 Swedish warfarin patients. However none of the 40 loci replicated for association with warfarin dose, the lowest p-value being 0.04 which is not significant after correction for 40 tests (Table S1). Having not found evidence for any additional genetic modulators of dose, we examined the entire data set (GWAS plus followup samples) for evidence of statistical interaction between pairs of the established dose predictors (VKORC1, CYP2C9, CYP4F2, age, sex). None of the pairs exhibited statistically significant interaction after p-values were corrected for the 15 interaction tests (Table S2).
We also performed a GWAS for a secondary trait (“over-anticoagulation”) which we previously found was associated with VKORC1 and CYP2C9 in a candidate gene study [3]. By titrating warfarin dose, physicians attempt to achieve a target level of anticoagulation determined by a reading of 2.0 to 3.0 for the prothrombin international normalized ratio (INR), which is the ratio of time required for a patient's blood to coagulate relative to that of a reference sample. However over-anticoagulation (defined as an INR above 4.0) sometimes occurs and, using Cox regression, our GWAS tested for SNP association with the occurrence of over-anticoagulation in patients during the first 5 weeks of treatment (see Materials and Methods: Association testing of SNPs and haplotypes). We observed genome-wide significant association (p<1.5×10−7) at several SNPs in and around VKORC1 including rs9923231 (P = 8.9×10−9), but no other SNPs achieved genome-wide significance including CYP2C9*3 (p<4.0×10−5), CYP2C9*2 (p = 0.93), or the “composite” *2*3 SNP rs4917639 (p<0.007) (Figure S2). However we note that our previous candidate gene study evaluated a larger sample set (1496 WARG subjects) which yielded genome-wide significant association with over-anticoagulation for both VKORC1 rs9923231 (P = 5.7×10−11) and CYP2C9*3 (P = 1.5×10−9) [3]. To explore whether these SNPs might cause over-anticoagulation independent of altering the required (i.e., administered) warfarin dose, we added required dose to the Cox regression model as a predictor of over-anticoagulation, and found that both VKORC1 and CYP2C9*3 have a significant effect independent of dose (P<0.05) (Table S3).
Discussion
We conducted the first GWAS sufficiently powered to detect DNA variants with a modest influence on the warfarin dose needed to achieve therapeutic anticoagulation. In univariate analysis of GWAS SNPs (Figure 1A), we identified extremely strong association signals (p = 10−78 to 10−13) at SNPs in and near VKORC1 and CYP2C9, two genes already known to explain ∼30% and ∼12% of warfarin dose variance, respectively. By applying multivariate regression adjusting for known genetic and non-genetic predictors of dose (Figure 1B), we also detected genome-wide significance of p<8.3×10−10 at CYP4F2 (rs2108622) that accounted for approximately 1.5% of dose variance. The increased power of multivariate regression to detect this modest effect is nicely illustrated in Table 1 which shows a higher univariate p-value for CYP4F2 (p<1.6×10−5) but progressively lower multivariate p-values as known predictors of dose are added to the regression model. We confirmed the CYP4F2 association in a second large sample set and the association was also reported by another group [12] during preparation of our work, thus fully establishing the genuine effect of CYP4F2 (see also [10] where CYP4F2 explained ∼1% dose variance with nominal p<0.043 significance). Although multivariate regression has not been widely used to increase power in other GWAS analyses because known genetic variants usually explain little phenotypic variance, the potential for power increase is perhaps obvious if known predictors do explain substantial variance. Thus multiple regression has, for example, been previously advocated for linkage analyses of line crosses [13],[14].
To estimate the multivariate regression power of our GWAS (1053 subjects), we used Equation 1 (see Materials and Methods) to calculate power to detect SNPs explaining specific magnitudes of variance ( ) for warfarin dose (see Table 3). The table shows that power to achieve genome-wide significance (p<1.5×10−7) is essentially 100% for VKORC1 rs9923231 (
) for warfarin dose (see Table 3). The table shows that power to achieve genome-wide significance (p<1.5×10−7) is essentially 100% for VKORC1 rs9923231 ( ), CYP2C9*3 (
), CYP2C9*3 ( ) and CYP2C9*2 (
) and CYP2C9*2 ( ), but power falls to ∼48% for CYP4F2 rs2108622 (
), but power falls to ∼48% for CYP4F2 rs2108622 ( ). The table also shows that when CYP4F2 is added to the multivariate model, a SNP accounting for 1.5% or 1.0% of the dose variance would have ∼82% or ∼41% power of being detected, respectively. Therefore we estimate that our GWAS had at least 80% power to detect warfarin-associated variants explaining at least 1.5% of the dose variance but 40% or less power to detect genome-wide significance if a variant accounts for 1% or less dose variance.
). The table also shows that when CYP4F2 is added to the multivariate model, a SNP accounting for 1.5% or 1.0% of the dose variance would have ∼82% or ∼41% power of being detected, respectively. Therefore we estimate that our GWAS had at least 80% power to detect warfarin-associated variants explaining at least 1.5% of the dose variance but 40% or less power to detect genome-wide significance if a variant accounts for 1% or less dose variance.
However it is important to emphasize that these power estimates assume that the dose-altering DNA variant is genotyped and tested directly or is indirectly detected through a marker in sufficiently high LD to the dose variant that the marker's  magnitude is detectable (Table 3). The assumption of directly testing the dose-altering variant is accurate for CYP2C9*2 and *3 which are each known to alter warfarin metabolism [15],[16] and is likely correct for CYP4F2 rs2108622 which, like *2 and *3, changes protein coding sequence. However, to explore whether other dose-altering variants might be undetected due to insufficient LD with genotyped GWAS SNPs, we determined the relationship between the variance observed at a marker (
magnitude is detectable (Table 3). The assumption of directly testing the dose-altering variant is accurate for CYP2C9*2 and *3 which are each known to alter warfarin metabolism [15],[16] and is likely correct for CYP4F2 rs2108622 which, like *2 and *3, changes protein coding sequence. However, to explore whether other dose-altering variants might be undetected due to insufficient LD with genotyped GWAS SNPs, we determined the relationship between the variance observed at a marker ( ) and at the causative variant (
) and at the causative variant ( ) assuming pairwise LD of r
2 between the two polymorphisms (see Materials and Methods: How Much Does Linkage Disequilibrium Attenuate Association with a Quantitative Trait?). The relationship is given by Equation 3 in Materials and Methods (
) assuming pairwise LD of r
2 between the two polymorphisms (see Materials and Methods: How Much Does Linkage Disequilibrium Attenuate Association with a Quantitative Trait?). The relationship is given by Equation 3 in Materials and Methods ( ) which is analogous to Pritchard and Prezworski's relationship (
) which is analogous to Pritchard and Prezworski's relationship ( ) for the number of cases (
) for the number of cases ( ) providing equal power in a case-control study that tests either the disease-causing SNP or a nearby marker [17]. To use the equation
) providing equal power in a case-control study that tests either the disease-causing SNP or a nearby marker [17]. To use the equation  to estimate
to estimate  magnitudes for variants that might be undetected by our GWAS, we note that ∼90% of the GWAS SNPs had a minor allele frequency (MAF) above 10% in our warfarin subjects implying that a “rare” dose-altering variant (MAF≈1%–5%) would be covered at a likely maximum r
2 of only ∼0.1 to ∼0.5. This low r
2 coverage implies that rare variants could have
magnitudes for variants that might be undetected by our GWAS, we note that ∼90% of the GWAS SNPs had a minor allele frequency (MAF) above 10% in our warfarin subjects implying that a “rare” dose-altering variant (MAF≈1%–5%) would be covered at a likely maximum r
2 of only ∼0.1 to ∼0.5. This low r
2 coverage implies that rare variants could have  values (0.05 to 0.02) easily detected by regression testing of the variant itself, but unlikely to be detected through a GWAS marker since maximum
values (0.05 to 0.02) easily detected by regression testing of the variant itself, but unlikely to be detected through a GWAS marker since maximum  values could drop to 0.01 or much lower (see Equation 3 and Table 3). By contrast, “common” SNPs (MAF≥5%), which might also be dose variants, are covered by GWAS SNPs of this study at reasonably high r
2 values in most instances (r
2>0.8 or r
2>0.5 for ∼60% or ∼80% respectively of common SNPs [18] and r
2>0.9 for ∼90% of non-synonymous common SNPs [19] in HapMap Caucasians). We therefore conclude that our GWAS probably detected most common SNP variants explaining 1.5% or more of the warfarin dose variance, but may have failed to detect rarer variants that could individually explain up to 5% of dose variance. We further note that the HumanCNV370 array used in this study does not have the required marker complement to undertake a comprehensive GWAS of common CNVs.
values could drop to 0.01 or much lower (see Equation 3 and Table 3). By contrast, “common” SNPs (MAF≥5%), which might also be dose variants, are covered by GWAS SNPs of this study at reasonably high r
2 values in most instances (r
2>0.8 or r
2>0.5 for ∼60% or ∼80% respectively of common SNPs [18] and r
2>0.9 for ∼90% of non-synonymous common SNPs [19] in HapMap Caucasians). We therefore conclude that our GWAS probably detected most common SNP variants explaining 1.5% or more of the warfarin dose variance, but may have failed to detect rarer variants that could individually explain up to 5% of dose variance. We further note that the HumanCNV370 array used in this study does not have the required marker complement to undertake a comprehensive GWAS of common CNVs.
As noted in the Introduction, the widely replicated warfarin dose associations with VKORC1 and CYP2C9 represent one of the most successful applications of pharmacogenetics to date. Our study together with that of Caldwell et al. [12] now also clearly demonstrates that CYP4F2 (rs2108622) is a third gene that influences warfarin dose, but our GWAS and statistical analysis also implies that additional common SNP variants that influence dose may not exist in Caucasian populations. However, Caucasians might carry common variants with effects smaller than CYP4F2 or rare variants whose effects are substantially larger than the ∼1% of dose variance explained by CYP4F2. Furthermore, other unidentified genes may influence warfarin dose in other ethnicities such as Asians or Africans, and some rare dose-altering variants in known genes such as VKORC1 may exist in only a subset of populations of European descent [20]. Hence, future research could address ethnic differences in the genetic variants that influence warfarin dose as well as subtle intra-ethnic differences and admixture that may exist in European or other populations.
In a recent study [3], we highlighted the potential benefit of pre-treatment forecasting of required warfarin dose based on patient genotypes at VKORC1 and CYP2C9 together with non-genetic predictors of dose. Indeed, in August 2007, the US Food and Drug Administration (FDA) updated warfarin labeling to recommend initiating lower warfarin dose in some patients based on VKORC1 and CYP2C9 genotypes. However this recommendation is not a requirement due to a lack of large trials demonstrating warfarin patient benefit from dose forecasting (though two small trials [21],[22] do support such benefit; see also [23]–[27] for reviews and other trials). The results of our GWAS provide further impetus for conducting large-scale dose-forecasting trials by identifying CYP4F2 as a third genetic predictor of dose and also by showing that additional major genetic predictors may not exist in Caucasians or may not emerge in the near-term. Hence, large-scale trials of patient benefit from dose forecasting based on VKORC1 and CYP2C9 (with possible inclusion of CYP4F2 as a minor predictor) are likely to provide state-of-the-art clinical benchmarks for warfarin use during the foreseeable future.
Materials and Methods
Subjects and Clinical Data
The study subjects were 1053 Swedish patients collected for the WARG study [3] (http://www.druggene.org/). This is a multi-centre study of warfarin bleeding complications and response to warfarin treatment [28]. Anticoagulant response is measured by INR, which is the ratio of the time required for a patient's blood to coagulate relative to that of a reference sample. By titrating warfarin dose, physicians aim for a therapeutic INR reading between 2.0 and 3.0; thus the primary quantitative outcome for the GWAS was the mean warfarin dose (mg/week) given to a patient during a minimum series of three consecutive INR measurements between 2 and 3 [3]. As a secondary GWAS outcome, we also catalogued each patient for the occurrence or non-occurrence of “over-anticoagulation” during the first 5 weeks of treatment (defined as an INR reading above 4.0) and tested for genetic association which adjusted for the treatment day (1 to 35) of the over-anticoagulation event (see “Association testing” below). The clinical data collected by the WARG protocol included gender and age since each is a known non-genetic predictor of warfarin dose but did not include bodyweight and dietary information (e.g. vitamin K intake). Regression analysis of prescribed medication which can potentiate or inhibit warfarin action was not a statistically significant predictor of warfarin dose in the 1053 WARG GWAS subjects and hence was not included as a predictor variable in the multivariate regression analyses. The WARG study samples were previously described elsewhere [3],[4],[28],[29] as were the Uppsala followup samples [8]. The WARG and Uppsala studies received ethical approval from the Ethics Committee of the Karolinska Institute and the Research Ethics Committee at Uppsala University, respectively.
Genotyping of SNPs and Sample Quality Control
From approximately 1500 WARG samples [3] examined for non-degradation and appropriate concentration of DNA (∼50 ng/µl), we randomly selected 1208 subjects for genotyping SNPs and CNV probes using the HumanCNV370 BeadChip array (Illumina). We excluded SNPs with MAF below 1%, call rate below 95%, or if call rate fell below 99% when MAF was below 5%. SNPs that departed from Hardy-Weinberg equilibrium (P<10−6) were also excluded. Subjects with genotyping call rate below 95% were also eliminated. Using iPLEX (Sequenom), subject identity (and associated phenotypic data) was cross-checked by genotyping four gender markers and 47 SNPs also carried on the HumanCNV370 array, enabling us to exclude ∼136 misidentified subjects. Sample quality (contamination) was further assessed by plotting each subject's genome-wide heterozygosity and eliminating outliers (with heterozygosity above or below the range of 0.312–0.372). After these quality control steps, a total of 1053 warfarin patients and 325,997 GWAS SNPs were retained for analysis. The GWAS SNPs included two SNPs not on the HumanCNV370 array but which are highly predictive of warfarin dose [rs9923231 (VKORC1) and rs1799853 (CYP2C9*2)] which we genotyped by TaqMan assay (Applied Biosystems).
Defining CNV Regions
Although we retained 325,997 GWAS SNPs for association testing of SNPs, it should be noted that all ∼370,000 probes on the Human CNV370 array were used to define CNVs. Log R ratio values of probes were output from the BeadStudio software [30]. A loess correction was applied to each sample to remove local correlations or genomic wave [31]. The resultant genomic copy number profiles were then segmented using Circular Binary Segmentation [32]. Some samples displayed abnormally high numbers of segments indicating problems in DNA quantity or quality or hybridization. Samples were removed until the number of segments across all samples was approximately normal. Using this technique, 143 (14%) of samples were flagged as problematic. These samples were excluded when CNV regions were defined but included for association testing. Putative CNV were defined from segments by applying a threshold on the segment log R ratio. This threshold was asymmetric allowing for a differing response for deletions and duplications. The central peak of the segment log R ratio distribution was fitted and the threshold values obtained by taking values at ±5 standard deviations from the centre.
In order to define regions for association testing, merging of CNV across samples was performed. This was achieved by merging two putative CNV into a region if there was greater than 40% reciprocal overlap. This procedure defined 2530 CNV regions in total. Of these, most were singletons (54%) or low frequency, <3% (93%), while 820 (70%) of the non-singleton regions overlapped CNVs from the Database of Genomic Variants [33]. We tested all 2530 CNVs for association, because a CNV discovered as a “singleton” might well include multiple copies of a rare CNV allele in the study samples.
Association Testing of SNPs and Haplotypes
At each SNP, genotypes were coded 0, 1 or 2 and the SNP was tested for association with the square root of warfarin dose [8] by either univariate or multivariate linear regression analysis conducted in PLINK [34] (http://pngu.mgh.harvard.edu/~purcell/plink/) or in R software (http://www.r-project.org/). We used the same regression analysis to test association with all HapMap SNPs not on the HumanCNV370 array by imputing ∼2.2 million SNPs using Beagle software [35] trained from genotypes of the 60 HapMap CEU parents [36]. We excluded SNPs whose imputed MAF was below 5% or differed by more than 5% with MAF of the CEU parents.
We also tested haplotypes for association with warfarin dose by two approaches: (1) each subject's warfarin dose residual (difference between observed and predicted dose based on the full multivariate regression model containing CYP4F2) was considered a quantitative trait value and tested for association with haplotypes defined across the genome in sliding windows of 2, 3 or 4 consecutive SNPs as implemented by PLINK software; (2) by scanning GWAS genotypes, Beagle software groups genetically related haplotypes into clusters which it then resolves into diallelic (SNP-like) “pseudo-markers” optimized for detecting phenotypic association. To test haplotypes, we evaluated the pseudo-marker genotypes of warfarin patients at 1.97 million pseudo-markers covering the genome by testing each pseudo-marker in the same multivariate regression framework used to test individual SNPs (as described in the preceding paragraph).
We tested for statistical interaction in modulating warfarin dose for each pair of established dose predictors (VKORC1 rs9923231, CYP2C9*2 and *3, CYP4F2 rs2108622, Age, Sex) using multivariate regression and R software as described above. An interaction term formed by multiplying the pair of predictor variables was added to the multivariate regression equation which contained only main effects of the 6 predictors, and standard ANOVA compared this main-effect model with the enhanced interaction model by testing for a statistically significant increase in explained dose variance. Interaction test p-values were considered statistically significant if below the Bonferroni cutpoint determined by correcting for the 15 interaction tests (i.e. p<0.0033≈0.05/15).
To test for association with over-anticoagulation (INR>4.0) during treatment days 1–35, we performed Cox proportional hazard regression on survival time (day of over-anticoagulation) using the survival library of R software. The GWAS data set of 1053 WARG subjects contained 215 subjects whose INR exceeded 4.0 during days 1–35 while the entire dataset of 1489 WARG subjects contained 312 such subjects.
Association Testing of CNVs
For each CNV locus, association was tested with square root of warfarin dose by multivariate regression analysis in which subject copy number intensity was the CNV predictor of dose. This analysis differs from association testing with SNP genotypes since the two CNV alleles on homologous chromosomes generate one copy number intensity rather than a separate allele for each chromosome. As a QC strategy, we determined each subject's rank in the dataset for copy number intensity at each CNV on chromosome 17. This enabled us to differentiate the majority of subjects (whose individual distribution of ranks were approximately random and uniform) from 174 obvious outliers due to poor quality DNA (whose ranking distributions were “U-shaped” since their intensities strongly clustered at both high and low ranks). These 174 subjects were excluded from the primary CNV association analysis (with further confirmation of lower quality DNA for these subjects being their rough correspondence to the subjects with lower (<99%) SNP call rates). However, we also crosschecked the primary CNV analysis by conducting association testing on the dataset without excluding the 174 subjects and found no statistically significant association with warfarin dose at any CNV whether the dataset excluded or included the subjects. Association testing of the CNVs was executed using R software [37].
Replication of CYP4F2
For the replication of CYP4F2 rs2108622, we genotyped a panel of 588 warfarin patients consisting of 410 subjects from the WARG cohort [3] and 178 from the Uppsala cohort [38]. Table 2 shows regression on this pooled sample of 588 subjects. Separate results for each of the two panels are given in Table S4.
Follow-Up of Moderately Significant SNPs
To possibly identify SNPs with genuine but weak associations to warfarin dose, we excluded VKORC1, CYP2C9, CYP4F2 and identified 40 other GWAS loci for follow-up genotyping exhibiting multivariate regression p-values below 0.0002, and selected 40 SNPs representing these loci for genotyping. Only genotyped (not imputed) SNPs were chosen for follow-up. We genotyped the same 558 patients as in the CYP4F2 replication using the iPLEX MassARRAY.
Power Calculation
Suppose multiple regression analysis is conducted in N total samples by testing a SNP with coefficient of determination (i.e., explained variance) R 2 test after adjustment for known predictors whose total of coefficient of determination is R 2 knw. The probability (power) to detect the tested SNP at a significance level α equals:
| (1) |
where F′(1, N–2, θ 2) is the probability density function for an F distribution with 1, N-2 degrees of freedom and non-centrality parameter θ 2 (Section 28.28 in [39], Example 8.4 in [40]). Here the constant c satisfies the equation:
| (2) |
where α is the significance level, and F(1, N–2) is the probability density function for a F-distribution of degree of freedom one and N–2.
How Much Does Linkage Disequilbrium (LD) Attenuate Association with a Quantitative Trait?
Association with a quantitative trait (QT) becomes weaker for a marker SNP in LD with a SNP that alters the QT, and hence the association becomes more difficult to detect at the marker than at the QT-altering SNP. Here we quantify the LD attenuation for a QT when testing for association by linear regression (which includes the Cochran-Armitage trend test for dichotomous traits), and we obtain a result analogous to the LD attenuation for the Pearson Chi-square test for allelic association to dichotomous traits as in cases and controls [17]. If a causative QT-altering SNP has a coefficient of determination (i.e., explained variance)  and is in pairwise LD of r
2 with a marker SNP, then the coefficient of determination for the marker SNP (
and is in pairwise LD of r
2 with a marker SNP, then the coefficient of determination for the marker SNP ( ) is approximated by:
) is approximated by:
| (3) |
In other words, when testing a marker, the proportion of explained variance decreases by a factor of r 2.
To begin the proof of Equation 3, let the QT be represented by the random variable “q”, and let “m” and “x” be SNP genotypes (coded 0, 1, or 2) representing the marker and causative (QT-altering) SNP, respectively. The coefficients of determination  are equal to the square of two correlation coefficients (denoted by “Corr”) measuring the correlation of m or x with q:
are equal to the square of two correlation coefficients (denoted by “Corr”) measuring the correlation of m or x with q:
| (4) |
| (5) |
Also note that correlation between genotypes at the marker and causative SNP is given by another correlation coefficient:
| (6) |
It is well known that the partial correlation coefficient of m and q conditioned on x is (equation 16.20, p. 649 in [41]):
 |
(7) |
However, conditional on genotype at the causative SNP, marker m and the QT q would be uncorrelated (assuming m is not in LD with a second causative polymorphism) and thus the numerator of Equation 7 would be zero implying that:
| (8) |
Based on prior work [42]–[44], we show in Text S1 that the squares of the genotypic correlation coefficient  and LD correlation coefficient r
2 are approximately equal if the population is in Hardy-Weinberg equilibrium. Therefore, substituting r
2 for
and LD correlation coefficient r
2 are approximately equal if the population is in Hardy-Weinberg equilibrium. Therefore, substituting r
2 for  in Equation 8 gives Equation 3.
in Equation 8 gives Equation 3.
Supporting Information
QQ plot for association of each GWAS SNP with warfarin dose. SNPs were tested for association with warfarin by regression analysis that adjusted for age, sex, and genotype at VKORC1, CYP2C9*2 and *3, CYP4F2. The QQ plot omits SNPs in loci already known to be associated with warfarin dose (VKORC1, CYP2C9, CYP4F2). The excess of SNPs with small p-values is minor: whereas 65.4 SNPs with p<0.0002 are expected, 70 were observed (1.069 times inflated).
(0.20 MB PDF)
Manhattan plot of GWAS results of testing for association with warfarin-induced over-anticoagulation. Horizontal axis is the genomic position, and vertical axis is minus log of p-value. Red dots above the gray line indicate association of genome-wide significance (p<1.5×10−7) at SNPs in the VKORC1 locus such as rs9923231 (p = 8.9×10−9). However, no other loci achieved genome-wide significance. See main text for more details.
(0.07 MB PDF)
Multivariate regression results for 40 SNPs followed-up after GWAS. Unlike rs2108622 of CYP4F2, none of these 40 SNPs exhibited statistical significance after correction for multiple testing. See main text for more details.
(0.03 MB XLS)
Testing for statistical interaction between predictors of warfarin dose. After correcting for the 15 interaction tests, no pair of predictors exhibited statistically significant interaction. Data is for the combined panel of subjects (N = 1641). See main text for more details.
(0.02 MB XLS)
Survival time analysis for incidence of over-anticoagulation within the first 5 weeks of treatment in the whole WARG cohort (N = 1489). See main text for more details.
(0.02 MB XLS)
Multiple regression analysis of warfarin dose in WARG replication samples, WARG GWAS plus replication, or Uppsala replication samples. This table shows the same results displayed in Table 2 of the main text except that WARG and Uppsala subjects are separated into different subsets.
(0.02 MB XLS)
For populations in Hardy-Weinberg Equilibrium, Linkage Disequilibrium r2 and Genotypic R2 are approximately equal.
(0.03 MB DOC)
Acknowledgments
We thank all doctors, nurses and patients who participated in this study.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by the Wellcome Trust. The work was also supported by the Swedish Science Council/Medicine 04496, Swedish Heart and Lung Foundation, the Swedish Society of Medicine, the Swedish Foundation for Strategic Research, the Soderberg, Thureus and Selander Foundations, Nycomed Ltd. of Sweden and the Clinical Research Support (ALF) at Uppsala University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Daly AK, King BP. Pharmacogenetics of oral anticoagulants. Pharmacogenetics. 2003;13:247–252. doi: 10.1097/00008571-200305000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi H, Echizen H. Pharmacogenetics of CYP2C9 and interindividual variability in anticoagulant response to warfarin. Pharmacogenomics J. 2003;3:202–214. doi: 10.1038/sj.tpj.6500182. [DOI] [PubMed] [Google Scholar]
- 3.Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJ, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113:784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindh JD, Lundgren S, Holm L, Alfredsson L, Rane A. Several-fold increase in risk of overanticoagulation by CYP2C9 mutations. Clin Pharmacol Ther. 2005;78:540–550. doi: 10.1016/j.clpt.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Wadelius M, Chen L, Downes K, Ghori J, Hunt S, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005;5:262–270. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 6.D'Andrea G, D'Ambrosio R, Di Perna P, Chetta M, Santacroce R, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105:645–649. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 7.Rettie AE, Tai G. The pharmocogenomics of warfarin: closing in on personalized medicine. Mol Interv. 2006;6:223–227. doi: 10.1124/mi.6.4.8. [DOI] [PubMed] [Google Scholar]
- 8.Wadelius M, Chen L, Eriksson N, Bumpstead S, Ghori J, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121:23–34. doi: 10.1007/s00439-006-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rieder M, Reiner A, Gage B, Nickerson D, Eby C, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 12.Caldwell M, Awad T, Johnson J, Gage B, Falkowski M, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haley C, Knott S. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- 14.Jansen R. Interval mapping of multiple quantitative trait loci. Genetics. 1993;135:205–211. doi: 10.1093/genetics/135.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stubbins MJ, Harries LW, Smith G, Tarbit MH, Wolf CR. Genetic analysis of the human cytochrome P450 CYP2C9 locus. Pharmacogenetics. 1996;6:429–439. doi: 10.1097/00008571-199610000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–719. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 17.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhangale TR, Rieder MJ, Nickerson DA. Estimating coverage and power for genetic association studies using near-complete variation data. Nat Genet. 2008;40:841–843. doi: 10.1038/ng.180. [DOI] [PubMed] [Google Scholar]
- 19.Evans DM, Barrett JC, Cardon LR. To what extent do scans of non-synonymous SNPs complement denser genome-wide association studies? Eur J Hum Genet. 2008;16:718–723. doi: 10.1038/sj.ejhg.5202011. [DOI] [PubMed] [Google Scholar]
- 20.Scott S, Edelman L, Kornreich R, Desnick R. Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am J Hum Genet. 2008;82:495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson JL, Horne BD, Stevens SM, Grove AS, Barton S, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–2570. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 22.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 23.Lenzini P, Grice G, Milligan P, Dowd M, Subherwal S, Deych E, Eby C, et al. Laboratory and clinical outcomes of pharmacogenetic vs. clinical protocols for warfarin initiation in orthopedic patients. J Thromb Haemost. 2008;6:1655–62. doi: 10.1111/j.1538-7836.2008.03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voora D, Eby C, Linder M, Milligan P, Bonny L, et al. Prospective dosing of warfarin based on cytochrome P-450 2C9 genotype. Thromb Haemost. 2005;93:700–705. doi: 10.1160/TH04-08-0542. [DOI] [PubMed] [Google Scholar]
- 25.Hynicka L, Cahoon W, Bukaveckas B. Genetic testing for warfarin therapy initiation. Ann Pharmacother. 2008;42:1298–1303. doi: 10.1345/aph.1L127. [DOI] [PubMed] [Google Scholar]
- 26.Limdi N, Veenstra D. Warfarin Pharmacogenetics. Pharmacotherapy. 2008;28:1084–1097. doi: 10.1592/phco.28.9.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillman M, Wilke R, Yale S, Vidaillet H, Caldwell M, et al. A prospective, randomized pilot trial of model-based warfarin dose initiation using CYP2C9 genotype and clinical data. Clinical Medicine & Research. 2006;3:137–145. doi: 10.3121/cmr.3.3.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindh JD, Holm L, Dahl ML, Alfredsson L, Rane A. Incidence and predictors of severe bleeding during warfarin treatment. J Thromb Thrombolysis. 2008;25:151–159. doi: 10.1007/s11239-007-0048-2. [DOI] [PubMed] [Google Scholar]
- 29.Lindh JD, Kublickas M, Westgren M, Rane A. Internet based clinical trial protocols – as applied to a study of warfarin pharmacogenetics. Br J Clin Pharmacol. 2004;58:482–487. doi: 10.1111/j.1365-2125.2004.02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marioni JC, Thorne NP, Valsesia A, Fitzgerald T, Redon R, et al. Breaking the waves: improved detection of copy number variation from microarray-based comparative genomic hybridization. Genome Biol. 2007;8:R228. doi: 10.1186/gb-2007-8-10-r228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 33.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 34.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barnes C, Plagnol V, Fitzgerald T, Redon R, Marchini J, et al. A robust statistical method for case-control association testing with copy number variation. Nat Genet. 2008;40:1245–1252. doi: 10.1038/ng.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadelius M, Sörlin K, Wallerman O, Karlsson J, Yue Q, et al. Warfarin sensitivity related to CYP2C9, CYP3A5, ABCB1 (MDR1) and other factors. Pharmacogenomics J. 2004;4:40–48. doi: 10.1038/sj.tpj.6500220. [DOI] [PubMed] [Google Scholar]
- 39.Stuart A, Ord J, Arnold S. Kendall's advanced theory of statistics. London: Arnold publishers; 1999. [Google Scholar]
- 40.Knight K. Mathematical statistics. Boca Raton: Chapman & Hall/CRC; 2000. [Google Scholar]
- 41.Sokal RR, Rohlf FJ. Biometry. New York: W. H. Freeman and Company; 1995. [Google Scholar]
- 42.Weir B. Genetic Data Analysis II. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- 43.Weir B. Inferences about linkage disequilibrium. Biometrics. 1979;35:235–254. [PubMed] [Google Scholar]
- 44.Zaykin D. Bounds and normalization of the composite disequilibrium coefficient. Genetic Epidemiology. 2004;271:252–257. doi: 10.1002/gepi.20015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
QQ plot for association of each GWAS SNP with warfarin dose. SNPs were tested for association with warfarin by regression analysis that adjusted for age, sex, and genotype at VKORC1, CYP2C9*2 and *3, CYP4F2. The QQ plot omits SNPs in loci already known to be associated with warfarin dose (VKORC1, CYP2C9, CYP4F2). The excess of SNPs with small p-values is minor: whereas 65.4 SNPs with p<0.0002 are expected, 70 were observed (1.069 times inflated).
(0.20 MB PDF)
Manhattan plot of GWAS results of testing for association with warfarin-induced over-anticoagulation. Horizontal axis is the genomic position, and vertical axis is minus log of p-value. Red dots above the gray line indicate association of genome-wide significance (p<1.5×10−7) at SNPs in the VKORC1 locus such as rs9923231 (p = 8.9×10−9). However, no other loci achieved genome-wide significance. See main text for more details.
(0.07 MB PDF)
Multivariate regression results for 40 SNPs followed-up after GWAS. Unlike rs2108622 of CYP4F2, none of these 40 SNPs exhibited statistical significance after correction for multiple testing. See main text for more details.
(0.03 MB XLS)
Testing for statistical interaction between predictors of warfarin dose. After correcting for the 15 interaction tests, no pair of predictors exhibited statistically significant interaction. Data is for the combined panel of subjects (N = 1641). See main text for more details.
(0.02 MB XLS)
Survival time analysis for incidence of over-anticoagulation within the first 5 weeks of treatment in the whole WARG cohort (N = 1489). See main text for more details.
(0.02 MB XLS)
Multiple regression analysis of warfarin dose in WARG replication samples, WARG GWAS plus replication, or Uppsala replication samples. This table shows the same results displayed in Table 2 of the main text except that WARG and Uppsala subjects are separated into different subsets.
(0.02 MB XLS)
For populations in Hardy-Weinberg Equilibrium, Linkage Disequilibrium r2 and Genotypic R2 are approximately equal.
(0.03 MB DOC)