Abstract
Corin is a cardiac serine protease that acts as the pro-atrial natriuretic peptide (ANP) convertase. Recently, two single nucleotide polymorphisms (SNPs) (T555I and Q568P) in the human corin gene have been identified in genetic epidemiological studies. The minor I555/P568 allele, which is more common in African-Americans, is associated with hypertension and cardiac hypertrophy. In this study, we examined the effect of T555I and Q568P amino acid substitutions on corin function. We found that corin frizzled-like domain 2, where T555I/Q568P substitutions occur, was required for efficient pro-ANP processing in functional assays. Mutant corin lacking this domain had 30 ± 5% (p<0.01) activity compared to that of wild-type. Similarly, corin variant T555I/Q568P had a reduced (38 ± 7%, p<0.01) pro-ANP processing activity compared to that of wild-type. The variant also exhibited a low activity (44 ± 15%, p<0.05) in processing pro-brain natriuretic peptide (BNP). We next examined the biochemical basis for the loss of activity in T555I/Q568P variant, and found that the zymogen activation of the corin variant was impaired significantly, as indicated by the absence of the activated protease domain fragment. This finding was confirmed in human embryonic kidney (HEK) 293 cells and murine HL-1 cardiomyocytes. Thus, our results show that the corin gene SNPs associated with hypertension and cardiac hypertrophy impair corin zymogen activation and natriuretic peptide processing activity. Our data suggest that corin deficiency may be an important mechanism in hypertensive and heart diseases.
Keywords: natriuretic peptides, genetic variants, protease
Introduction
Atrial natriuretic peptide (ANP) is a cardiac hormone that regulates blood pressure and salt-water balance 1–3. The ANP pathway also has an antihypertrophic function in the heart, which is independent of its systemic action on blood pressure 4–7. Corin is a cardiac enzyme of the type II transmembrane serine protease family 8–10. It has a cytoplasmic tail and a transmembrane domain near the N-terminus. In its extracellular region, there are two frizzled-like domains, eight LDL receptor repeats, a scavenger receptor-like domain, and a C-terminal trypsin-like protease domain. Corin converts pro-ANP into active ANP 11, 12. In mice, corin deficiency prevented pro-ANP activation 13, demonstrating that corin is the physiological pro-ANP convertase. Corin null mice developed hypertension and cardiac hypertrophy 13. Corin also cleaved pro-brain natriuretic peptide (BNP) in vitro, although the cleavage was less efficient than that for pro-ANP 12.
Hypertension is the most common cardiovascular disease and its prevalence is even higher in African-Americans but the underlying mechanism is unclear 14, 15. Recently, two non-synonymous and non-conservative single nucleotide polymorphisms (SNPs) (T555I and Q568P) are found in the human corin gene. These two SNPs are in complete linkage disequilibrium in the population and, as a result, are co-localized in a minor allele (I555/P568) 16. Epidemiological studies of large population-based cohorts, including the Dallas Heart Study, the Multi-Ethnic Study of Atherosclerosis (MESA), and the Chicago Genetics of Hypertension Study, have shown that the minor corin I555/P568 allele is more common in African-Americans than in Caucasians (~12% vs. <0.2% carrying one or more copies of the allele) and associated with an increased risk for hypertension 16. Moreover, this corin minor allele is associated with an enhanced concentric cardiac hypertrophic response to high systolic blood pressure in African-Americans from the Dallas Heart and MESA cohorts 17, 18. The results suggest that the SNPs may impair corin function, thereby attenuating the antihypertensive and antihypertrophic actions of the natriuretic peptide system.
T555I and Q568P amino acid changes caused by the SNPs occur in the second frizzled-like domain in the corin propeptide 16, 19. Both T555 and Q568 residues are conserved in corin molecules of most vertebrates. To date, the functional significance of T555I and Q568P changes has not been determined. Here, we examined the effect of T555I and Q568P, either individually or together, on corin biological activity. We show that corin variant T555I/Q568P, but not variants T555I or Q568P, had a markedly reduced natriuretic peptide processing activity. We further identified impaired zymogen activation as a molecular basis for the defect of T555I/Q568P variant.
Materials and Methods
Cell Culture
Human embryonic kidney (HEK) 293 cells were cultured in Dulbeco’s Modified Eagle’s medium with 10% fetal bovine serum (FBS). The murine HL-1 cells 20, a generous gift from Dr. William Claycomb (Louisiana State University Medical Center), were cultured in Claycomb medium (JRH Biosciences) with 10% FBS, 100 μM norepinephrine, and 4 mM L-glutamine in gelatin/fibronectin-coated flasks.
Expression Plasmid Constructs
Plasmids expressing human wild-type corin and active site mutant S985A were described previously 21. Recombinant corin proteins contained a C-terminal V5 tag to facilitate protein detection. Plasmids expressing human corin variants T555I, Q568P and T555I/Q568P were made by site-directed mutagenesis using plasmid pcDNAhumanCorin 21 as a template with the following oligonucleotide primers: sense 5′-GTG GCC TGA AGA CAT AGA TTG-3′ and anti-sense 5′-CAA TCT ATG TCT TCA GGC CAC-3′ for mutant T555I; sense 5′-CAG ACA ATC CAA CCT GCC TG-3′ and anti-sense 5′-CAG GCA GGT TGG ATT GTC TG-3′for mutant Q568P. To express corin proteins (WT-EK and I/P-EK) with an enteropeptidase (EK) cleavage site, plasmids were made by mutagenesis to replace the original amino acid sequence RMNKR at the zymogen activation cleavage site with an EK recognition sequence DDDDK.
Plasmids expressing human corin frizzle domain 1 deletion mutant (ΔFz1), and LDL receptor repeats 1–4 deletion mutant (ΔR1–4) were described previously 22. Plasmid expressing human corin frizzled domain 2 deletion mutant (ΔFz2) was made by site-directed mutagenesis. All recombinant corin proteins from this series contained an N-terminal Xpress tag and were detected by an anti-Xpress antibody (Invitrogen) 22.
Transfection, Immunoprecipitation and Western blotting
HEK 293 and HL-1 cells were transfected with plasmids using FuGENE reagent (Roche Diagnostics). Conditioned medium was collected overnight. Cells were lysed in a buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100 (v/v), and a protease inhibitor cocktail (1:100 dilution, Sigma). Immunoprecipitation and Western blotting were done, as described previously 21.
Natriuretic Peptide Processing
Conditioned medium with pro-ANP was prepared in transfected cells. The medium was then added to HEK 293 cells expressing corin proteins, and incubated at 37°C for 1 h. Pro-ANP and ANP in the conditioned medium were analyzed by immunoprecipitation and Western blotting, as described previously 23. To quantify pro-ANP processing, Western blots were scanned by a densitometer (The PharosFx System, Bio-Rad). The percentage of pro-ANP to ANP conversion was calculated using the Quantity One 1-D Analysis software (Bio-Rad). For pro-BNP processing assays, a stable cell line was established expressing human pro-BNP. Plasmids expressing wild-type corin and variants were transfected in the cells. Conditioned medium was collected overnight. Pro-BNP processing was analyzed similarly as for pro-ANP processing.
Cell Surface Protein Expression
HEK 293 cells expressing corin proteins were incubated with phosphate-buffered saline (PBS), pH 8.0, containing 1 mM sulfo-NHS-biotin (Pierce) at room temperature for 30 min. Reactions were quenched by adding PBS containing 100 mM glycine. The cells were washed with PBS and lysed in a buffer 21. Streptavidin-Sepharose beads (30 μL) were added to cell lysate (300 μL) and mixed at 4°C for 2 h. The beads were washed with the buffer, and boiled in a sample buffer. Proteins were analyzed by Western blotting using an anti-V5 antibody.
Statistical Analysis
All data are presented as means ± S.D. Multigroup comparisons were done by ANOVA followed by post hoc least significant difference and Turkey honestly significant difference tests. A p value of <0.05 was considered to be statistically significant.
Results
Importance of Corin Frizzled-like Domains for Pro-ANP Processing
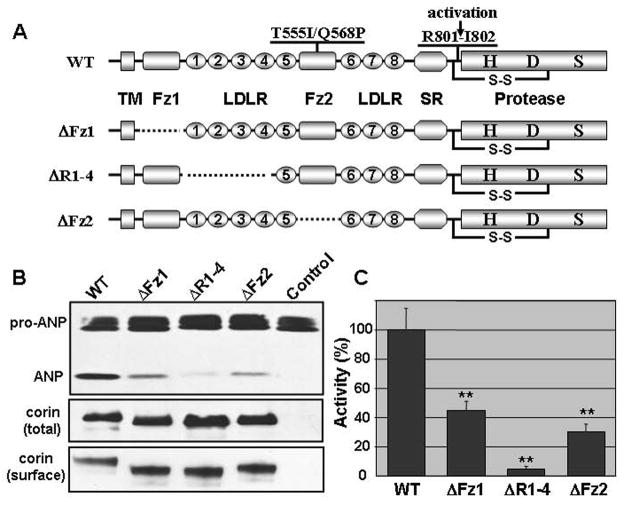
The two SNPs found in patients with hypertension are located in corin gene exon 12 16, 19, causing T555I and Q568P changes in frizzled-like domain 2. Previously, we found that propeptide domains such as frizzled-like domain 1 and LDLR repeats 1–4 were required for corin-mediated pro-ANP processing 22, but the functional importance of corin frizzled-like domain 2 was not examined. In this study, we made a mutant corin without frizzled-like domain 2 (ΔFz2) and tested its activity. We used wild-type corin and mutants lacking either frizzled-like domain 1 (ΔFz1) or LDLR repeats 1–4 (ΔR1–4) as positive and negative controls, respectively (Fig. 1A). Recombinant pro-ANP was incubated with HEK 293 cells expressing wild-type and corin mutants. Pro-ANP processing was analyzed by immunoprecipitation and Western blotting. As shown in Figs. 1B (top panel) and 1C, ΔFz2 mutant had 30 ± 5% of activity compared to that of wild-type. Consistent with our previous results 22, ΔFz1 and ΔR1–4 mutants had 44 ± 7 and 4 ± 3% of activities, respectively (Figs. 1B and 1C). In controls, comparable levels of total recombinant corin proteins were confirmed in the transfected cells (Fig. 1B, middle panel). By biotin-labeling of cell surface proteins followed by immunoprecipitation and Western blotting, we also confirmed that these corin proteins were present on the cell surface (Fig. 1B, bottom panel). The results show that corin frizzled-like domain 2 is important for pro-ANP processing.
Fig. 1.
A. Domain structures of corin and deletion mutants. The transmembrane domain (TM), frizzled-like domains (Fz), LDL receptor repeats (LDLR), scavenger receptor-like domain (SR), and protease domain (Protease) with active residues His (H), Asp (D), and Ser (S) are indicated. The activation cleavage site is shown by an arrow. In mutants, deleted domains are shown by dashed lines. B. Pro-ANP processing by corin mutants. Pro-ANP was incubated with cells expressing wild-type (WT) corin and mutants (ΔFz1, ΔR1–4, and ΔFz2) or vector-transfected control cells. Pro-ANP processing was analyzed by Western blotting (top panel). Total (middle panel) and cell surface (bottom panel) corin protein expression in the transfected cells was verified by Western blotting. C. Pro-ANP processing activities of corin mutants. Pro-ANP processing activity was quantified by densitometric analysis of Western blots. The data represent mean ± S.D. from three independent experiments. **p<0.01 vs. WT.
Pro-ANP Processing Activity of T555I/Q568P Variant
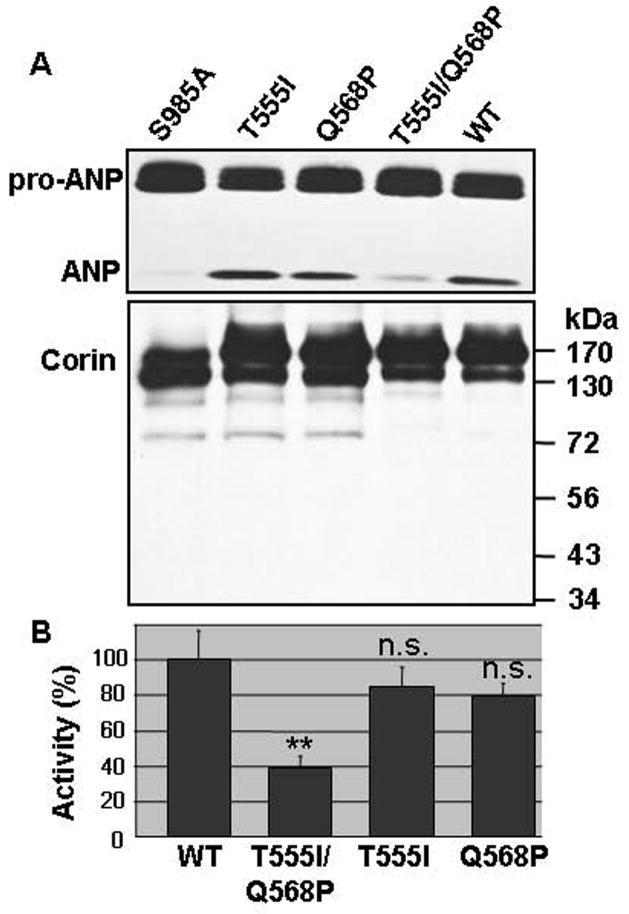
To examine if T555I/Q568P changes alter corin function, we transfected HEK 293 cells with plasmids expressing human pro-ANP or corin variants in separate plates. Conditioned medium containing recombinant pro-ANP then was collected and added to the cells expressing corin proteins. After 1-h at 37°C, the conditioned medium was analyzed for pro-ANP processing. As shown in Fig. 2A (top), pro-ANP was processed similarly by wild-type corin and variants T555I and Q568P. In contrast, the activity of variant T555I/Q568P was greatly reduced. In these experiments, a negative control, corin S985A, in which the active site Ser was mutated to Ala, was used. This inactive mutant produced little ANP (Fig. 2A, top). The Western blots were quantified by densitometry. Values from S985A negative control were used as background and subtracted in calculating values for wild-type and variants. As shown in Fig. 2B, T555I/Q568P variant had 38 ± 7% of activity compared to that of wild-type (n=6, p<0.01). Variants T555I (84 ± 12%) and Q568P (80 ± 8%) had lower activities than that of wild-type, but the differences were not statistically significant (n=6, p>0.05). We verified corin proteins in the transfected cells by Western blotting using an anti-C-terminal V5 tag antibody and found comparable levels of corin proteins, which appeared as two band of ~150 and ~170 kDa (Fig. 2A, lower panel), representing differentially N-glycosylated molecules 21. These results indicate that T555I/Q568P variant had a markedly reduced pro-ANP processing activity.
Fig. 2.
Processing pro-ANP by corin variants. A. Human pro-ANP was expressed in HEK 293 cells and then added to the cells expressing wild-type corin and variants. Pro-ANP processing was analyzed (top panel). Corin protein expression in the transfected cells was verified by Western blotting (lower panel). B. Pro-ANP processing activities of corin variants. Pro-ANP processing activity was quantified by densitometric analysis of Western blots. The data represent mean ± S.D. from six independent experiments. **p<0.01 vs. WT. n.s., not statistically significant vs. WT.
Pro-BNP Processing by Corin Variants
Previously, we showed that corin also cleaved pro-BNP, although the reaction was less efficient than that for pro-ANP 12. To test pro-BNP processing by corin variants, we transfected plasmids for corin variants in HEK 293 cells stably expressing human pro-BNP. The conditioned medium was collected and pro-BNP processing was analyzed. As shown in Fig. 3A, a small fraction of pro-BNP was processed in control 293 cells (control), possibly by an unknown protease. The processing was enhanced in cells expressing wild-type corin and variants T555I and Q568P, but was barely increased in cells expressing variant T555I/Q568P. By densitometric analysis of Western blots (Fig. 3B), variant T555I/Q568P had 44 ± 15% of activity (n=3, p<0.05) compared to that of wild-type, whereas variants T555I and Q568P had 87 ± 22% and 81 ± 6% of the activity, respectively (n=3, p values >0.05). The results support that variant T555I/Q568P had a reduced biological activity.
Fig. 3.
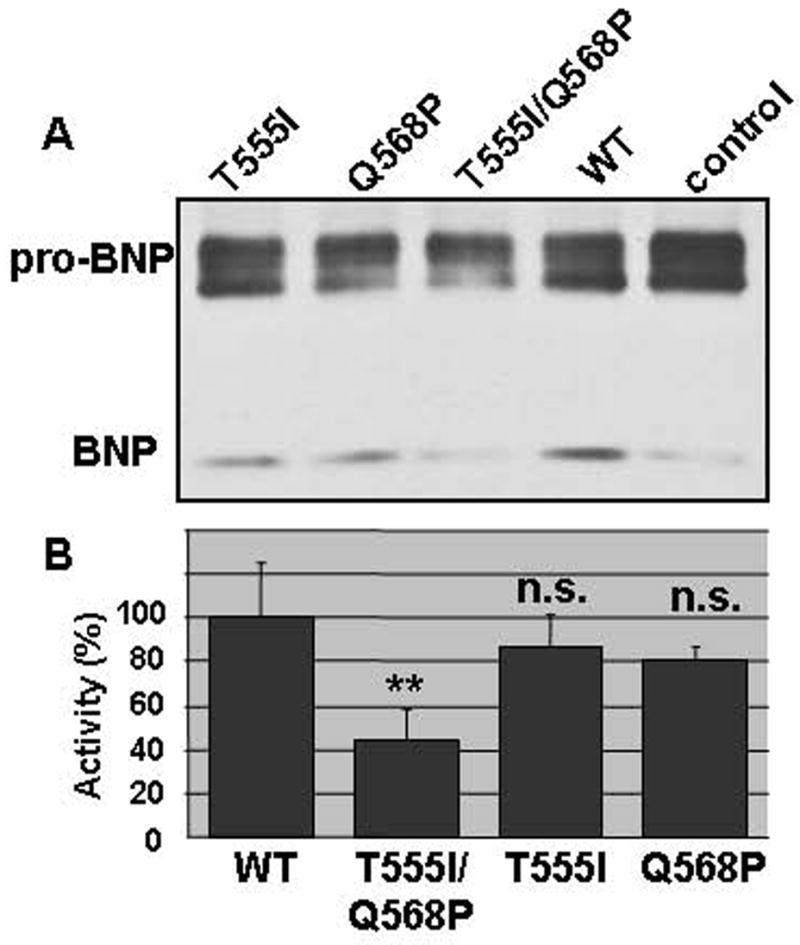
Processing pro-BNP by corin variants. A. Human pro-BNP was stably expressed in HEK 293 cells. The cells were transfected with plasmids expressing wild-type corin or variants. Conditioned medium was collected and pro-BNP processing was analyzed by immunoprecipitation and Western blotting. B. Pro-ABP processing activity was quantified by densitometric analysis of Western blots. The data represent mean ± S.D. from three independent experiments. **p<0.05 vs. WT. n.s., not statistically significant vs. WT.
Cell Surface Expression of Corin Variants
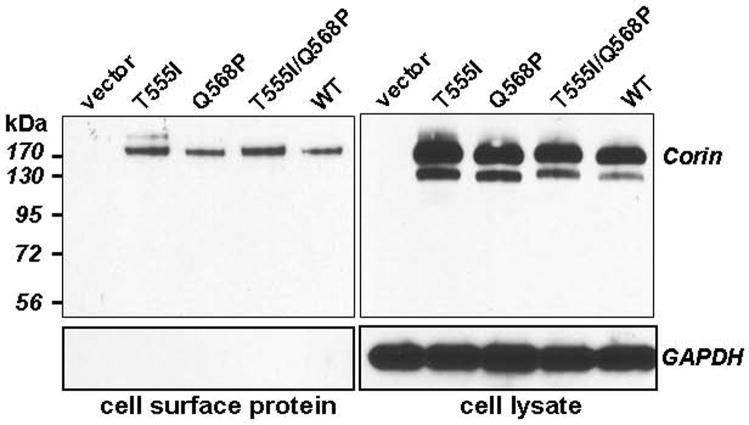
Corin is a transmembrane protein. We next examined if the reduced activity of variant T555I/Q568P was caused by low cell surface expression. Total surface proteins in transfected cells were labeled by biotinylation and precipitated using Streptavidin-Sepharose beads followed by Western blotting. Fig. 4 (top left) shows similar levels of biotin-labeled corin proteins in HEK 293 cells expressing wild-type corin and variants. Densitometric analysis of four independent experiments showed that the expression levels for variants T555I, Q568P, and T555I/Q568P were 120 ± 13, 114 ± 8, and 96 ± 8% of the wild-type (p values >0.05). In controls, similar levels of corin proteins in cell lysates were found by Western analysis (Fig. 4, top right). As another control for the specificity of cell surface labeling, an antibody against intracellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) detected the protein in cell lysate but not in Streptavidin precipitates (Fig. 4, lower panels). The results show that T555I/Q568P changes did not prevent corin expression on the cell surface.
Fig. 4.
Cell surface expression of corin proteins. Cell surface proteins in HEK 293 cells expressing corin proteins or control cells were biotinylated and analyzed by Streptavidin precipitation followed by Western blotting using an anti-V5 antibody (top left). As a control, total corin proteins in cell lysates were verified using the same antibody (top right). As another control for cell surface protein labeling, the same Western blots were analyzed by an antibody against GAPDH (lower panels). The results shown were representative of four independent experiments.
Impaired Zymogen Activation for Variant T555I/Q568P
Like most trypsin-like proteases, corin is made as a zymogen, which is activated proteolytically at the conserved site, Arg-801↓Ile-802 24, 25 (Fig. 1A). A disulfide bond links the activated protease fragment to the propeptide. On Western blots under reducing conditions, this fragment is expected to migrate as a band of ~35–40 kDa whereas it should attach to the propeptide under non-reducing conditions. In transfected cells, corin had activity but the activated protease fragment was not readily detectable (Fig. 2A, lower panel), indicating that only a small fraction of the total corin molecules was activated, making it difficult to assess the status of corin activation. To circumvent this problem, we used immunoprecipitation to isolate all recombinant corin proteins in cell lysates instead of analyzing only a fraction of samples. The rationale was to enrich corin proteins and hence detect the activated protease fragment.
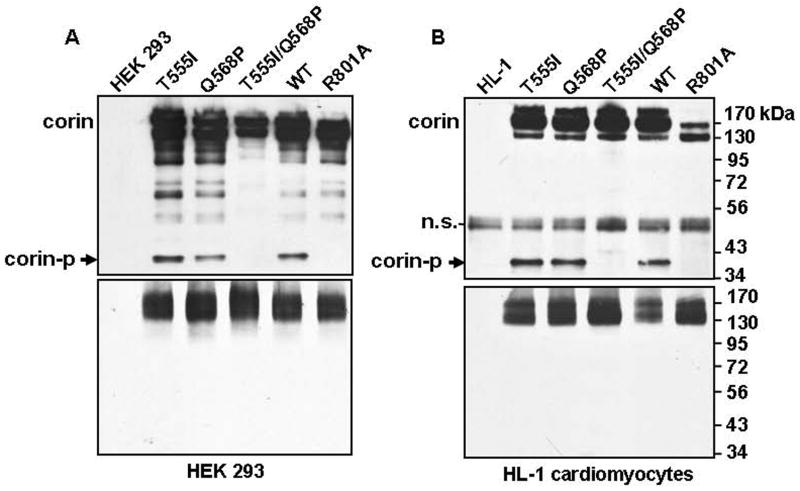
We transfected HEK 293 cells with plasmids expressing wild-type corin and variants. As a control, we included a plasmid encoding corin mutant R801A, in which the activation site Arg-801 was replaced by Ala (Fig. 1A). This mutation prevents corin activation cleavage and, as a result, mutant R801A had no detectable pro-ANP processing activity 21, 24. Lysates from the transfected cells were immunoprecipitated by an anti-V5 antibody to pull down recombinant corin proteins and then analyzed by Western blotting under reducing and non-reducing conditions. As shown in Fig. 5A (top), an ~40-kDa band was detected in samples containing wild-type corin and variants T555I and Q568P, but not variant T555I/Q568P. This band also was absent in mutant R801A sample, indicating that it was the activated protease fragment. Consistent with the predicted corin structure, the protease fragment was absent under non-reducing conditions (Fig. 5A, bottom). Analysis of five independent experiments by densitometry showed that the activated protease fragment represented 20 ± 3, 22 ± 3, and 20 ± 4%, respectively, of total corin protein for the wild-type and variants T555I and Q568P. This fragment was undetectable in variant T555I/Q568P and mutant R801A samples, suggesting that T555I/Q568P amino acid changes impaired corin zymogen activation.
Fig. 5.
Analysis of corin zymogen activation. Recombinant corin proteins were expressed in HEK 293 (A) or HL-1 (B) cells. Corin proteins in cell lysates were immunoprecipitated and analyzed by Western blotting using an anti-V5 tag antibody under reducing (top panels) and non-reducing (lower panels) conditions. Cell lysates from parental HEK 293 cells or HL-1 cells were included as controls. The corin protease fragment (corin-p), represented by an ~40-kDa band, was indicated by an arrow. A non-specific (n.s.) band was present in all samples from HL-1 cells under reducing conditions (B, top panel). Data shown were representative of at least three independent experiments.
We verified these results in physiologically relevant murine HL-1 cardiomyocytes, which retain phenotypic characteristics of adult cardiomyocytes 20, 26. A recent study indicates that corin activation in HL-1 cells is similar to that in the heart 27. HL-1 cells were transfected with corin expressing plasmids. Recombinant corin proteins in cell lysates were analyzed by immunoprecipitation and Western blotting. As shown in Fig. 5B (top), the activated corin protease fragment, represented by the ~40-kDa band, was detected under reducing conditions in samples of wild-type and variants T555I and Q568P, but not variant T555I/Q568P or mutant R801A. A non-specific band of ~50 kDa was present in all samples including parental HL-1 cells under reducing conditions (Fig. 5B, top). Similar to the results from HEK 293 cells, the activated corin protease fragment was not detected under non-reducing conditions (Fig. 5B, bottom). Analysis of three independent experiments by densitometry showed that the activated protease fragment represented 16 ± 1, 19 ± 1, and 17 ± 1%, respectively, of total wild-type and variants T555I and Q568P proteins. The fragment was undetectable for variant T555I/Q568P and mutant R801A. The results show that impaired zymogen activation of variant T555I/Q568P occurred not only in HEK 293 cells but also in cardiomyocytes.
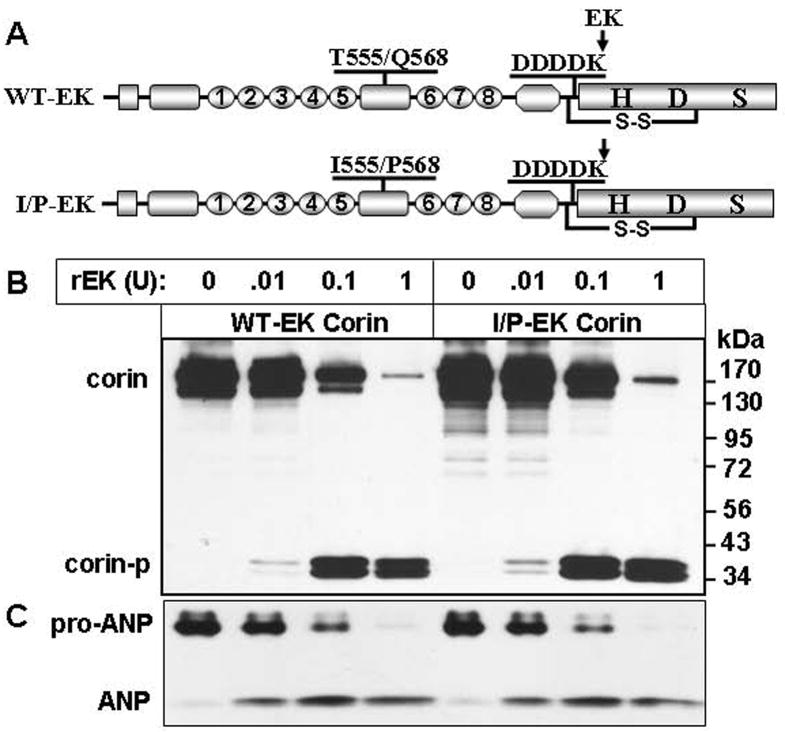
Restoration of Variant T555I/Q568P Activity by An EK Cleavage Site
To confirm that T555I/Q568P changes alter only corin zymogen activation but not substrate recognition, we designed recombinant corin proteins that can be activated in a controlled manner. Mutagenesis was made to replace the corin activation sequence RMNKR at residues 797–801 with a specific enteropeptidase (EK) cleavage sequence DDDDK 24 (Fig. 6A). Wild-type and variant T555I/Q568P versions of corin with the EK site were expressed in HEK 293 cells. Cell lysates were prepared and incubated with increasing concentrations of recombinant human EK. Western blotting showed that WT-EK and variant T555I/Q568P-EK proteins were activated similarly by EK in a dose-dependent manner (Fig. 6B). More importantly, the EK-activated corin variant T555I/Q568P converted pro-ANP to ANP as efficiently as WT-EK in functional assays (Fig. 6C). In controls, recombinant EK did not convert pro-ANP to ANP (data not shown). These results show that T555I/Q568P variant, once activated, functioned normally, indicating that the impaired zymogen activation is responsible primarily for the reduced activity of the variant.
Fig. 6.
Activation and activity of WT-EK and T555I/Q568P-EK corin proteins. A. Schematic illustrations of WT (WT-EK) and T555I/Q568P (I/P-EK) versions of corin that contain an EK recognition sequence (DDDDK) at the conserved activation site. B. Recombinant WT-EK and I/P-EK were activated by increasing concentrations of recombinant EK (rEK), as shown by Western analysis under reducing conditions. C. The activity of EK-activated WT-EK and I/P-EK was tested in pro-ANP processing assays by Western blotting. The data shown were representative of three independent experiments.
Discussion
The ANP pathway is important in regulating blood pressure 1–3. Knockout mice lacking either ANP or its receptor are hypertensive 28, 29. Corin is an essential enzyme for ANP production 13. Recently, human corin gene SNPs have been found to be associated with an increased risk for hypertension and cardiac hypertrophy 16, 18, suggesting that gene variants involved in the natriuretic peptide system may contribute to hypertensive disease. Further studies are needed to determine if the gene variants alter corin function.
In this report, we characterized human corin variants in functional studies. We showed that frizzled-like domain 2, where the naturally occurring variants occur, was required for corin to process pro-ANP efficiently (Fig. 1). Previously, we showed that corin frizzled-like domain 1 and LDLR 1–4 repeats are important for pro-ANP processing 24. The new data indicate that other domains such as frizzled-like domain 2 are also important for corin function. Consistently, in other transmembrane serine proteases such as enteropeptidase and matriptase, structural elements in the propeptide region are critical for their interactions with substrates and inhibitors 30, 31. We also showed that corin variant T555I/Q568P had significantly lower activities in processing pro-ANP and pro-BNP (Figs. 2 and 3) than that of wild-type, indicating that the amino acid substitutions caused by the SNPs altered corin protein structure, thereby lowering its biological activity.
To understand the molecular basis for the reduced activity in variant T555I/Q568P, we examined the cell surface expression of corin variants and found similar expression levels for wild-type and the variants (Fig. 4), indicating that the cell surface expression is not altered by the amino acid changes. We next examined the zymogen activation of corin variants. As a trypsin-like protease, corin is made as a zymogen that is activated by cleavage at a conserved site 24, 25. For many proteolytic enzymes, zymogen activation represents a critical step in regulating their activities. Our recent data indicated that a protease present in HEK 293 cells and cardiomyocytes was responsible for initial activation of corin, although the identity of this enzyme remains unknown 21.
In transfected HEK 293 cells and HL-1 cardiomyocytes, we showed that corin variant T555I/Q568P activation was impaired, as indicated by the absence of the activated protease fragment (Fig. 5). Because T555I/Q568P variant still exhibited some activities, it is likely that some variant molecules were activated but not enough to be detected by the methods used in our studies. To confirm that impaired zymogen activation is the principal mechanism responsible for the reduced activity of the corin variant, we made wild-type and T555I/Q568P versions of corin that can be activated by EK. We showed that, once activated, the variant and wild-type versions of corin had similar activities toward pro-ANP (Fig. 6). Together, the results indicate that the naturally occurring variants reduce corin function by impairing zymogen activation. Our data are consistent with findings from other trypsin-like proteases in which mutations in the propeptide cause disease. For example, mutations in prothrombin propeptide are shown to prevent the zymogen activation, resulting in bleeding in patients 32. In TMPRSS3, another membrane protease involved in hearing, mutations in its propeptide prevent zymogen activation and cause autosomal recessive deafness 33. Because frizzled-like domains are present in many biologically important proteins including Wnt signaling molecules, mutations in different frizzled-like domains may also cause other human diseases.
Interestingly, the activity and zymogen activation of corin variants with single amino acid change, either T555I or Q568P, did not appear to be reduced significantly. In functional assays, T555I and Q568P variants had slightly lower activities than that of wild-type but the difference was not statistically significant. At this time, the corin crystal structure is not available. Based on a computer model using mouse Frizzled 8 structure as a template 34, single amino acid change, either T555I or Q568P, could alter the corin frizzled-like domain structure (data not shown). It appears, however, that conformational changes induced by single amino acid change are not sufficient to disrupt corin function, suggesting that more profound structural alterations occur when both T555 and Q568 residues are substituted. In mouse Frizzled 8, the frizzled domain is involved in protein dimerization and amino acid changes in this domain abolish its function 34. At this time, we do not have experimental evidence that corin protein dimerizes or its frizzled domains interact, but such possibilities can not be excluded. The population-based genetic studies have shown that T555I and Q568P SNPs are in complete linkage disequilibrium 16. The minor allele, which is associated with hypertension and cardiac hypertrophy, encodes both I555 and P568 16. As a result, the corin protein encoded by this allele is expected to have a lower activity. It will be interesting to know if corin alleles with one SNP, either T555I or Q568P, exist in the general population and if such alleles are associated with a hypertensive phenotype.
Hypertension is a major cardiovascular disease but its pathogenesis remains unknown in most patients with the disease 35. Genome-wide searches for SNPs associated with hypertension yielded few candidate genes 36, which may reflect the fact that blood pressure is regulated by many biological pathways. Our studies have established corin as the long-sought pro-ANP convertase 13. The identification of the naturally occurring corin variants in patients with hypertension links the corin gene to human disease 16–18. Interestingly, I555/P568 corin allele is found mostly in African-Americans, a population known for its high prevalence of salt-sensitive hypertension 37, 38. In mice, corin deficiency causes salt-sensitive hypertension and cardiac hypertrophy 13. Here we show that the naturally occurring corin variant had a markedly reduced activity in vitro, which was most likely caused by impaired zymogen activation. At this time, we do not know if corin T555I/Q568P variant is associated with impaired pro-ANP/pro-BNP processing in vivo, nor do we know if this variant directly causes hypertension and cardiac hypertrophy in patients. Further studies also will be important to examine if corin expression, activity, and regulation in the myocardium are affected in individuals carrying this polymorphism. Our findings should encourage more genetic studies to screen additional corin mutations in patients with hypertension and heart disease. The results will help to understand the role of this newly-discovered enzyme in cardiovascular disease.
Acknowledgments
We thank Dr. William Claycomb for providing HL-1 cells, and Drs. Edward Plow and Martin Schreiber for their support.
Sources of funding: This work was supported in part by a start-up fund from the Cleveland Clinic and grants from Ralph Wilson Medical Research Foundation and the National Institutes of Health.
Footnotes
Disclosures: None
Publisher's Disclaimer: This is an un-copyedited author manuscript accepted for publication in Circulation Research, copyright The American Heart Association. This may not be duplicated or reproduced, other than for personal use or within the "Fair Use of Copyrighted Materials" (section 107, title 17, U.S. Code) without prior permission of the copyright owner, The American Heart Association. The final copyedited article, which is the version of record, can be found athttp://circres.hajournals.org/. The American Heart Association disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by the National Institutes of Health or other parties.
References
- 1.Lee CY, Burnett JC., Jr Natriuretic peptides and therapeutic applications. Heart Fail Rev. 2007;12:131–142. doi: 10.1007/s10741-007-9016-3. [DOI] [PubMed] [Google Scholar]
- 2.McGrath MF, de Bold ML, de Bold AJ. The endocrine function of the heart. Trends Endocrinol Metab. 2005;16:469–477. doi: 10.1016/j.tem.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev. 2006;27:47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- 4.Calderone A, Thaik CM, Takahashi N, Chang DL, Colucci WS. Nitric oxide, atrial natriuretic peptide, and cyclic GMP inhibit the growth-promoting effects of norepinephrine in cardiac myocytes and fibroblasts. J Clin Invest. 1998;101:812–818. doi: 10.1172/JCI119883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holtwick R, van Eickels M, Skryabin BV, Baba HA, Bubikat A, Begrow F, Schneider MD, Garbers DL, Kuhn M. Pressure-independent cardiac hypertrophy in mice with cardiomyocyte-restricted inactivation of the atrial natriuretic peptide receptor guanylyl cyclase-A. J Clin Invest. 2003;111:1399–1407. doi: 10.1172/JCI17061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knowles JW, Esposito G, Mao L, Hagaman JR, Fox JE, Smithies O, Rockman HA, Maeda N. Pressure-independent enhancement of cardiac hypertrophy in natriuretic peptide receptor A-deficient mice. J Clin Invest. 2001;107:975–984. doi: 10.1172/JCI11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silberbach M, Gorenc T, Hershberger RE, Stork PJ, Steyger PS, Roberts CT., Jr Extracellular signal-regulated protein kinase activation is required for the anti-hypertrophic effect of atrial natriuretic factor in neonatal rat ventricular myocytes. J Biol Chem. 1999;274:24858–24864. doi: 10.1074/jbc.274.35.24858. [DOI] [PubMed] [Google Scholar]
- 8.Hooper JD, Clements JA, Quigley JP, Antalis TM. Type II transmembrane serine proteases. J Biol Chem. 2001;276:857–860. doi: 10.1074/jbc.R000020200. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q. Type II transmembrane serine proteases. Curr Top Dev Biol. 2003;54:167–206. doi: 10.1016/s0070-2153(03)54009-1. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q. The serine protease corin in cardiovascular biology and disease. Front Biosci. 2007;12:4179–4190. doi: 10.2741/2379. [DOI] [PubMed] [Google Scholar]
- 11.Wu F, Yan W, Pan J, Morser J, Wu Q. Processing of pro-atrial natriuretic peptide by corin in cardiac myocytes. J Biol Chem. 2002;277:16900–16905. doi: 10.1074/jbc.M201503200. [DOI] [PubMed] [Google Scholar]
- 12.Yan W, Wu F, Morser J, Wu Q. Corina transmembrane cardiac serine protease, acts as a pro-atrial natriuretic peptide-converting enzyme. Proc Natl Acad Sci USA. 2000;97:8525–8529. doi: 10.1073/pnas.150149097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan JC, Knudson O, Wu F, Morser J, Dole WP, Wu Q. Hypertension in mice lacking the proatrial natriuretic peptide convertase corin. Proc Natl Acad Sci U S A. 2005;102:785–790. doi: 10.1073/pnas.0407234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields LE, Burt VL, Cutler JA, Hughes J, Roccella EJ, Sorlie P. The burden of adult hypertension in the United States 1999 to 2000: a rising tide. Hypertension. 2004;44:398–404. doi: 10.1161/01.HYP.0000142248.54761.56. [DOI] [PubMed] [Google Scholar]
- 15.Hajjar I, Kotchen TA. Trends in prevalence, awareness, treatment, and control of hypertension in the United States, 1988–2000. JAMA. 2003;290:199–206. doi: 10.1001/jama.290.2.199. [DOI] [PubMed] [Google Scholar]
- 16.Dries DL, Victor RG, Rame JE, Cooper RS, Wu X, Zhu X, Leonard D, Ho SI, Wu Q, Post W, Drazner MH. Corin gene minor allele defined by 2 missense mutations is common in blacks and associated with high blood pressure and hypertension. Circulation. 2005;112:2403–2410. doi: 10.1161/CIRCULATIONAHA.105.568881. [DOI] [PubMed] [Google Scholar]
- 17.Burnett JC, Jr, Olson TM. Natriuretic peptides and myocardial structure: insights from population genetics. Hypertension. 2007;49:765–766. doi: 10.1161/01.HYP.0000258567.67263.96. [DOI] [PubMed] [Google Scholar]
- 18.Rame JE, Drazner MH, Post W, Peshock R, Lima J, Cooper RS, Dries DL. Corin I555(P568) allele is associated with enhanced cardiac hypertrophic response to increased systemic afterload. Hypertension. 2007;49:857–864. doi: 10.1161/01.HYP.0000258566.95867.9e. [DOI] [PubMed] [Google Scholar]
- 19.Pan J, Hinzmann B, Yan W, Wu F, Morser J, Wu Q. Genomic structures of the human and murine corin genes and functional GATA elements in their promoters. J Biol Chem. 2002;277:38390–38398. doi: 10.1074/jbc.M205686200. [DOI] [PubMed] [Google Scholar]
- 20.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao X, Wang W, Chen S, Wu Q. Role of glycosylation in corin zymogen activation. J Biol Chem. 2007;282:27728–27735. doi: 10.1074/jbc.M703687200. [DOI] [PubMed] [Google Scholar]
- 22.Knappe S, Wu F, Madlansacay MR, Wu Q. Identification of domain structures in the propeptide of corin essential for the processing of proatrial natriuretic peptide. J Biol Chem. 2004;279:34464–34471. doi: 10.1074/jbc.M405041200. [DOI] [PubMed] [Google Scholar]
- 23.Wu C, Wu F, Pan J, Morser J, Wu Q. Furin-mediated processing of Pro-C-type natriuretic peptide. J Biol Chem. 2003;278:25847–25852. doi: 10.1074/jbc.M301223200. [DOI] [PubMed] [Google Scholar]
- 24.Knappe S, Wu F, Masikat MR, Morser J, Wu Q. Functional analysis of the transmembrane domain and activation cleavage of human corin. J Biol Chem. 2003;278:52363–52370. doi: 10.1074/jbc.M309991200. [DOI] [PubMed] [Google Scholar]
- 25.Yan W, Sheng N, Seto M, Morser J, Wu Q. Corin, a mosaic transmembrane serine protease encoded by a novel cDNA from human heart. J Biol Chem. 1999;274:14926–14935. doi: 10.1074/jbc.274.21.14926. [DOI] [PubMed] [Google Scholar]
- 26.White SM, Constantin PE, Claycomb WC. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol. 2004;286:H823–H829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 27.Gladysheva IP, Robinson BR, Houng AK, Kovats T, King SM. Corin is co-expressed with pro-ANP and localized on the cardiomyocyte surface in both zymogen and catalytically active forms. J Mol Cell Cardiol. 2008;44:131–142. doi: 10.1016/j.yjmcc.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 28.John SW, Krege JH, Oliver PM, Hagaman JR, Hodgin JB, Pang SC, Flynn TG, Smithies O. Genetic decreases in atrial natriuretic peptide and salt-sensitive hypertension. Science. 1995;267:679–681. doi: 10.1126/science.7839143. [DOI] [PubMed] [Google Scholar]
- 29.Lopez MJ, Wong SK, Kishimoto I, Dubois S, Mach V, Friesen J, Garbers DL, Beuve A. Salt-resistant hypertension in mice lacking the guanylyl cyclase-A receptor for atrial natriuretic peptide. Nature. 1995;378:65–68. doi: 10.1038/378065a0. [DOI] [PubMed] [Google Scholar]
- 30.Oberst MD, Williams CA, Dickson RB, Johnson MD, Lin CY. The activation of matriptase requires its noncatalytic domains, serine protease domain, and its cognate inhibitor. J Biol Chem. 2003;278:26773–26779. doi: 10.1074/jbc.M304282200. [DOI] [PubMed] [Google Scholar]
- 31.Zheng X, Sadler JE. Mucin-like domain of enteropeptidase directs apical targeting in Madin-Darby canine kidney cells. J Biol Chem. 2002;277:6858–6863. doi: 10.1074/jbc.m109857200. [DOI] [PubMed] [Google Scholar]
- 32.Miyata T, Morita T, Inomoto T, Kawauchi S, Shirakami A, Iwanaga S. Prothrombin Tokushima, a replacement of arginine-418 by tryptophan that impairs the fibrinogen clotting activity of derived thrombin Tokushima. Biochemistry. 1987;26:1117–1122. doi: 10.1021/bi00378a020. [DOI] [PubMed] [Google Scholar]
- 33.Guipponi M, Vuagniaux G, Wattenhofer M, Shibuya K, Vazquez M, Dougherty L, Scamuffa N, Guida E, Okui M, Rossier C, Hancock M, Buchet K, Reymond A, Hummler E, Marzella PL, Kudoh J, Shimizu N, Scott HS, Antonarakis SE, Rossier BC. The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum Mol Genet. 2002;11:2829–2836. doi: 10.1093/hmg/11.23.2829. [DOI] [PubMed] [Google Scholar]
- 34.Dann CE, Hsieh JC, Rattner A, Sharma D, Nathans J, Leahy DJ. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature. 2001;412:86–90. doi: 10.1038/35083601. [DOI] [PubMed] [Google Scholar]
- 35.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 36.Consortium TWTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calhoun DA, Oparil S. Racial differences in the pathogenesis of hypertension. Am J Med Sci. 1995;310:S86–S90. doi: 10.1097/00000441-199512000-00016. [DOI] [PubMed] [Google Scholar]
- 38.Manatunga AK, Jones JJ, Pratt JH. Longitudinal assessment of blood pressures in black and white children. Hypertension. 1993;22:84–89. doi: 10.1161/01.hyp.22.1.84. [DOI] [PubMed] [Google Scholar]