Abstract
Cytochrome c (Cyt c) is part of the mitochondrial electron transport chain (ETC), accepting electrons from bc1 complex and transferring them to cytochrome c oxidase (CcO). The ETC generates the mitochondrial membrane potential, which is used by ATP synthase to produce ATP. In addition, the release of Cyt c from the mitochondria often commits a cell to undergo apoptosis. Considering its central role in life (respiration) and death (apoptosis) decisions one would expect tight regulation of Cyt c function. Reversible phosphorylation is a main cellular regulatory mechanism, but the effect of cell signaling targeting the mitochondrial oxidative phosphorylation system is not well understood, and only a small number of proteins that can be phosphorylated have been identified to date. We have recently shown that Cyt c isolated from cow heart tissue is phosphorylated on tyrosine 97 in vivo, which leads to inhibition of respiration in the reaction with CcO. In this study we isolated Cyt c from a different organ, cow liver, under conditions preserving the physiological phosphorylation state. Western analysis with a phospho-tyrosine specific antibody suggested that liver Cyt c is phosphorylated. Surprisingly, the phosphorylation site was unambiguously assigned to Tyr-48 by immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry (IMAC/nano-LC/ESI-MS), and not to the previously identified phospho-Tyr-97 in cow heart. As is true of Tyr-97, Tyr-48 is conserved in eukaryotes. As one possible consequence of Tyr-48 phosphorylation we analyzed the in vitro reaction kinetics with isolated cow liver CcO revealing striking differences. Maximal turnover of Tyr-48 phosphorylated Cyt c was 3.7 s−1 whereas dephosphorylation resulted in a 2.2 fold increase in activity to 8.2 s−1. Effects of Tyr-48 phosphorylation based on the Cyt c crystal structure are discussed.
1. Introduction
Cytochrome c (Cyt c) is a small globular protein with a covalently linked heme group. The mature protein, which lacks the start methionine, contains 104 amino acids in humans and other mammals and is highly positively charged with a pI of 9.6. As part of the electron transport chain Cyt c is located in the mitochondrial intermembrane space and transfers electrons from bc1 complex to cytochrome c oxidase (CcO). Cyt c is essential for aerobic energy production and Cyt c knockout mice die around midgestation [1], when metabolism switches from the glycolytic pathway to aerobic energy production [2].
Cyt c plays a second role in type II apoptosis, which involves the release of Cyt c into the cytosol, binding to Apaf-1, and activation of procaspase-9 to caspase-9 [3]. While the release of Cyt c from the mitochondria is mechanistically unclear, it is considered a crucial proapoptotic signal [4, 5]. Recently, Cyt c was shown to function as a cardiolipin peroxidase at early stages during apoptosis, where it selectively oxidizes the mitochondrial inner membrane lipid cardiolipin [6], which binds to Cyt c. Another function as a reactive oxygen species scavenger has been proposed for Cyt c under normal (non-apoptotic) conditions [7].
Cyt c has a testes-specific isoform in rodents [8], but the syntenic region of testes Cyt c in the human genome contains a non-transcribed pseudogene [9]. Testes-specific Cyt c has an interesting feature in that it shows a threefold increased activity to reduce hydrogen peroxide compared with the somatic isoform, but also shows a three to fivefold activity to induce apoptosis [10]. In addition to regulation of Cyt c function via the expression of tissue-specific isoforms, it can be regulated allosterically through binding of ATP affecting the binding of Cyt c to CcO, leading to an inhibition of the reaction between Cyt c and CcO [11]. A similar regulatory mechanism is present in CcO that also leads to an inhibition of respiration (reviewed in [12]).
Important metabolic enzymes can be regulated by a third mechanism in addition to isoform expression and allosteric control –– via reversible phosphorylation. Until recently, not much attention has been devoted to cell signaling targeting the mitochondrial oxidative phosphorylation (OxPhos) complexes, and only 14 epitopes that can be phosphorylated have been mapped in mammals (reviewed in [13]). Among these phosphorylation sites, one was recently identified in cow heart Cyt c [14]. Using tandem-mass spectrometry tyrosine 97 was shown to be phosphorylated, and the reaction with CcO was inhibited in the presence of tyrosine 97-phosphorylation and showed pronounced sigmoidal kinetics [14].
In this study we purified Cyt c from cow liver under conditions that preserve the physiological phosphorylation state. Western blot analysis indicated tyrosine phosphorylation. Surprisingly, analysis of isolated Cyt c by tandem mass spectrometry revealed a novel phosphorylation site on tyrosine 48 of the mature protein. This phosphorylation reduces maximal turnover in the reaction with CcO by more than 50%.
2. Materials and methods
2.1. Isolation of liver cytochrome c
Chemicals were purchased from Sigma (St. Luis, MO) unless otherwise stated. Cow liver tissue (3 kg) was ground using a commercial meat grinder and supplemented with 2 L of ice-cold buffer A (250 mM sucrose, 2 mM EDTA, 20 mM Tris-Cl (pH 7.4)), containing protease inhibitor phenylmethylsulfonyl fluoride (PMSF; 1 mM), and unspecific tyrosine phosphatase inhibitor sodium vanadate (1 mM), unspecific serine/threonine phosphatase inhibitor KF (10 mM), and calcium chelator EGTA (2 mM) to maintain the physiological phosphorylation status. To remove hemoglobin and soluble proteins released from broken cells that may interfere with Cyt c isolation the suspension was centrifuged (13,000 × g, 8 min) and the supernatant discarded. To extract Cyt c the combined pellets were immersed in 7 L of dd H2O supplemented with sodium vanadate, KF, and EGTA, and the pH was immediately adjusted to 4.5 with acetic acid under stirring. Cells were broken up and homogenized using a commercial blender at maximum speed for 20 sec. The pH was readjusted to 4.5 and the homogenate was incubated for 12 h on ice. The suspension was centrifuged (27,000 × g, 40 min), the combined pellets were re-extracted with 3 L ddH2O supplemented with 1 mM sodium vanadate, 10 mM KF, and 2 mM EGTA (pH 4.5 adjusted with acetic acid), and centrifuged as above. The combined supernatants containing Cyt c were adjusted to pH 7.5 and centrifuged once more as above to remove proteins that aggregate after pH adjustment. The Cyt c-containing supernatant was purified via two step ion exchange chromatography modified from ref. [15]. Briefly, the solution was diluted with water until a conductance of 4.8 mS/cm was reached, adjusted to pH 7.5, and applied to a DE52 anion exchange column (Whatman, Florham Park, NJ) equilibrated with KH2PO4, pH 7.5, of similar conductance (about 20 mM). Under those conditions more than 80% of all proteins bind to the column [15]. The flowthrough-containing Cyt c was diluted to a conductance of 2 mS/cm, the pH was adjusted to 6.5, and applied to a CM52 sepharose cation exchange column (Whatman, Florham Park, NJ) equilibrated with KH2PO4, pH 6.5, 2 mS/cm conductance. Cyt c binds to the column, which was then washed with 4 column bed volumes of equilibration buffer. A three step gradient was applied (90, 120, and 180 mM KH2PO4, pH 6.5) in which Cyt c eluted in the last fraction. To increase purity, the DE52 and CM52 ion exchange steps were repeated. Cyt c was concentrated under vacuum to 2 mL and desalted using Sephadex G50 gel filtration equilibrated with 10 mM KH2PO4, pH 7.0. Finally, Cyt c was subjected to HPLC purification (Jasco Inc., Easton, MD) using a Macrosphere™ GPC column (7.5×300 mm; 100 Å, Grace, Deerfield, IL). The column was equilibrated with 150 mM KH2PO4, pH 7.4. After elution, Cyt c was concentrated and desalted as above, and stored at −80 °C.
2.1.1 Concentration determination of cytochrome c
Cyt c was oxidized with 100 mM potassium ferrocyanide (K3Fe(CN)6) and desalted using NAP-10 columns (GE Healthcare, Piscataway, NJ) equilibrated with 20 mM Tris-Cl (pH 7.5) and analyzed on a Jasco V-570 double beam spectrophotometer (2 nm bandwidth). Reduced Cyt c was generated by addition of 100 mM sodium dithionite (Na2S2O4) and spectra were acquired after desalting the samples as above. The concentration of reduced Cyt c was determined by the difference spectra at 550 nm of the dithionite reduced minus potassium ferrocyanide oxidized form, and calculated via ε(red. – ox.)550nm = 21 mM−1 cm−1.
2.2. Cytochrome c dephosphorylation
Unspecific phosphatase shrimp alkaline phosphatase (10 units, Roche, Indianapolis, IN) was employed to dephosphorylate Cyt c as previously described [14] using 300 μL of a 40 μM Cyt c solution and overnight incubation at 30 °C. The sample was desalted using NAP-10 columns (GE Healthcare, Piscataway, NY) equilibrated with 50 mM Tris-Cl (pH 7.5). Cyt c was separated from the phosphatase using a Microcon YM-100 centrifugal device (Upstate, Bilerica, MA) and obtained as the flow-through.
2.3. Western blot analysis
Western analysis—SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of Cyt c was carried out using a 4–12% gradient NuPAGE Bis-Tris gel (Invitrogen, Carlsbad, CA). Protein bands were visualized using the Silver Stain Plus kit (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. For Western analysis, protein transfer time on a nitrocellulose membrane (0.2 μm, GE Healthcare, Piscataway, NJ) was 30 min at 10 V in 25 mM Tris base, 192 mM glycin, 20% methanol. Small and highly charged proteins can easily be lost during the multiple washing steps. Thus, to prevent Cyt c dissociation we incubated the membrane in a 1% glutaraldehyde solution for 10 min, and subsequently applied two UV-crosslinking pulses (0.12 joules each, UV Stratalinker 1800, Stratagene, Cedar Creek, TX). This combination was found to be optimal with respect to Cyt c retention on the membrane. Because amines, such as Tris, compete with the chemically formed crosslinks (Schiff base), all solutions subsequently used contained 20 mM HEPES (pH 7.5) instead of an amine-based buffering reagent. Western analysis was performed with a 1:5,000 dilution of anti-phospho tyrosine (4G10, Upstate, Bilerica, MA) followed by a 1:10,000 dilution of anti-mouse IgG alkaline phosphatase-conjugated secondary antibody (Invitrogen, Carlsbad, CA). EGF stimulated A431 cell lysate (Upstate, Bilerica, MA) was included as a positive control and ovalbumin as a negative control. Signals were detected via the colorimetric method using nitro blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Roche, Indianapolis, IN). Anti-phosphoserine and anti-phosphothreonine antibodies were sets of four (1C8, 4A3, 4A9, and 16B4), and three (1E11, 4D11, and 14B3) individual monoclonal antibodies (EMD Biosciences, Gibbstown, NJ) were used as described above for the anti-phospho-tyrosine antibody.
2.4. Activity measurements of cytochrome c with cytochrome c oxidase
Cytochrome c oxidase (CcO) was isolated from cow liver under standard conditions as previously described [16]. To remove cholate and to replace potentially damaged cardiolipin, CcO was dialyzed in the presence of 0.5 mM ATP and a 40-fold molar excess of cardiolipin in 10 mM K-HEPES (pH 7.4), 40 mM KCl, 1% Tween 20, 2 mM EGTA, and 10 mM KF. CcO activity was analyzed in a closed 200 μL chamber containing a micro Clark-type oxygen electrode (Oxygraph system, Hansatech, Amesbury, MA). Measurements were performed using 45 nM CcO at 25 °C after the addition of ascorbic acid (20 mM) and increasing amounts of purified Cyt c from 0–25 μM. Oxygen consumption was recorded on a computer and analyzed with the Oxygraph software. Turnover number (TN) is defined as oxygen consumed [μ moles]/(sec · CcO [μmoles]).
2.5. Mass spectrometry
2.5.1. Protein reduction, alkylation, and digestion
Protein samples (40 μg of purified and desalted Cyt c) were reduced with 10 mM DTT for 1 h in a 56 °C water bath followed by alkylation with 55 mM indoacetamide for 1 h at room temperature in the dark. Samples were diluted five times with 100 mM NH4HCO3 (pH 8.9). Proteins were digested with affinity purified and TPCK treated trypsin (Promega, Madison, WI) at a trypsin:protein ratio of 1:100 (w/w) overnight at 37 °C. Tryptic peptides were desalted using Sep-Pak C18 Cartridges (Waters, Milford, MA) as described [17] and dried in a Speed Vac plus (Thermo Savant, Holbrook, NY).
2.5.2. Peptide Immunoprecipitation
Dry peptides were reconstituted in 1 mL cold IP buffer (30 mM Tris-Cl, 30 mM NaCl, and 0.3% NP-40, pH 7.4) and 10 pmol synthetic peptide LIEDAEpYTAK was added to each time point as a control for label-free quantitation, accompanying the cellular phosphopeptides through the peptide immunoprecipitation, subsequent peptide purification steps, and reversed-phase elution of peptides into the mass spectrometer. Bead-conjugated anti-phosphotyrosine antibody was prepared by coupling monoclonal anti-phosphotyrosine antibody clone pTyr100 (Cell Signaling Technology, Danvers, MA) to protein G agarose beads (Roche, Basel, Switzerland) noncovalently at 2 mg/mL overnight at 4 °C. Anti-phosphotyrosine beads were then added at 15 μL resin per sample overnight at 4 °C with gentle shaking. Beads were washed and eluted as described [18].
2.5.3. Automated desalt/immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry (IMAC/nano-LC/ESI-MS)
Tryptic peptides were analyzed by a fully automated phosphoproteomic technology platform incorporating peptide desalting via reversed-phase chromatography, gradient elution to an Fe3+-loaded IMAC column, reversed-phase separation of peptides followed by tandem mass spectrometry with static peak parking as described previously [19]. Briefly, tryptic peptides were loaded onto a desalting reversed-phase column (360 μm OD × 200 μm ID fused silica (Polymicro Technologies, Phoenix, AZ), fitted with an inline microfilter (M520, Upchurch Scientific, Oak Harbor, WA), packed on a pressure bomb (GNF Commercial Systems, San Diego, CA) with 12 cm SelfPack POROS 10 R2 resin (Applied Biosystems, Foster City, CA). Peptides were rinsed with 0.1 M acetic acid/MilliQ water (solvent A) at a flow rate of 10 μL/min and then eluted to an Fe3+-activated IMAC column (360 μm OD × 200 μm ID fused silica, bomb packed with 15 cm SelfPack POROS 20 MC resin (Applied Biosystems) with a HPLC gradient of 0–70% solvent B (0.1 M acetic acid/acetonitrile) in 17 min at a flow rate of 1.8 μL/min. The column was washed with ~40 μL of a 25:74:1 acetonitrile/water/acetic acid mixture containing 100 mM NaCl followed by 4 min solvent A at a flow rate of 35 μL/min. Enriched phosphopeptides were eluted to a precolumn (360 μm OD × 75 μm ID) containing 2 cm of 5 μm Monitor C18 resin (Column Engineering, Ontario, CA) with 40 μl of 25 mM potassium phosphate (pH 9.0) and rinsed with solvent A for 30 min. Peptides were eluted into the mass spectrometer (LTQ-FT, Thermo Electron, San Jose, CA) through an analytical column (360 μm OD × 75 μm ID fused silica with 12 cm of 5 μm Monitor C18 particles with an integrated ~4 μm ESI emitter tip fritted with 3 μm silica; Bangs Labs, Fishers, IN) with an HPLC gradient (0–70% solvent B in 30 min). Static peak parking was performed via flow rate reduction from the initial 200 nL/min to ~20 nL/min when peptides began to elute as judged from a BSA peptide scouting run as described previously [19]. The electrospray voltage of 2.0 kV was applied in a split flow configuration as described [19]. Spectra were collected in positive ion mode and in cycles of one full MS scan in the FT (m/z: 400–1800) (~1 s each) followed by MS/MS scans in the LTQ (~0.3 s each) sequentially of the five most abundant ions in each MS scan with charge state screening for +1, +2, and +3 ions and dynamic exclusion time of 30 s. The AGC was 1,000,000 for the FTMS MS scan and 10,000 for the LTMS MS/MS scans. The max ion time was 100 ms for the LTMS MS/MS scan and 500 ms for the FTMS full scan. FTMS resolution was set at 100,000.
2.5.4. Database analysis
MS/MS spectra were assigned to peptide sequences from the NCBI non-redundant protein database sliced in Bioworks 3.1 for bovine proteins and searched with the SEQUEST algorithm [20]. SEQUEST search parameters designated variable modifications of +79.9663 Da on Ser, Thr, and Tyr (phosphorylation). Phosphopeptide spectra were manually verified.
2.6. Molecular Modeling
High resolution crystallographic data from horse Cyt c (1hrc; [21]) was used, processed with the program Swiss PDB viewer (version 3.7), and rendered with the program POV-Ray 3.6.
3. Results
3.1. Isolation of phosphorylated cytochrome c and Western blot analysis
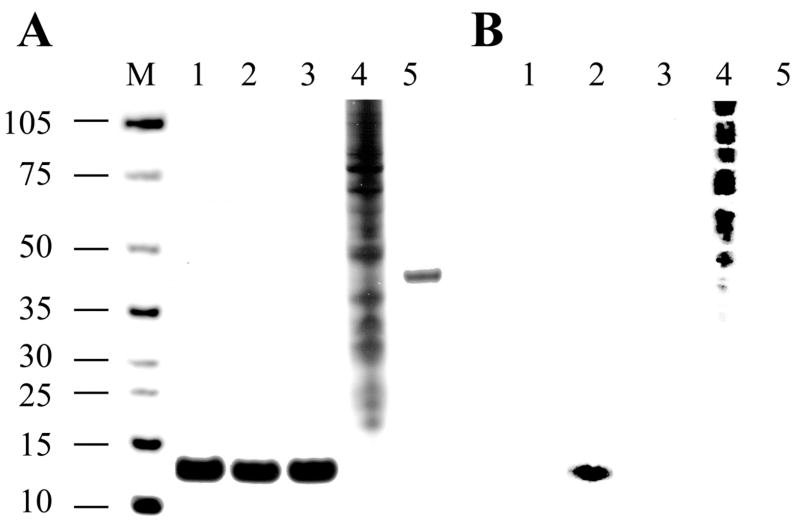
Using conditions in all buffers that preserve protein phosphorylation through addition of vanadate and fluoride, two unspecific phosphatase inhibitors, and calcium chelator EGTA to minimize activation of calcium-dependent protein phosphatases, we employed a modified acidic extraction of Cyt c from total tissue without prior isolation of mitochondria (Materials and methods). Extracted proteins were subjected to weak anion followed by weak cation exchange chromatography, and this sequence was repeated to increase purity. Cyt c was further purified by HPLC gel filtration, resulting in a highly purified Cyt c fraction (Fig. 1A, lane 2).
Fig. 1.
Gel and Western analysis of isolated cow liver Cyt c. A, Cyt c was purified under conditions preserving the in vivo phosphorylation status (lane 2) and treated with shrimp alkaline phosphatase (SAP, lane 3). Samples were applied to a 4–12% SDS-PAGE gradient gel and protein bands were detected using the silver staining method. Lane M, protein size marker (kDa); lane 1, Sigma Cyt c; lane 4, EGF stimulated A431 cell lysate (positive control for Western analysis); lane 5, ovalbumin (negative control for Western analysis). B, for Western analysis, protein samples were applied to SDS-PAGE and subsequently transferred to a nitrocellulose membrane and analyzed using an anti-phosphotyrosine antibody (4G10, Upstate). Purified Cyt c (lane 2) produces a strong signal with the anti-phosphotyrosine antibody, whereas overnight treatment with SAP abolishes the signal (lane 3). Samples in lanes 1 – 5 as denoted in A. Protein sizes are indicated on the left.
To test possible phosphorylation of Cyt c we performed Western analysis with phospho-Ser/Thr/Tyr specific antibodies. Phospho-Ser and –Thr antibodies did not show a signal (not shown) whereas the phospho-Tyr antibody produced a strong signal (Fig. 1B, lane 2). This signal was specific since it was eliminated after treatment with shrimp alkaline phosphatase, which nonspecifically removes phosphate groups (Fig. 1B, lane 3).
3.2. Cow liver Cyt c is phosphorylated on Tyr-48
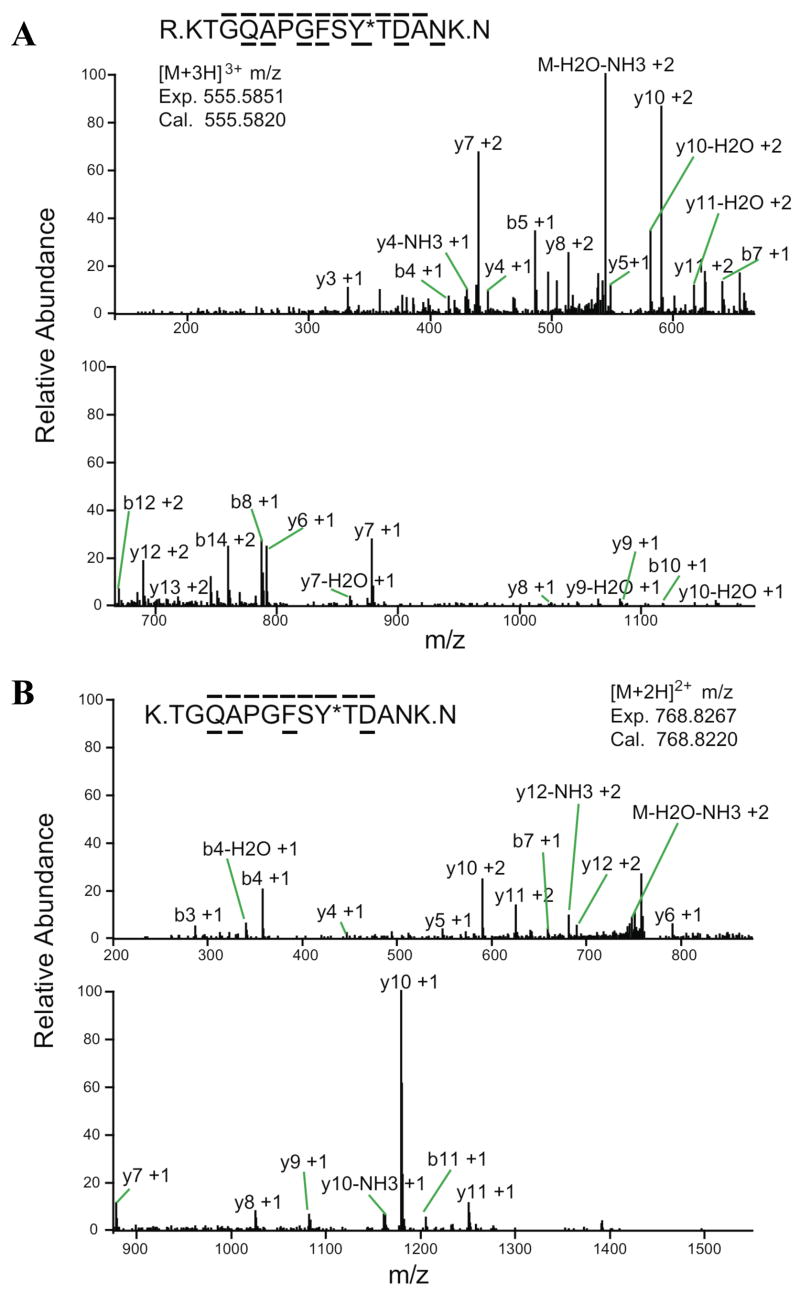
In order to determine the phosphorylation site isolated Cyt c was digested with trypsin and analyzed by immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry (IMAC/nano-LC/ESI-MS/MS). This method first enriches phosphorylated peptides, which are then subjected to tandem-MS for fragment examination. This analysis unambiguously revealed Tyr-48 phosphorylation of Cyt c on the two peptides KTGQAPGFSpYTDANK (peptide 1, Fig. 2A) and TGQAPGFSpYTDANK (peptide 2, Fig. 2B) that differ only in the presence or absence of the first lysine residue due to alternative tryptic digestion after R or K in the sequence 34-GLFGR ↓ K ↓ TGQAP-44. The phosphorylation site was unambiguously assigned by fragment ions b10, and y5, y6, y7 on peptide 1 and y5, y6, and y7 on peptide 2 (Fig. 2). The sequences were assigned by fragment ions b4, b5, b7, b8, b10, b122+, b142+, y3, y4, y5, y6, y7, y82+, y9, y102+, y112+, y12+2, and y13+2 in peptide 1 and b3, b4, b7, b11, y4, y5, y6, y7, y8, y9, y10, y11, and y122+ in peptide 2 (Fig. 2).
Fig. 2.
Nano-LC/ESI/MS/MS spectrum of KTGQAPGFSpYTDANK and TGQAPGFSpYTDANK. Peptides were eluted into the mass spectrometer by applying a HPLC gradient of 0–70% 0.1 M acetic acid/acetonitrile in 30 minutes. The mass spectrometer acquired top 3 data dependent ESI MS/MS spectra. A, on peptide KTGQAPGFSpYTDANK the phosphorylation site was unequivocally assigned by fragment ions b10, and y5, y6, y7. The sequence of the peptide was definitively assigned by b4, b5, b7, b8, b10, b122+, b142+, y3, y4, y5, y6, y7, y82+, y9, y102+, y112+, y12+2, and y13+2. B, the same phosphorylation site was further confirmed on fragment TGQAPGFSpYTDANK by fragment ions y5, y6, and y7, and the sequence was identified by fragment ions b3, b4, b7, b11, y4, y5, y6, y7, y8, y9, y10, y11, and y122+.
3.3 Tyr-48 phosphorylation of Cyt c leads to strong inhibition in the reaction with cytochrome c oxidase (CcO)
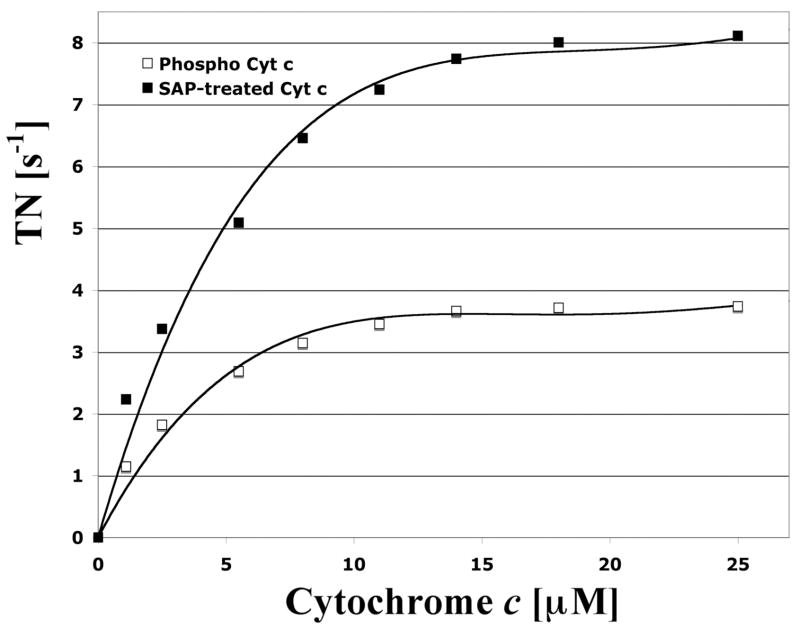
We analyzed the effect of Tyr-48 phosphorylation of Cyt c on respiration with isolated cow liver CcO by measuring oxygen consumption with in vivo phosphorylated and SAP-treated dephosphorylated Cyt c, and observed striking differences. Maximal turnover number of phosphorylated Cyt c was 3.7 s−1 whereas dephosphorylation resulted in a 2.2 fold increase in activity to 8.2 s−1 (Fig. 3, compare open and closed squares, respectively). Both Cyt c species produced a hyperbolic response with a similar Km of CcO for Cyt c of 3 μM for Tyr-48 phosphorylated and 3.8 μM for the SAP-treated Cyt c.
Fig. 3.
Kinetics analysis of Tyr-48 phosphorylated and dephosphorylated cytochrome c with cytochrome c oxidase (CcO). Increasing amounts of isolated phosphorylated (open squares) and shrimp alkaline phosphatase-treated (closed squares) cytochrome c were added to solubilized CcO. CcO activity was measured with the polarographic method at 25 °C. CcO activity (turnover number, TN) is defined as consumed O2 [μmoles]/(s · CcO [μmoles]).
Discussion
Among the many protocols available for the purification of Cyt c, the two main approaches differ in the first step, which involves either the purification of mitochondria or, alternatively, a direct extraction of Cyt c from total cells or tissues. The latter protocol as employed here commonly involves the release of Cyt c from tissue and mitochondria under acidic conditions, such as at pH 4.5. It has the advantage that all cellular Cyt c can be extracted whereas the isolation of mitochondria produces lower yields due to loss of mitochondria during the repetitive centrifugation steps and further loss of Cyt c due to some breakage of mitochondria. The acidic extraction protocol may have the disadvantage of partial deamidation of glutamine and especially asparagine residues. Such deamidated forms have been described for Cyt c (as an example see ref. [22]), but one might speculate that perhaps some of those previously described derivatives might represent phosphorylated forms, in which — similar to deamidation chemistry that introduces a negative charge after hydrolysis of the amide into a carboxo group — polar amino acid side chains are changed into negatively charged side chains through the attachment of the phosphate moiety.
Using cow heart tissue we have previously shown that purification of mitochondria and subsequently isolation of Cyt c produced a Tyr-97 phosphorylated protein [14]. Here we employed the acidic extraction protocol on cow liver tissue because mitochondria are less abundant in liver and this method generally produces a better yield. Importantly, for the isolation of Cyt c with both methods we included unspecific protein phosphatase inhibitors vanadate and fluoride in the isolation buffers to preserve the physiological phosphorylation status during protein purification. In addition, we also included calcium chelator EGTA, since calcium is believed to be the most potent activator of mitochondrial function, and it has recently been shown that under conditions where calcium is elevated, calcium activated phosphatases appear to prevail, leading to dephosphorylation of most mitochondrial proteins [23].
Following purification of liver Cyt c we observed tyrosine phosphorylation after Western analysis (Fig. 1B, lane 2). Treatment of phosphorylated Cyt c with shrimp alkaline phosphatase abolished the signal (Fig. 1B, lane 3). Unexpectedly, using mass spectrometry we unambiguously identified Tyr-48 as the site of phosphorylation, and there was no indication of a second phosphorylation site. We also examined extracted ion chromatogram (XIC) mass chromatograms of both +1 and +2 charged fragments of GEREDLIAYLK and EDLIAYLK (including phosphotyrosine) corresponding to the previously identified phospho-Tyr-97 epitope from heart Cyt c. XIC data of the phosphorylated forms of GEREDLIAYLK and EDLIAYLK using a 50 ppm mass window showed no significant peaks (not shown), further suggesting that Tyr-97 phosphorylation is not present in significant amounts in the Cyt c sample purified from liver tissue.
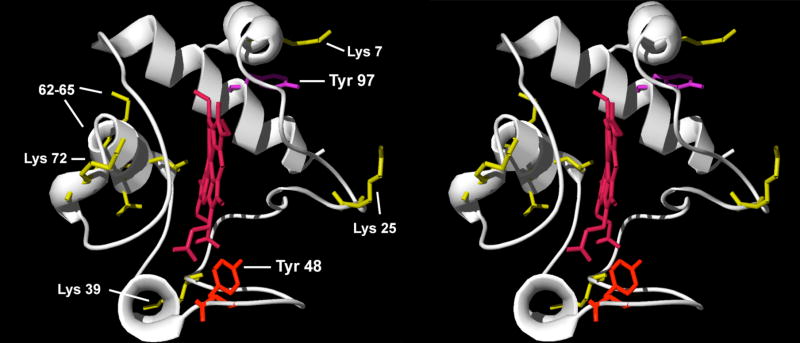
The novel phosphorylation site on Tyr-48 is distinct from the previously identified phospho-Tyr-97 in cow heart tissue (Fig. 5): Tyr-48 is located on the lower median frontal area, whereas Tyr-97 is located on the upper distal area of the conventional right side of the molecule. In contrast to Tyr-97 where a phosphate group can be easily attached via computer modeling without major spatial restraints [14], Tyr-48 points inward and we hypothesize that phosphorylation at this site may result in a profound structural change. Future structure determination of the two phosphorylated variants would address this issue adequately.
Fig. 5.
Stereo-view of Cyt c. Horse Cyt c [21] is shown in ribbon presentation. The newly identified residue Tyr-48 in liver and the previously identified residue Tyr-97 in heart that can be phosphorylated are highlighted as sticks in red and purple, respectively. The heme group is shown in dark red. Key amino acids required for Apaf-1 binding and subsequent caspase activation [24] are shown in yellow.
We addressed one possible effect of this phosphorylation and analyzed the reaction kinetics with CcO of Tyr-48 phosphorylated and SAP-dephosphorylated Cyt c. Supporting our hypothesis that such phosphorylation may result in structural changes affecting the function of Cyt c we observed a more than twofold increase in respiration after dephosphorylation at maximal (3.7 s−1 versus 8.2 s−1) and half-maximal (1.9 s−1 versus 4.1 s−1) turnover. These kinetic data are distinct from those of Tyr-97 phosphorylated Cyt c, which showed a sigmoidal response but a similar maximal turnover compared to the dephosphorylated form [14]. The data presented here may also suggest that the majority of isolated Cyt c is present in the phosphorylated form.
Several questions related to Cyt c phosphorylation have to be addressed in yet to be done experiments. Previously, we proposed that Tyr-97 phosphorylation may affect the role of Cyt c in apoptosis [14] because Lys-7, an essential residue for the interaction with Apaf-1 [24], is spatially located next to Tyr-97 and may possibly form a salt bridge neutralizing the positive charge on the lysine residue (Fig. 4), which may alter or prevent induction of apoptosis. In addition, Tyr-48 phosphorylation might affect the cardiolipin peroxidase activity of Cyt c. The Kagan group has recently shown that this mechanism involves a tyrosine radical [6], and if Tyr-48 is involved in this process phosphorylation of this residue may affect radical stability and also spatially interfere with lipid binding due to the presence of the bulky phosphate group.
Fig. 4.
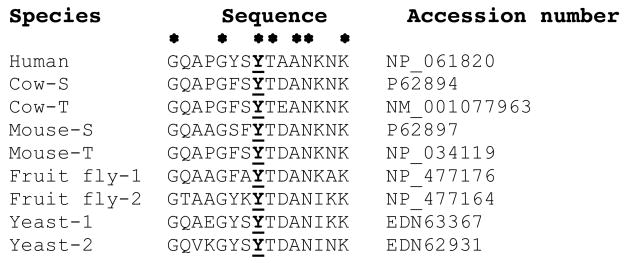
Cytochrome c sequence alignment reveals that Tyr-48 is conserved in eukaryotes. The epitope surrounding Tyr-48 was aligned with the program MegAlign. In addition to humans, which only express a single Cyt c isoform, selected eukaryotic representatives of the somatic (S) and testes-specific Cyt c (T), as well as other species with two Cyt c isoforms (1 and 2), are shown. The phosphorylation site (bold and underlined) is conserved in all species analyzed. Identical amino acids are indicated with an asterisk.
Although humans express only a single Cyt c, most other eukaryotes have two actively transcribed Cyt c genes. In mammals, the resulting isoforms are expressed in a tissue-specific manner, and the so-called somatic isoform is expressed in all tissues except for several testicular cell types whereas the other isoform is testes-specific and is the exclusive isoform found in sperm. Interestingly, Tyr-48 is conserved in all eukaryotes we analyzed (Fig. 4), and is also present in both the somatic and testes-specific isoform, suggesting that this residue may also be targeted by a cell signaling pathway in the male reproductive system. Bacterial Cyt c species belonging to the same group (class 1B) as the mitochondrial homologues are more diverse and Tyr-48 is not conserved (not shown). This makes sense because tyrosine phosphorylation is a specialized regulatory mechanism mainly found in higher organisms.
What lies ahead? Including this study, 15 phosphorylation sites have been identified in mammalian OxPhos components to date (reviewed in [13]), most likely representing the tip of the iceberg given the novelty of this research direction. Among those, in addition to Tyr-48 and -97 on Cyt c, there are two further tyrosine phosphorylation sites, Tyr-304 on CcO catalytic subunit I [16] and Tyr-75 of the ATP synthase δ-subunit [25]. The phospho epitope encompassing Tyr-48 does not show any sequence homology with any of the other three epitopes, and the Scansite Motif Scan web tool (http://scansite.mit.edu/motifscan_seq.phtml) did not suggest any kinase candidate even under lowest stringency settings. Identification of additional phosphorylation sites and the underlying signaling pathways including kinases and phosphatases acting on OxPhos complexes will ultimately reveal how mitochondrial activity and function is integrated into various cellular and organismal processes.
Acknowledgments
The Center for Molecular Medicine and Genetics, Wayne State University School of Medicine, Detroit is gratefully acknowledged for supporting this work. We thank Drs. Jeffrey W. Doan and Lawrence I. Grossman for suggestions on the manuscript.
Footnotes
Abbreviations: CcO, cytochrome c oxidase; Cyt c, cytochrome c; ETC, electron transport chain; IMAC/nano-LC/ESI-MS, immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry; OxPhos, oxidative phosphorylation; SAP, shrimp alkaline phosphatase.
References
- 1.Li K, Li Y, Shelton JM, Richardson JA, Spencer E, Chen ZJ, Wang X, Williams RS. Cytochrome c deficiency causes embryonic lethality and attenuates stress-induced apoptosis. Cell. 2000;101:389–99. doi: 10.1016/s0092-8674(00)80849-1. [DOI] [PubMed] [Google Scholar]
- 2.Morriss GM, New DA. Effect of oxygen concentration on morphogenesis of cranial neural folds and neural crest in cultured rat embryos. J Embryol Exp Morphol. 1979;54:17–35. [PubMed] [Google Scholar]
- 3.Green DR. Apoptotic pathways: paper wraps stone blunts scissors. Cell. 2000;102:1–4. doi: 10.1016/s0092-8674(00)00003-9. [DOI] [PubMed] [Google Scholar]
- 4.Kroemer G, Dallaporta B, Resche-Rigon M. The mitochondrial death/life regulator in apoptosis and necrosis. Annu Rev Physiol. 1998;60:619–42. doi: 10.1146/annurev.physiol.60.1.619. [DOI] [PubMed] [Google Scholar]
- 5.Skulachev VP. Cytochrome c in the apoptotic and antioxidant cascades. FEBS Lett. 1998;423:275–80. doi: 10.1016/s0014-5793(98)00061-1. [DOI] [PubMed] [Google Scholar]
- 6.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritow VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nature Chem Biol. 2005;1:223–32. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 7.Pereverzev MO, Vygodina TV, Konstantinov AA, Skulachev VP. Cytochrome c, an ideal antioxidant. Biochem Soc Trans. 2003;31:1312–5. doi: 10.1042/bst0311312. [DOI] [PubMed] [Google Scholar]
- 8.Hennig B. Change of cytochrome c structure during development of the mouse. Eur J Biochem. 1975;55:167–83. doi: 10.1111/j.1432-1033.1975.tb02149.x. [DOI] [PubMed] [Google Scholar]
- 9.Hüttemann M, Jaradat S, Grossman LI. Cytochrome c oxidase of mammals contains a testes-specific isoform of subunit VIb--the counterpart to testes-specific cytochrome c? Mol Reprod Dev. 2003;66:8–16. doi: 10.1002/mrd.10327. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z, Lin H, Ye S, Liu QY, Meng Z, Zhang CM, Xia Y, Margoliash E, Rao Z, Liu XJ. Remarkably high activities of testicular cytochrome c in destroying reactive oxygen species and in triggering apoptosis. Proc Natl Acad Sci U S A. 2006;103:8965–70. doi: 10.1073/pnas.0603327103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson-Miller S, Brautigan DL, Margoliash E. Correlation of the kinetics of electron transfer activity of various eukaryotic cytochromes c with binding to mitochondrial cytochrome c oxidase. J Biol Chem. 1976;251:1104–15. [PubMed] [Google Scholar]
- 12.Ludwig B, Bender E, Arnold S, Hüttemann M, Lee I, Kadenbach B. Cytochrome C oxidase and the regulation of oxidative phosphorylation. Chembiochem. 2001;2:392–403. doi: 10.1002/1439-7633(20010601)2:6<392::AID-CBIC392>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 13.Hüttemann M, Lee I, Samavati L, Yu H, Doan JW. Regulation of mitochondrial oxidative phosphorylation through cell signaling. Biochim Biophys Acta. 2007;1773:1701–20. doi: 10.1016/j.bbamcr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Lee I, Salomon AR, Yu K, Doan JW, Grossman LI, Hüttemann M. New prospects for an old enzyme: mammalian cytochrome c is tyrosine-phosphorylated in vivo. Biochemistry. 2006;45:9121–8. doi: 10.1021/bi060585v. [DOI] [PubMed] [Google Scholar]
- 15.Kadenbach B, Urban PF. Application of a quantitative chromatographic method of purification in the study of the biosynthesis of cytochrome c. Zeitschrift für analytische Chemie. 1968;243:542–54. [Google Scholar]
- 16.Lee I, Salomon AR, Ficarro S, Mathes I, Lottspeich F, Grossman LI, Hüttemann M. cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity. J Biol Chem. 2005;280:6094–100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4:1240–50. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Kim JE, White FM. Quantitative analysis of phosphotyrosine signaling networks triggered by CD3 and CD28 costimulation in Jurkat cells. J Immunol. 2006;176:2833–43. doi: 10.4049/jimmunol.176.5.2833. [DOI] [PubMed] [Google Scholar]
- 19.Ficarro SB, Salomon AR, Brill LM, Mason DE, Stettler-Gill M, Brock A, Peters EC. Automated immobilized metal affinity chromatography/nano-liquid chromatography/electrospray ionization mass spectrometry platform for profiling protein phosphorylation sites. Rapid Commun Mass Spectrom. 2005;19:57–71. doi: 10.1002/rcm.1746. [DOI] [PubMed] [Google Scholar]
- 20.Eng J, McCormack A, Yates JR. An approach to correlate tandem mass spectral Data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spec. 1994;5:976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 21.Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. J Mol Biol. 1990;214:585–95. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 22.Flatmark T. Multiple molecular forms of bovine heart cytochrome c. V. A comparative study of their physicochemical properties and their reactions in biological systems. J Biol Chem. 1967;242:2454–9. [PubMed] [Google Scholar]
- 23.Hopper RK, Carroll S, Aponte AM, Johnson DT, French S, Shen RF, Witzmann FA, Harris RA, Balaban RS. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry. 2006;45:2524–36. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu T, Wang X, Purring-Koch C, Wei Y, McLendon GL. A mutational epitope for cytochrome C binding to the apoptosis protease activation factor-1. J Biol Chem. 2001;276:13034–8. doi: 10.1074/jbc.M009773200. [DOI] [PubMed] [Google Scholar]
- 25.Ko YH, Pan W, Inoue C, Pedersen PL. Signal transduction to mitochondrial ATP synthase: evidence that PDGF-dependent phosphorylation of the delta-subunit occurs in several cell lines, involves tyrosine, and is modulated by lysophosphatidic acid. Mitochondrion. 2002;1:339–48. doi: 10.1016/s1567-7249(01)00036-8. [DOI] [PubMed] [Google Scholar]