Summary
The promyelocytic leukemia protein PML and its associated nuclear bodies are hot topics of investigation. This interest arises for multiple reasons including the tight link between the integrity of PML nuclear bodies and several disease states and the impact of the PML protein and PML nuclear bodies on proliferation, apoptosis and viral infection. Unfortunately, an understanding of the molecular underpinnings of PML nuclear body function remains elusive. Here, a general overview of the PML field is provided and is extended to discuss whether some of the basic tenets of “PML-ology” are still valid. For instance, recent findings suggest that some components of PML nuclear bodies form bodies in the absence of the PML protein. Also, a new model for PML nuclear body function is proposed which provides a unifying framework for its effects on diverse biochemical pathways such as Akt signaling and the p53-Mdm2 axis. In this model, the PML protein acts as an inhibitor of gene expression post-transcriptionally via inhibiting a network node in the eIF4E RNA regulon. An example is given for how the PML RNA regulon model provided the basis for the development of a new anti-cancer strategy being tested in the clinic.
Keywords: PML, ND10, POD, eIF4E, RNA regulon/operon
Brief overview of the PML protein and PML nuclear bodies
In the last almost 20 years, many groups have focused their attention on the structure and function of the promyelocytic leukemia protein PML and its associated nuclear structures referred to as PML nuclear bodies (PML NBs), PML oncogenic domains (PODs), Kremer bodies or ND10s (nuclear dot 10). The PML protein was the first identified component of these nuclear structures and remains their defining feature. General interest in the PML field has arisen due to the disruption/alteration of PML nuclear bodies in several pathogenic conditions including acute promyelocytic leukemia (APL)[1–3] and polyglutamine repeat neurodegenerative diseases [4–6], as well as a wide variety of viral infections including HIV and lymphocytic choriomeningitis virus (LCMV) [7–10]. The PML protein is expressed in all mammalian tissue types reported thus far [11]. Importantly, the PML protein appears to be conserved in mammals but seems to be absent from lower eukaryotes and plants [12].
What has captured the attention of many investigators is the potent growth suppressive and apoptotic roles of the PML protein, and the correlation of these activities to the structural integrity of the nuclear bodies. When overexpressed, the PML protein inhibits cell cycle progression leading to G1/S arrest, suppresses some forms of oncogenic transformation, promotes apoptosis as well as Ras induced senescence [12–15]. Recent reports suggest that the PML protein may even play roles in the suppression of angiogenesis thereby impacting on metastases [16]. How does the PML protein achieve these physiological affects? At the molecular level, PML nuclear bodies have been suggested to play roles in mRNA export [17–21], DNA repair [2, 22–25], DNA replication [22, 26, 27], transcription [28–30], and PML nuclear bodies may act as sites for post-translational modifications (e.g. SUMO modification, acetylation and phosphorylation) [31–34], to name a few. There is no framework postulated that could provide a molecular basis for these diverse biochemical activities and thereby unite these seemingly unrelated functionalities. This may be difficult as it seems likely that all PML nuclear bodies will not be functionally equivalent in all contexts [12]. For instance, PML nuclear bodies are dynamic structures, which differ in position, size, number, motional state and composition as a function of conditions [12, 35–37].
Although the physiological impact of PML expression is reasonably well described, the molecular and biochemical basis for the physiological activities associated with the PML protein and PML nuclear bodies is poorly defined. Ultimately, one would like to determine PML nuclear body function at the same level that one understands how other cellular organelles work. With such an understanding one can begin to infer how these structures impact on cell physiology and how their dysregulation contributes to cancer and neurodegeneration.
When considering PML nuclear body function, a few facts should be kept in mind. These will be discussed in greater detail throughout this review. The first issue is the lack of a significant phenotype in PML−/− mice [12]. These mice apparently develop normally and have only slightly increased cancers later in life relative to controls [38]. Second, the PML gene is not evolutionarily conserved in eukaryotes, being absent from D. melanogaster, S. cerivisiae and A. thaliana where the sequencing of these genomes has been completed [12]. In contrast to the PML protein, some PML body constituents are highly conserved being found in at least a subset of these organisms. Finally, the PML protein was thought to be required for the organization of PML nuclear bodies i.e. for the organization of any of the components into spherical nuclear structures. However, this is not the case for at least endogenous eIF4E [17, 39, 40] and when human Sp100 is overexpressed in murine PML knock-out cells (Staege and Will, in preparation). These findings with eIF4E suggest that other evolutionarily conserved proteins could form prototypic nuclear bodies in the absence of the PML protein. In this way, the PML protein may not be a required element for the formation of the nuclear structures it is associated with.
Scope of this review
Many important issues relevant to determining the biochemical and molecular basis for PML function were discussed in our previous reviews [12, 14]. In particular, general experimental limitations for many areas of “PML-ology” were described. There are several areas that will not be discussed as excellent reviews are available, with topics including the role of PML in acute promyelocytic leukemia (APL), in viral infection, the role of sumoylation, as well as other areas [1–3, 8, 10, 13, 34, 36, 37, 41, 42].
In this review, general features of the PML protein and PML nuclear body function will be discussed and a potential framework for unifying PML’s disparate functions will be outlined. As stated in our previous reviews [12, 14], studies into the function of the PML protein and PML nuclear body have relied on answering the following questions: what nuclear structures are the bodies near to, what other macromolecules co-localize with the nuclear bodies, and what are the effects of disrupting the nuclear bodies? These strategies have not changed significantly in the last 5 years and so will only be discussed briefly here. A model that could provide a unifying framework for the disparate biochemical effects associated with PML is presented. In the PML RNA regulon model, the PML protein modulates gene expression in a combinatorial and coordinated fashion by inhibiting the eIF4E RNA regulon. As one will see, the PML protein may modulate other regulons as well and its assembly into nuclear bodies may enhance this activity. This model provides a molecular basis for the disparate affects of the PML protein on gene expression and its activities in what seem to be unrelated biochemical pathways. This model may provide a starting point for understanding the molecular underpinnings of the effects of the PML protein and its related nuclear bodies on proliferation and tumor suppression.
PML’s place in the nucleus
PML nuclear bodies are spherical structures generally 0.1–1.0 µm in size which are distributed throughout the nucleus and are generally excluded from the nucleolus. PML nuclear bodies are found in the inter-chromatin space [43, 44]. There are, on average, 10–30 bodies per nucleus. These structures are multi-protein complexes. Electron microscopy (EM) studies suggest that in many cases these bodies are donut shaped, i.e. that the PML protein is found in an outer ring, with the centre of the body “hollow,” or negative for the PML protein (reviewed in [12]). The PML protein is also found in track-like structures throughout the nucleus in an isoform dependent manner [45]. All PML isoforms reported thus far have the RING B-box coiled coil (RBCC or TRIM) motif [46]. Mutation of the RING or B-boxes leads to disruption of PML nuclear bodies [47, 48]. This disruption is coupled with a loss of the physiological functions associated with the PML protein [12]. Treatment of permeabilized cells with RNAse or DNAse does not alter the morphology of these bodies, suggesting that nucleic acid is not required for their structural integrity [12]. The integrity of PML nuclear bodies is also disrupted by the addition of the guanosine analogue, m7GpppG, and another cap analogue, ribavirin [17, 49]. m7GpppG or ribavirin treatment also leads to disruption of Sp100, another established PML body component, but this treatment does not disrupt other nuclear organelles indicating their effects are specific [17, 50]. Both of these small molecules bind a direct protein partner of PML, eIF4E (see below), leading to a conformational change in eIF4E that may contribute to the disruption of PML nuclear bodies [17, 49].
Both the PML protein and PML nuclear bodies respond to extracellular stimuli [51, 52]. For instance, treatment of cells with γ-IFN leads to increased expression of the PML protein due to the presence of a GAS element in the PML promoter and further, PML nuclear bodies double in number and size [21, 51, 52]. Treatment of cells with heavy metals, such as cadmium, leads to a redistribution of the PML protein from the nucleus to the cytoplasm [21, 51, 52]. Obviously, these are only a few, of many, examples of how extracellular stimuli can modulate PML nuclear bodies.
The relationship between structural integrity of PML nuclear bodies and sumoylation of the PML protein has been studied by several groups (see reviews [13, 34, 36, 42, 53, 54]). There are PML nuclear bodies in the absence of sumoylation and thus SUMO is not a nuclear body targeting signal [13, 53]. However, nuclear bodies in the absence of SUMO appear as “primitive bodies” and do not appear to have the full complement of PML body components associated with these [13, 53]. Additionally, PML harbours a SUMO binding motif [55] which may play a role in assembly. Further, detailed discussion of the different isoforms of PML and their functionalities have been covered in recent reports [45, 53].
What do neighbours tell us about function?
The relative position of PML nuclear bodies to other nuclear structures could give some insight into the functionalities of these structures. For instance, there is a correlation between the spatial position of PML nuclear bodies with Cajal bodies, cleavage bodies and splicing speckles [43]. Importantly, these structures do not co-localize with PML nuclear bodies, but rather are adjacent to PML nuclear bodies i.e. neighbours [43]. For instance, studies in T24 cells indicate that one of the four Cajal bodies in each cell is adjacent to a PML nuclear body [43]. Cajal bodies are thought to be involved in assembly of spliceosomes and the transcriptome [56], cleavage bodies are involved in mRNA 3’end processing [57] and Sc35 is involved in pre-mRNA processing [58]. Thus, it is possible that the PML protein could play a role in some sort of RNA processing events (for more in depth discussion see [12]).
PML nuclear bodies appear to have a spatial relationship with some genomic loci. For instance, one PML nuclear body per cell is found adjacent to a gene-rich major histocompatibility complex (MHC) locus on chromosome 6 [59, 60]. This association was independent of the transcriptional status of the cell or of progression through the cell cycle [59]. Thus, PML nuclear bodies can have specific genomic associations independent of transcription [59]. Of course, it is not known whether there is a functional relationship between PML nuclear bodies and MHC, or other loci located near to PML bodies. Early reports suggested there was a link [61]. However, loss of PML nuclear bodies does not affect the expression of the majority of MHC-I genes involved in antigen presentation despite its localization to this loci [60, 62, 63]. Equally well, these spatial associations could mark a relationship with other nearby, but as-yet-unidentified, compartments with which PML nuclear bodies interact. Some studies have suggested that PML nuclear bodies are close to sites of active transcription. For instance, up to 30% of PML nuclear bodies partially overlap with a marker of nascent RNA implicating PML bodies in a role in transcription [64]. Importantly, approximately 30% of Cajal bodies and 70% of splicing speckles were found to associate with nascent RNA in the same study. Thus, PML could function adjacent to active transcription sites or act in post-transcriptional functions as do splicing speckles. Importantly, RNA Polymerase II does not immunoprecipitate with the PML protein indicating that the protein is unlikely to act directly in transcription [20, 43]. However in general, no detailed study controlling for the relative crowding in the nuclear space, and thus what is the likelihood of random association, has been reported (to our knowledge). Such a study would be extremely informative in terms of formulating theories for the connection between PML nuclear bodies and transcription.
If this type of “neighbour” analysis is used to infer the function of PML nuclear bodies, one must take into account their motional status. Interestingly, PML nuclear bodies can be rapidly mobile organelles in some cellular contexts. A subset of PML nuclear bodies move within the nucleus in an energy dependent fashion [35]. These studies used the SP100 protein fused to the yellow fluorescent protein (YFP) in living BHK cells [35]. Sp100 is a well-established component of PML nuclear bodies. Three populations of nuclear bodies were observed: 25% of nuclear bodies were stationary, 63% showed minor motions and 12% showed rapid motions [35]. The minor motions, which describes the majority of PML nuclear bodies, include localized movements similar to those observed in other nuclear organelles such as Cajal bodies [35]. PML nuclear bodies in the rapid class were typically the smaller nuclear bodies, where different PML nuclear bodies moved at different times in the nucleus. Motions included not only translational movements but also the coalescence of small bodies into larger bodies and conversely, the budding off of small bodies from larger ones [35]. Movement was not sensitive to RNA Polymerase II activity but was sensitive to depletion of ATP and was myosin dependent [35]. Surprisingly, these motions were cell type specific, occurring readily in BHK cells but not in HeLa cells [35].
Aside from its nuclear distribution, a substantial fraction of the PML protein is found in the cytoplasm. In fact, some isoforms of PML lack the C-terminal nuclear localization signal (NLS) [46, 65]. PML is also found in the cytoplasm in certain pathogenic conditions including hepatocellular carcinomas [11] and in cells infected with certain viruses such as HIV and LCMV (reviewed in [12]).
Taken together, these findings strongly suggest that there are multiple classes of PML nuclear bodies. For instance not all PML nuclear bodies in a given nucleus have the same preference for their neighbours, (e.g. some are near to Cajal bodies or the MHC locus), or are of the same size. Further, they can have different compositions and motional properties. The PML protein within structures morphologically similar to PML nuclear bodies can be found in the cytoplasm, further demonstrating the challenge for investigators trying to define molecular functions for the PML protein and its associated supermolecular assemblies.
It is important to remember that the nucleus is a crowded place, and living next door to the fire station, does not necessarily make you a fireman. Thus, interpretation of these nearest neighbour studies require careful analysis before they can give us genuine insight into function(s) of the PML protein and PML nuclear bodies.
PML is not evolutionary conserved
Given the central role often purported for the function of the PML protein and PML nuclear bodies, one would assume that the PML protein is evolutionarily conserved. However, analysis of several genomes where the sequencing is complete indicates that this is not the case [12]. Sequence based analogues of the PML protein are not found in S. cerevisiae, A. thaliana, D. melanogaster or bacteria [12]. One study suggested that the PML protein was an analogue of the COP1 plant protein [12]. However, more detailed analysis indicates that COP1 has a different domain structure (with the exception of the shared RBCC motif) [12] and that there is a human homologue of this plant protein named hCOP1 [66]. Of course, there could be functional homologues of the PML protein with no sequence homology in these species. However, at least at the sequence level, PML expression seems to be limited to higher eukaryotes [12]. These observations lead to some basic questions which remain to be answered- do organisms lacking the PML protein have some sort of allied nuclear structures and if so, do these have similar functions to PML nuclear bodies in mammals? Further, it would appear that mammals have evolved an additional regulator, the PML protein, to regulate the function of an evolutionarily more conserved nuclear structure.
Interestingly, there are proteins that are associated with PML nuclear bodies and that are evolutionarily more conserved than the PML protein. This includes eIF4E which forms bodies in the absence of the PML protein (see below). Perhaps a subset of these evolutionarily more conserved proteins form the structural/functional core or are organizers of PML nuclear bodies.
What’s in a name- is the PML protein required for the formation of PML nuclear bodies?
Traditionally, PML nuclear bodies are defined by the presence of the PML protein. Several reports claim that the PML protein is the central organizer of PML nuclear bodies and thus in its absence, there are no such nuclear structures [38, 67]. For instance, in PML−/− cells, the PML nuclear body components Daxx and p53 are found distributed diffusely throughout the nucleus [54, 67]. These studies suggest that these proteins thereby lose substantial functionality in the absence of being organized into bodies. Re-introduction of PML leads to formation of PML containing structures and to the recruitment of Daxx and p53 (and others) to these bodies [54, 67]. Typically, the traditional philosophy would suggest that in the absence of the PML protein, these proteins are not functional (or as functional) as in the presence of PML.
However, exciting findings from Staege and Will (unpublished observations) indicate that in murine PML−/− cells human Sp100 forms structures that are morphologically indistinguishable from PML nuclear bodies and in addition is able to recruit human overexpressed p53 and endogenous murine Daxx protein. Therefore, PML is not the only protein that can recruit PML nuclear body components into nuclear structures. Whether such bodies are functionally distinct from bodies which also contain the PML protein is yet to be determined. However, these findings questions one of the basic tenets of “PML-ology”, that PML is required for the organization of all the components into these nuclear structures.
In support of these findings, earlier studies show that in PML−/− cells, another PML body component eIF4E, is also found in nuclear body-type structures [17, 21]. In PML−/− cells, eIF4E seems to be more active than in controls suggesting that the absence of the PML protein leads to dysregulation of eIF4E [17, 21]. Importantly, this provides an example of a protein that is, in fact, both in nuclear structures and is more active in the absence of PML. If PML nuclear body content is analyzed in terms of evolutionary conservation, one finds that the eIF4E protein is more evolutionarily conserved than the PML protein. This would suggest that some components form some type of “PML” nuclear bodies in these “older” organisms. In this way, the PML protein could be a late arrival (in the evolution context) evolved to regulate a much older set of structures. For instance, eIF4E forms nuclear body structures in D. melanogaster, where no sequence homologue of the PML protein is observed [12]. Similarly, eIF4E nuclear bodies are observed in X. laevis, [14] where no sequence homologue of PML is obvious. However, there appears to be a sequence homologue of PML in chicken (with 30% homology to human PML), suggesting that it can be found in other vertebrates (Skrabanek and Borden, unpublished observations). Thus, one might expect that proteins which are more evolutionarily conserved would form structures in the absence of the PML protein [12]. In this way, organelle function may be somewhat conserved, even if the PML protein is not. Whether such bodies have related or distinct functions to their PML containing counterparts in mammalian cells, remains to be determined.
In summary, there are components of nuclear bodies that do not require the PML protein to form nuclear body type structures. This is in contrast to one of the basic tenets of the field: that the PML protein is the component required for the formation of these nuclear structures. Only further studies will elucidate whether evolutionary conservation will be a good predictor of this body-forming ability. It may be that identifying the structural underpinnings of PML nuclear bodies is akin to peeling an onion, the PML protein may be just one layer in the middle and PML-ologists must continue peeling until they find the centre. Clearly, these findings unleash a semantics argument-what should these structures be called in the absence of the PML protein?
Biochemical functions of PML and its associated bodies
Establishing the composition of PML nuclear bodies is an area of both great interest and substantial technical difficulty. To date, over 70 proteins have been reported to localize with PML nuclear bodies (Nuclear Protein Database http://npd.hgu.mrc.ac.uk). Identification of proteins which bind the PML protein is used as a means to assign function to the PML nuclear body itself [12]. This is mainly due to the fact that the PML protein does not have many defined biochemical activities associated with it. Two direct and distinct biochemical activities have been reported to date: 1. SUMO modification where the RING domain of PML interacts with the SUMO E2, Ubc9, acting in auto-sumoylation [68–71], and 2. reduction of the affinity of eIF4E for its ligand, the m7GpppG cap, by about 100 fold [17, 71]. This latter activity will be discussed at length below. Potentially, direct interactions might give one unique insight into the function of the PML protein, and thus the body. To date, there are only a few components of PML nuclear bodies that are known to directly interact with the PML protein, for example: eIF4E, SUMO/PIC1, Ubc9, PRH/Hex (the proline rich homeoprotein) and the viral protein Z [68, 69]. Importantly, PML contains a SUMO binding motif [55], and thus could potentially recruit other SUMO modified proteins to the body, this possibility would be a interesting future direction of investigation. There have been other proteins suggested to directly bind the PML protein, but these assays are typically done in reticulocyte lysates, rather than using two purified proteins. Thus, they are not established direct targets. However, subsequent experiments with purified proteins may demonstrate they are also direct partners.
Many groups have attempted to purify PML nuclear bodies. The tight association of PML nuclear bodies with the nuclear “matrix” and the fact that the PML protein is found both in the nucleoplasm as well as in nuclear bodies have presented confounding issues in the purification of intact nuclear bodies and thus, in the determination of their composition. Further, PML nuclear bodies are heterogeneous in nature, thus not all PML nuclear bodies are likely to be equivalent in terms of structure, composition, function, spatial distribution and motional properties. This heterogeneity even occurs in the same cell. Clearly, it seems likely that not all components will be present at all PML nuclear bodies all of the time. Further, not all PML nuclear bodies may be structurally or functionally equivalent.
The physicality of PML nuclear bodies?
The wide variety of partner proteins, and the compositional heterogeneity of PML nuclear bodies, even in the same nucleus, has led to the suggestion that PML nuclear bodies are some sort of nuclear storage facility [8, 34]. Consistent with this model, many foreign and inappropriately expressed proteins are found accumulated at PML nuclear bodies. For instance, when transfected into cells, the lac repressor localizes to PML nuclear bodies [72]. In addition, several proteins involved in polyglutamine repeat diseases such as huntingtin, ataxin 1 and ataxin 3 (in their expanded forms) localize to these bodies [4, 5, 35, 73]. In these neurological disorders, PML nuclear bodies appear to be much larger than normal nuclear bodies and more inclusion-like [4, 5, 73]. PML nuclear bodies are sometimes adjacent to proteosomal components [53]. Their presence adjacent to PML nuclear bodies is consistent with this role as a sensor of foreign/inappropriately expressed proteins and salso with many other potential functions of PML nuclear bodies. Additionally, it has been proposed that PML nuclear bodies store normal, functional proteins. In this way, the body could serve to titrate levels of the active forms of these proteins in the nucleoplasm and thus modulate biochemistries there [8]. This is consistent with the proposition that other types of nuclear bodies are storage areas for certain types of components.
It has also been hypothesized that PML nuclear bodies could form catalytic surfaces with specific biochemistries occurring on the surface of the body [74, 75]. RING domains from several proteins, including the PML protein, self assemble into spherical structures visible by EM [74]. PML and the viral protein Z self assemble into structures and are better inhibitors of eIF4E in their assembled state than in their monomeric state [74]. Mdm2 and BRCA1/BARD1 act similarly, where they assemble into spherical structures with their corresponding E2 proteins assembling on to the spheres [74, 76]. Again, the assembled Mdm2 and BRCA1/BARD1 forms are better E3 ligases than their corresponding monomeric forms [74, 76].
Whether or not PML nuclear bodies are nuclear storage facilities or catalytic surfaces (or both) remains an open question. Both of these would provide a molecular basis for the specific biochemistries associated with PML nuclear bodies.
The PML protein and PML nuclear bodies - all things to all men?
The PML protein and consequently, PML nuclear bodies, have been attributed functions in diverse, and apparently unrelated biochemical activities such as DNA repair, DNA replication, transcription, mRNA export, and sites of post-translational modification (reviewed in [12, 34]). The data supporting these models has been described elsewhere in detail, and thus will not be discussed here (e.g. in [12, 34]). Generally, these functions are attributed to PML nuclear bodies based on its proximity to, or interactions with, proteins or structures of known function, as discussed above. Such disparate activities could arise from a number of factors (which are not mutually exclusive). For instance, PML nuclear bodies could be so heterogeneous that bodies in different cell types, under different physiological conditions, or variance of bodies within the same nucleus, mean that they simply function in different biochemistries. Alternatively, it could be that the PML protein and thus PML nuclear bodies function in some process(es) so basic, that it impacts on all of these biochemical pathways. We propose that these structures impact coordinately and combinatorially on gene expression by inhibiting a key node in an RNA regulon governing cell proliferation and survival [15, 19, 77]. This PML RNA regulon model provides a theoretical basis to understand many PML associated phenomenon. The model is described below.
RNA regulons- a unifying framework for PML nuclear body function?
The PML protein and PML nuclear bodies were shown to interact with eIF4E by several groups [20, 71, 78–80]. eIF4E functions in cap dependent translation in the [17]cytoplasm and mRNA export of specific transcripts in the nucleus [77]. In fact, eIF4E is a central node in an RNA regulon which governs proliferation and survival [15, 19, 77]. Our studies show that the PML protein is a potent inhibitor of this regulon [15, 19, 77].
An RNA regulon is a theoretical construct proposed by Keene and colleagues as a means to explain how post-transcriptional regulation could be coordinated in eukaryotes [81–83]. This parallels the operon model in prokaryotes. Here, all the genes for the same biochemical pathway, such as tryptophan biogenesis, are adjacent to each other and are co-regulated by the same promoter. Further, transcription and translation occur at the same time on the transcript. In this way, prokaryotes can readily coordinate production of all of the proteins necessary for a given biochemical pathway. The situation in eukaryotes is vastly different. Genes coding for proteins in the same biochemical pathway do not appear to be spatially organized, in general. Further, once the genes are transcribed, even if transcription is coordinated by the use of common promoter elements, the resulting mRNAs may not have, for instance, similar stabilities, nuclear export rates or translation efficiencies. The RNA regulon model proposed that RNAs in a given pathway are co-regulated by the presence of distinct sequences in their UTRs referred to as USER codes. These codes enable them to be regulated at particular levels of gene expression e.g. mRNA export, translation, stability etc. The combination of USER codes in the RNA would ultimately determine its fate. RNAs with the same combination of USER codes would be coordinately regulated permitting the production of, for instance, all the proteins in some metabolic pathway such as tryptophan biosynthesis.
Our studies suggest that the PML protein is a potent inhibitor of eIF4E, which is a key node in an RNA regulon governing proliferation and survival [18, 19, 77]. eIF4E promotes proliferation and rescues cells from apoptosis. Although eIF4E acts in the cap dependent translation of all mRNAs, its overexpression only leads to enhanced translational efficiency of a subset of transcripts usually referred to as eIF4E sensitive mRNAs [84]. Translational efficiency refers to the observation that these transcripts are loaded onto heavier polysomes (which are more efficient) without changing the amount of mRNA present [84]. In addition, the nuclear fraction of eIF4E acts in mRNA export providing more mRNA for translation and thus, increased production of these proteins without necessarily having these transcripts loaded preferentially onto heavier polysomes [77]. In particular, it promotes the nuclear export of mRNAs involved in proliferation and survival via the presence of a ~50 nucleotide sequence found in the UTR of these RNAs [19]. This sequence is referred to as the eIF4E sensitivity element (4E-SE)[19]. RNAs can be sensitive to eIF4E at the export level (e.g. cyclin D1), translation efficiency level (e.g. VEGF) or both (e.g. ODC and Pim1) [19, 77]. Our studies show that the PML protein directly binds eIF4E and thereby inhibits nuclear export of these mRNAs [71,77]. The PML protein achieves this by reducing the affinity of eIF4E for the m7G cap moiety on these RNAs by about 100 fold [17, 71].
The ability of the PML protein, and presumably PML nuclear bodies, to inhibit a network node in an RNA regulon potentially provides an explanation for several of the physiological and biochemical phenomenon associated with the PML protein (Figure 1 and Figure 2). For instance, several groups have shown that PML overexpression promotes apoptosis in a variety of contexts [85–87]. Similarly, eIF4E is known to rescue cells from apoptosis in several contexts e.g. [88, 89]. Our studies demonstrate that PML overexpression dramatically inhibits eIF4E mediated apoptotic rescue during serum deprivation in fibroblasts (the only context examined thus far) [15]. We propose that the PML protein does this by inhibiting the eIF4E regulon [77].
Figure 1.
The PML regulon model is used to explain the effects of the PML protein and PML nuclear bodies on survival signaling as described in the text. This is the first model that provides an explanation for the transcriptional independence of PML promoted apoptosis [86]. Recent data indicate that eIF4E is a node in a regulon governing cellular proliferation and survival [15]. eIF4E can promote Akt activation in a NBS1 dependent manner [15]. Furthermore, eIF4E promotes the mRNA export (and in some cases also the translational efficiency) of several downstream effectors of Akt as shown. Proteins in blue have their levels upregulated at the mRNA export level by eIF4E [18]. Conversely, PML overexpression inhibits the production of these proteins by impairing eIF4E dependent mRNA export of their corresponding mRNAs [18]. Further, PML overexpression impairs Akt activation by inhibiting production of NBS1 through inhibiting eIF4E activity [15]. Thus PML overexpression impairs Akt signaling at (at least) two levels [15]. Green arrows indicate that the activity of the given factor is enhanced by Akt and conversely, red indicates that it is inhibited. For clarity of presentation, the complete Akt pathway and other possible feedback loops are not depicted.
Figure 2.
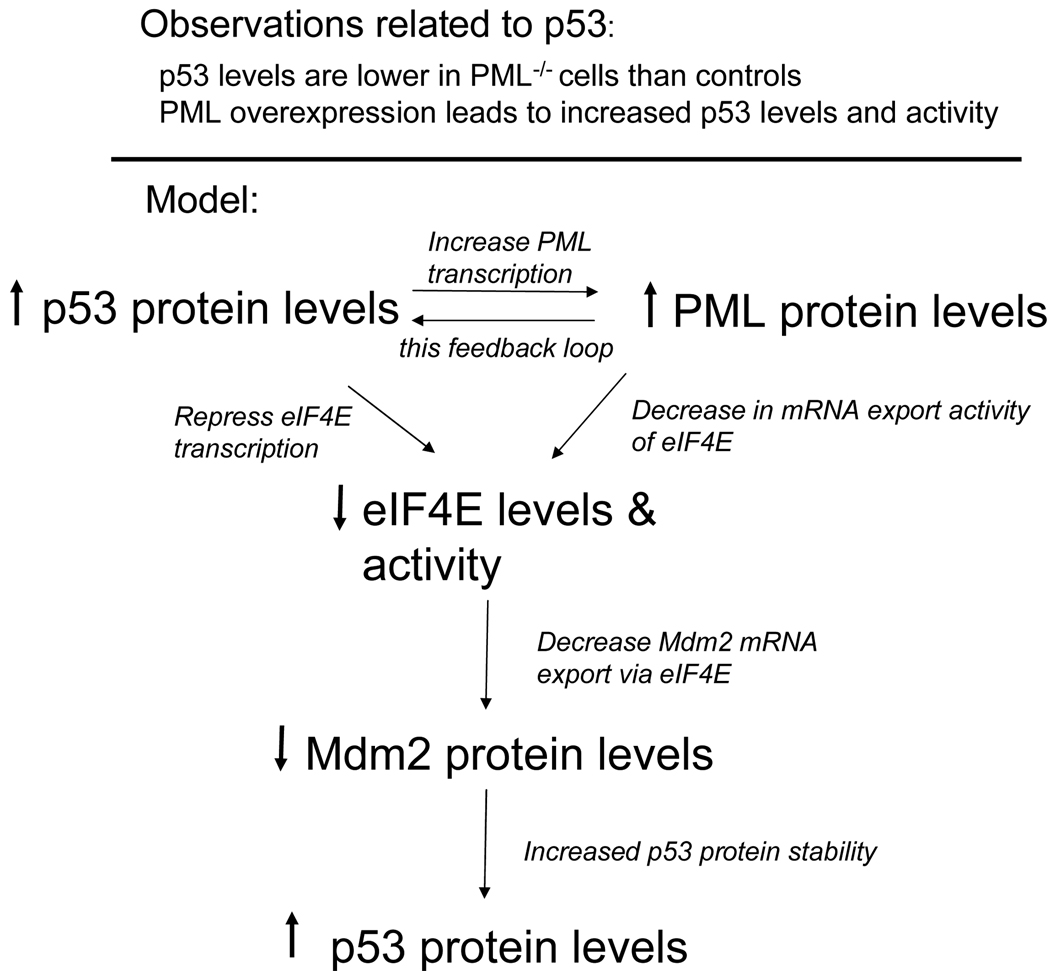
The PML regulon model is used to explain the effects of the PML protein on p53 signaling. As described in the text, PML overexpression leads to reduced eIF4E activity which in turn leads to reduced Mdm2 mRNA export. Note that eIF4E promotes Mdm2 protein production at the level of mRNA export, not at the level of translation [19, 101, 120]. In this way, overexpression of PML leads to enhanced p53 levels which in turn lead to enhanced PML levels given observations that PML is a downstream effector of p53 [98]. For clarity of presentation, all the interactions described in the text (and others reported in the literature) are not depicted.
The positioning of eIF4E in a survival network including Akt is central to its survival function [15] (summarized in Figure 1). Recent studies indicate that eIF4E activates the Akt pathway at two levels [15]. First, overexpression of eIF4E leads to increased phosphorylation of Akt via increased expression of NBS1, a protein involved in activation of Akt through an interaction with PI3K [90, 91]. Here, eIF4E overexpression leads to enhanced NBS1 mRNA export, and thus elevated protein levels, of NBS1. eIF4E overexpression is correlated with increased phosphorylation of Akt and also of BP1 and S6 [15]. These activities of eIF4E are PI3K and NBS1 dependent [15]. Importantly, eIF4E cannot rescue Akt1−/− cells from serum deprivation induced apoptosis, thus its activity is Akt1 dependent in this context. The second level by which eIF4E activates Akt signaling is by increasing the expression of downstream effectors of Akt such as cyclin D1 and c-myc [19, 92] (see Figure 1). Thus eIF4E can impact on Akt signaling both upstream of Akt through NBS1 and by increasing the concentrations of downstream effectors of Akt.
The PML protein is a potent inhibitor of these eIF4E-mediated effects [17–19, 71, 77]. For instance, PML overexpression leads to inhibition of Akt phosphorylation, at least in part, via reduction in mRNA export of NBS1, thus leading to reduced levels of this upstream Akt activator [15, 91]. PML nuclear bodies also interact with NBS1 [93], thus it may have yet another level of control on the eIF4E regulon [77]. This may also segue into understanding the link between PML nuclear bodies, NBS1 and DNA repair. Further, PML overexpression leads to reduction in the levels of downstream effectors of Akt such as cyclin D1 and c-myc [18–20, 92]. Again, this is due, at least in part, to reduced mRNA export and thus expression, of these genes. Thus, the PML protein potently impedes the apoptotic rescue function of eIF4E, at least in part, through these activities [15]. These findings are consistent with observations that PML overexpression leads to decreased phosphorylation of Akt [15 94]. Further, it is consistent with the observation by another group that PML−/− cells have increased levels of activated Akt [94]. Clearly, different apoptotic stimuli will utilize different genetic pathways. In summary, the role for the PML protein and PML nuclear bodies as inhibitors of an RNA regulon provides a theoretical context in which to understand the mechanism by which PML overexpression promotes apoptosis during serum deprivation of fibroblasts. In support of our model, the PML protein requires its RING domain to promote apoptosis, to bind eIF4E, to impair eIF4E activity and to form nuclear bodies [17, 71, 85]. Further, these data provide an explanation for the previous observation that PML overexpression promotes apoptosis independent of ongoing transcription [86].
Another (not mutually exclusive) explanation has been proposed to understand the effects of the PML protein on mTOR and Akt activation [16, 94]. One study suggests that mTOR and Akt may co-localize with PML nuclear bodies [94]. In this study, the authors suggest that the PML protein inactivates Akt by recruiting phosphatase PP2a into PML nuclear bodies. Importantly, this model would require novel modes for mTOR and Akt function, since these proteins are usually associated with the plasma membrane.
The RNA regulon also provides a theoretical context in which to understand the relationship between PML, Mdm2 and p53 protein levels (Figure 2). Several models have been described to explain the following observations. In some conditions, PML and p53 co-localize at PML nuclear bodies [24]. Other studies suggested that PML nuclear bodies could modulate the acetylation of p53 by recruiting CBP, an acetyltransferase, as well as p53, to the bodies [32]. PML nuclear bodies may also act in the recruitment of Chk2 and HIPK2 kinases to facilitate phosphorylation and activation of p53 under stress conditions [95–97]. PML itself is a target gene of p53 and may be a key downstream effector of p53 activity [98]. In PML−/− cells, there is less p53 protein than in controls both under steady state conditions and during γ-irradiation [99, 100]. Introduction of PML into PML−/− cells leads to increased levels of p53 [99, 100]. These changes in gene expression are paralleled by expected changes in cell physiology.
The role of the PML protein as an inhibitor of the eIF4E RNA regulon provides a basis for understanding the relationship between PML and p53 expression. Previous studies showed that eIF4E enhances the expression of Mdm2, a negative regulator of p53 [19, 101]. eIF4E enhances the mRNA export, but not the translational efficiency, of Mdm2 mRNA [19, 101]. PML inhibits eIF4E dependent mRNA export of Mdm2, leading to reduced levels of Mdm2 protein [19]. Consistently, PML overexpression leads to elevated levels of p53 [99]. Interestingly, p53 overexpression leads to transcriptional repression of eIF4E and thus, decreased eIF4E protein levels [102]. Mdm2 overexpression leads to reduced p53 and increased eIF4E levels [102]. This could constitute a feedback loop, whereby increased eIF4E levels correlate with enhanced Mdm2 mRNA export. The interaction between PML and Mdm2 proteins provides more intricate feedback loops [99, 103, 104]. Co-expression of Mdm2 and PML lead to the redistribution of PML to the cytoplasm leading to destabilization of p53 [105]. Further, increased p53 levels result in reduced translation initiation via dephosphorylation of 4EBP-1, which consequently causes formation of translationally impaired eIF4E complexes [106]. Thus, the role of the PML protein as an inhibitor of the RNA regulon provides a theoretical basis to understand its effects on p53 activity.
The growth and transformation suppressive activities of the PML protein and PML nuclear bodies can also be understood in the regulon context. PML overexpression leads to a G1/S arrest in several cell types (reviewed in [12]). PML overexpression leads to reduction in several proteins involved in cell cycle progression including: cyclin D1, cyclin B1, cyclin E1, c-myc and cyclin A2 (to name a few) [19, 20, 107, 108]. Consistently, loss of PML expression alters c-myc target gene expression [109]. All of these mRNAs are eIF4E mRNA export targets and their mRNA export is inhibited by PML [18, 19]. The PML protein requires its RING domain for both its growth suppressive activities and its ability to impair production of these proteins [12, 17, 19]. Interestingly, cyclin D1 mRNA export and cyclin D1 protein levels are elevated in PML−/− cells relative to controls [18, 21]. γ-IFN does not reduce cyclin D1 mRNA export or protein levels in PML−/− cells, but does so in wildtype cells, where PML protein levels are elevated due to γ-IFN dependent transcription of PML [21]. Re-introduction of PML into PML−/− cells leads to reduced cyclin D1 mRNA export and protein levels [21]. In this way, the PML protein can coordinately impact on cellular proliferation in response to extracellular stimuli.
PML suppresses eIF4E mediated oncogenic transformation [17]. The mRNA export function of cyclin D1 contributes to the oncogenic potential of eIF4E [18]. The presence of the 4E-SE USER code in cyclin D1 mRNA substantially promotes the ability of eIF4E to transform cyclin D1−/− cells when cyclin D1 constructs are re-introduced [18]. Clearly, as an inhibitor of eIF4E dependent mRNA export, PML’s transformation suppressive activities (in this context) can be understood using the RNA regulon model.
Several studies suggest that PML activity may be lost as part of the metastatic process through reduced levels of the PML protein or relocalization of the PML protein to the cytoplasm [11, 45, 110, 111]. Tumour angiogenesis may be increased in a prostate cancer model using PML−/− mice [16]. It was suggested that the PML protein reduces HIF1α protein synthesis without modulating HIF1α RNA levels [16]. This reduction in HIF1α correlates with reduced VEGF levels. Note that increased VEGF levels correlate with increased metastasis [112, 113]. How can the PML protein impact on protein synthesis, a cytoplasmic event? The PML protein could impact on mTOR and thereby reduce HIF1α, although deletion of the 5’UTR of HIF1α did not modulate its synthesis in the presence or absence of the PML protein [16]. This is somewhat surprising given that translational control is usually through this region, as discussed by the authors. The authors suggest the possibility that the PML protein impacts on translation of HIF1α through sequestering mTOR in PML nuclear bodies during hypoxia [16]. Additionally, it is possible that the PML protein could impact on the mRNA export of HIF1α (this has yet to be examined). Further, the PML protein inhibits Akt phosphorylation (which thereby impacts on mTOR activation) via its effects on the RNA regulon [15]. The PML protein could also impact on metastasis by reducing the production of ODC, another protein important for this process, given that ODC mRNA export is impeded by PML [19]. Thus, the PML protein could also reduce VEGF levels independently of HIF1α, via its effects on eIF4E and Akt. Further, PML could affect metastasis via its effects on other gene products such as ODC.
In summary, the PML protein is positioned to coordinately and combinatorially modulate gene expression through its inhibitory effects on eIF4E, a network node in an RNA regulon. This model for function of the PML protein and PML nuclear bodies provides a circuit diagram level of understanding for PML associated phenomenon. Clearly, PML is likely to have eIF4E independent functions, and these may be by acting on other RNA regulons. For the regulon model described here, the question is an obvious one: why don’t PML−/− mice have aggressive cancers? The answer appears to be redundancy in the regulation of eIF4E. For instance, in PML−/− cells, eIF4E associates with other negative regulators including the proline rich homeodomain protein PRH/Hex [70, 114, 115]. This is addressed below.
Why are the PML−/− mice well?
One of the nagging issues in “PML-ology” is the lack of a more striking phenotype in the PML−/− mice [38, 116, 117]. The PML−/− mice appear morphologically normal, where loss of the gene does not impact on survival [38]. PML−/− mice are ~2 fold more resistant to γ-irradiation than normal mice [38, 117]. PML−/− mice treated with DMBA and tetradecanoyl phorbol acetate developed about twice as many papillomas than controls and these mice have a ~2 fold reduction in TNFα response [38, 87]. However, many reports describe a wide variety of activities that absolutely require the PML protein. Thus, one is faced with a choice: either that activity is not critical for the health of the cell/animal, or that the PML protein is one of many redundant regulators that can modulate a given process. Additionally, these factors may suffer from a loss of efficiency when not organized into nuclear bodies (as discussed for the catalytic surface hypothesis section above). In this case, the factors may function well enough for most situations and only in stress situations is the reduced efficiency phenotype more obvious.
It is clear that redundancy plays a major role with regard to the activities of the PML protein and PML nuclear bodies in the eIF4E RNA regulon. Over 200 homeoproteins are positioned to modulate eIF4E dependent changes in gene expression [70, 118]. These homeoproteins bind the same region of eIF4E that is recognized by the PML protein [70, 114, 118]. These homeoproteins can either stimulate or repress eIF4E dependent gene expression [70, 118]. These homeoprotein regulators of eIF4E are phylogenetically more conserved than the PML protein, allowing modulation of the eIF4E regulon in a wide variety of evolutionary contexts. A specific example of a redundant regulator is found in the proline rich homeodomain PRH, which negatively regulates eIF4E in APL cells [70] (where the PML protein is displaced from nuclear bodies) [70]. In APL cells prior to ATRA treatment, PRH and eIF4E form nuclear structures indistinguishable from PML nuclear bodies despite the micro-particulate distribution of the PML protein in these cells [70]. Consistently, APL cells do not have dysregulated eIF4E activity [17], but of course, they do have a variety of other unrelated problems that contribute to their oncogenic nature. It is likely that redundancy in control of the regulon is not complete and thus under certain conditions, the redundant factors may not be as effective as the PML protein.
Insights from the PML Regulon model into drug design
Given the medical relevance of PML, interest in the PML protein and PML nuclear bodies goes well beyond understanding basic questions in cell biology. It is clear that understanding the function of these entities could aid in the design and development of novel therapeutic strategies. The proposed PML regulon model appears to explain many of the observations regarding the effects of PML on certain pathways and its physiological effects. A disadvantage of this model is that it does not describe a physical picture of how PML nuclear bodies may work (see below). However, an advantage of this model is that it has allowed specific insights into how one might predict other physiological effects of the PML protein and PML nuclear bodies. Further, it provides a model for drug design because it strongly suggests that finding a means to inhibit eIF4E (as the PML protein does), will enable one to shutdown its associated proliferation/transformation/survival activities at a network level. In this case, the PML protein inhibits eIF4E by reducing its affinity for the m7G cap. We used a parallel strategy to the PML protein, whereby we found a small molecule mimic of the m7G cap, ribavirin, which inhibits eIF4E and the regulon similarly to the PML protein ([50, 77, 119] and Tan et al., in preparation)). Thus the precise mechanisms by which the PML protein and ribavirin inhibit eIF4E are different, but the overall strategy of interfering with m7G cap binding is the same. Importantly, ribavirin inhibits eIF4E mediated transformation and apoptotic rescue by targeting the eIF4E regulon [15]. This has led to a phase II clinical trial in acute myeloid leukemias with dysregulated eIF4E (www.ribatrial.com; [50, 77, 119]). This phase II trial, the first to look at the efficacy of inhibiting eIF4E and the regulon (that we are aware of), started enrolling patients in 2007 and shows promising results with regard to this strategy (our unpublished observations).
The missing pieces of the puzzle
Clearly there are limitations to this, as with any, model. For instance, this model only describes PML protein and nuclear body function at the level of a circuit diagram. This is useful in generally understanding why PML affects certain pathways and has particular biological effects. This is the first model proposed that could provide a framework to understand why the PML protein and PML nuclear bodies can act in so many different pathways. It also could be useful in predicting new pathways and functionalities for both the PML protein and its associated nuclear bodies. However, generally, one cannot build a transistor with only a circuit diagram. In this way, the regulon model does not address the issue of the “nuts and bolts” of how PML nuclear bodies function, how they are structured/organized, or how nuclear body heterogeneity impacts on PML function (to name a few). Equally well, there are likely functions of the PML protein and PML nuclear bodies which are independent of eIF4E, and in this way, the PML regulon may, in part, reflect the heterogeneity of this system. It is interesting to note that some PML partner proteins, such as c-myc, Mdm2 and NBS1 [93, 104, 109],are also regulated by the PML protein at the RNA regulon level, suggesting that there could be some sort of intricate feedback loop on this PML circuit.
How do we move forward to find the missing pieces of the PML puzzle? Can we fit the pieces we have together? To date, composition alone has not provided a unifying framework to understand the diverse phenomenon associated with PML nuclear bodies. The development of new strategies to micro-dissect composition and thus account for body heterogeneity would give clearer insight into PML nuclear body functions. Additionally, high-resolution ultra-structure analysis would be extremely useful. In summary, the PML field continues to struggle with the identification of a unifying function for these bodies. We present the PML RNA regulon model, which may serve to provide an understanding of PML associated phenomenon at the circuit diagram level. To our knowledge, such a model would be the first step in describing a unifying framework for PML function. However, the one constant in “PML-ology” remains, there are more questions than answers.
Acknowledgements
I am very grateful to Hans Will and Hannah Staege for sharing data prior to publication and for critical reading of the manuscript. I am grateful to Biljana Culjkovic, Slobodanka Orolicki and Nadeem Siddiqui for critical reading of the manuscript. K.L.B.B is supported by the NIH (RO1 98571 and 80728) and the Leukemia and Lymphoma Society Translational Research Program. She holds a Canada Research Chair in Molecular Biology of the Cell Nucleus.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 2.Grimwade D, Solomon E. Characterisation of the PML/RAR alpha rearrangement associated with t(15;17) acute promyelocytic leukaemia. Curr Top Microbiol Immunol. 1997;220:81–112. doi: 10.1007/978-3-642-60479-9_6. [DOI] [PubMed] [Google Scholar]
- 3.de The H, Chelbi-Alix MK. APL, a model disease for cancer therapies? Oncogene. 2001;20:7136–7139. doi: 10.1038/sj.onc.1204851. [DOI] [PubMed] [Google Scholar]
- 4.Chai Y, Koppenhafer SL, Koppenhafer SL, Shoesmith SJ, Perez MK, Paulson HL. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum Mol Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 5.Yasuda S, Inoue K, Hirabayashi M, Higashiyama H, Yamamoto Y, Fuyuhiro H, Komure O, Tanaka F, Sobue G, Tsuchiya K, Hamada K, Sasaki H, Takeda K, Ichijo H, Kakizuka A. Triggering of neuronal cell death by accumulation of activated SEK1 on nuclear polyglutamine aggregations in PML bodies. Genes Cells. 1999;4:743–756. doi: 10.1046/j.1365-2443.1999.00294.x. [DOI] [PubMed] [Google Scholar]
- 6.Janer A, Martin E, Muriel MP, Latouche M, Fujigasaki H, Ruberg M, Brice A, Trottier Y, Sittler A. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J Cell Biol. 2006;174:65–76. doi: 10.1083/jcb.200511045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borden KLB, Campbell Dwyer EJ, Carlile GW, Djavani M, Salvato MS. Two RING finger proteins, the oncoprotein PML and the Aren avirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J Virol. 1998;72:3819–3826. doi: 10.1128/jvi.72.5.3819-3826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maul GG, Negorev D, Bell P, Ishov AM. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 9.Turelli P, Doucas V, Craig E, Mangeat B, Klages N, Evans R, Kalpana G, Trono D. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol Cell. 2001;7:1245–1254. doi: 10.1016/s1097-2765(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 10.Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819–830. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Henin D, Dejean A. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 1995;55:1590–1597. [PubMed] [Google Scholar]
- 12.Borden KL. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol Cell Biol. 2002;22:5259–5269. doi: 10.1128/MCB.22.15.5259-5269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y, Lallemand-Breitenbach V, Zhu J, de H. The, PML nuclear bodies and apoptosis. Oncogene. 2004;23:2819–2824. doi: 10.1038/sj.onc.1207533. [DOI] [PubMed] [Google Scholar]
- 14.Strudwick S, Borden KL. The emerging roles of translation factor eIF4E in the nucleus. Differentiation. 2002;70:10–22. doi: 10.1046/j.1432-0436.2002.700102.x. [DOI] [PubMed] [Google Scholar]
- 15.B.T. Culjkovic K, Orolicki S, Amri A, Meloche S, Borden KLB. The eIF4E RNA regulon promotes the Akt signaling pathway. The Journal of Cell Biology. 2008;181:51–63. doi: 10.1083/jcb.200707018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernardi R, I. Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, Cordon-Cardo C, Simon MC, Rafii S, Pandolfi PP. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442:779–785. doi: 10.1038/nature05029. [DOI] [PubMed] [Google Scholar]
- 17.Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. Embo J. 2001;20:4547–4559. doi: 10.1093/emboj/20.16.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E promotes nuclear export of cyclin D1 mRNAs via an element in the 3'UTR. J Cell Biol. 2005;169:245–256. doi: 10.1083/jcb.200501019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–426. doi: 10.1083/jcb.200607020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai HK, Borden KL. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene. 2000;19:1623–1634. doi: 10.1038/sj.onc.1203473. [DOI] [PubMed] [Google Scholar]
- 21.Topisirovic I, Capili AD, Borden KL. Gamma Interferon and Cadmium Treatments Modulate Eukaryotic Initiation Factor 4E-Dependent mRNA Transport of Cyclin D1 in a PML-Dependent Manner. Mol Cell Biol. 2002;22:6183–6198. doi: 10.1128/MCB.22.17.6183-6198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dellaire G, Ching RW, Ahmed K, Jalali F, Tse KC, Bristow RG, Bazett-Jones DP. Promyelocytic leukemia nuclear bodies behave as DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1 and the kinases ATM, Chk2, and ATR. J Cell Biol. 2006;175:55–66. doi: 10.1083/jcb.200604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carbone R, Pearson M, Minucci S, Pelicci PG. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene. 2002;21:1633–1640. doi: 10.1038/sj.onc.1205227. [DOI] [PubMed] [Google Scholar]
- 25.Boe SO, Haave M, Jul-Larsen A, Grudic A, Bjerkvig R, Lonning PE. Promyelocytic leukemia nuclear bodies are predetermined processing sites for damaged DNA. J Cell Sci. 2006;119:3284–3295. doi: 10.1242/jcs.03068. [DOI] [PubMed] [Google Scholar]
- 26.Jul-Larsen A, Visted T, Karlsen BO, Rinaldo CH, Bjerkvig R, Lonning PE, Boe SO. PML-nuclear bodies accumulate DNA in response to polyomavirus BK and simian virus 40 replication. Exp Cell Res. 2004;298:58–73. doi: 10.1016/j.yexcr.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 27.Burkham J, Coen DM, Weller SK. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boisvert FM, Hendzel MJ, Bazett-Jones DP. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ching RW, Dellaire G, Eskiw CH, Bazett-Jones DP. PML bodies: a meeting place for genomic loci? J Cell Sci. 2005;118:847–854. doi: 10.1242/jcs.01700. [DOI] [PubMed] [Google Scholar]
- 30.Eskiw CH, Bazett-Jones DP. The promyelocytic leukemia nuclear body: sites of activity? Biochem Cell Biol. 2002;80:301–310. doi: 10.1139/o02-079. [DOI] [PubMed] [Google Scholar]
- 31.Quimby BB, Yong-Gonzalez V, Anan T, Strunnikov AV, Dasso M. The promyelocytic leukemia protein stimulates SUMO conjugation in yeast. Oncogene. 2006;25:2999–3005. doi: 10.1038/sj.onc.1209335. [DOI] [PubMed] [Google Scholar]
- 32.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann H, Sindre H, Stamminger T. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J Virol. 2002;76:5769–5783. doi: 10.1128/JVI.76.11.5769-5783.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003;10:1290–1299. doi: 10.1038/sj.cdd.4401313. [DOI] [PubMed] [Google Scholar]
- 35.Muratani M, Gerlich D, Janicki SM, Gebhard M, Eils R, Spector DL. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat Cell Biol. 2001;21:21. doi: 10.1038/ncb740. [DOI] [PubMed] [Google Scholar]
- 36.Eskiw CH, Dellaire G, Mymryk JS, Bazett-Jones DP. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J Cell Sci. 2003;116:4455–4466. doi: 10.1242/jcs.00758. [DOI] [PubMed] [Google Scholar]
- 37.Hodges M, Tissot C, Howe K, Grimwade D, Freemont PS. Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am J Hum Genet. 1998;63:297–304. doi: 10.1086/301991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 39.Lang V, Zanchin NI, Lunsdorf H, Tuite M, McCarthy JE. Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA, and consequences of its overproduction. J Biol Chem. 1994;269:6117–6123. [PubMed] [Google Scholar]
- 40.Lejbkowicz F, Goyer C, Darveau A, Neron S, Lemieux R, Sonenberg N. A fraction of the mRNA 5' cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc Natl Acad Sci U S A. 1992;89:9612–9616. doi: 10.1073/pnas.89.20.9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Everett RD. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene. 2001;20:7266–7273. doi: 10.1038/sj.onc.1204759. [DOI] [PubMed] [Google Scholar]
- 42.Zhong S, Muller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- 43.Grande MA, van der Kraan I, van Steensel B, Schul W, de The H, van der Voort HT, de Jong L, van Driel R. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J Cell Biochem. 1996;63:280–291. doi: 10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 44.Ascoli CA, Maul GG. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Condemine W, Takahashi Y, Zhu J, Puvion-Dutilleul F, Guegan S, Janin A, de The H. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 2006;66:6192–6198. doi: 10.1158/0008-5472.CAN-05-3792. [DOI] [PubMed] [Google Scholar]
- 46.Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 2001;20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 47.Borden KLB, Lally JM, Martin SR, O'Reilly NJ, Solomon E, Freemont PS. In vivo and in vitro characterization of the B1 and B2 zinc-binding domains from the acute promyelocytic leukemia protooncoprotein PML. Proc Natl Acad Sci U S A. 1996;93:1601–1606. doi: 10.1073/pnas.93.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borden KLB, Boddy MN, Lally J, O'Reilly NJ, Martin S, Howe K, Solomon E, Freemont PS. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. Embo J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004 doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101:18105–18110. doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maul GG, Yu E, Ishov AM, Epstein AL. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J Cell Biochem. 1995;59:498–513. doi: 10.1002/jcb.240590410. [DOI] [PubMed] [Google Scholar]
- 52.Korioth F, Gieffers C, Maul GG, Frey J. Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J Cell Biol. 1995;130:1–13. doi: 10.1083/jcb.130.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de The H. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, 3rd, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- 57.Schul W, van Der Kraan I, Matera AG, van Driel R, de Jong L. Nuclear domains enriched in RNA 3'-processing factors associate with coiled bodies and histone genes in a cell cycle-dependent manner. Mol Biol Cell. 1999;10:3815–3824. doi: 10.1091/mbc.10.11.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu XD, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3' splice site. Proc Natl Acad Sci U S A. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shiels C, Islam SA, Vatcheva R, Sasieni P, Sternberg MJ, Freemont PS, Sheer D. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J Cell Sci. 2001;114:3705–3716. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- 60.Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 61.Zheng P, Guo Y, Niu Q, Levy DE, Dyck JA, Lu S, Sheiman LA, Liu Y. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature. 1998;396:373–376. doi: 10.1038/24628. [DOI] [PubMed] [Google Scholar]
- 62.Bruno S, Ghiotto F, Fais F, Fagioli M, Luzi L, Pelicci PG, Grossi CE, Ciccone E. The PML gene is not involved in the regulation of MHC class I expression in human cell lines. Blood. 2003;101:3514–3519. doi: 10.1182/blood-2002-11-3335. [DOI] [PubMed] [Google Scholar]
- 63.Larghero J, Zassadowski F, Rousselot P, Padua RA, Degos L, Chomienne C. Alteration of the PML proto-oncogene in leukemic cells does not abrogate expression of MHC class I antigens. Leukemia. 1999;13:1295–1296. doi: 10.1038/sj.leu.2401464. [DOI] [PubMed] [Google Scholar]
- 64.Kiefllich A, von Mikecz A, Hemmerich P. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. Journal of Structural Biology. 2002;140:167–179. doi: 10.1016/s1047-8477(02)00571-3. [DOI] [PubMed] [Google Scholar]
- 65.Flenghi L, Fagioli M, Tomassoni L, Pileri S, Gambacorta M, Pacini R, Grignani F, Casini T, Ferrucci PF, Martelli MF, et al. Characterization of a new monoclonal antibody (PG-M3) directed against the aminoterminal portion of the PML gene product: immunocytochemical evidence for high expression of PML proteins on activated macrophages, endothelial cells, and epithelia. Blood. 1995;85:1871–1880. [PubMed] [Google Scholar]
- 66.Wang H, Kang D, Deng XW, Wei N. Evidence for functional conservation of a mammalian homologue of the light-responsive plant protein COP1. Curr Biol. 1999;9:711–714. doi: 10.1016/s0960-9822(99)80314-5. [DOI] [PubMed] [Google Scholar]
- 67.Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S, Higashimoto Y, Appella E, Minucci S, Pandolfi PP, Pelicci PG. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 68.Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC 1,a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 69.Duprez E, Saurin AJ, Desterro JM, Lallemand-Breitenbach V, Howe K, Boddy MN, Solomon E, de The H, Hay RT, Freemont PS. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J Cell Sci. 1999;112:381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- 70.Topisirovic I, Culjkovic B, Cohen N, Perez JM, Skrabanek L, Borden KL. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. Embo J. 2003;22:689–703. doi: 10.1093/emboj/cdg069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kentsis A, Dwyer EC, Perez JM, Sharma M, Chen A, Pan ZQ, Borden KL. The RING Domains of the Promyelocytic Leukemia Protein PML and the Arenaviral Protein Z Repress Translation by Directly Inhibiting Translation Initiation Factor eIF4E. J Mol Biol. 2001;312:609–623. doi: 10.1006/jmbi.2001.5003. [DOI] [PubMed] [Google Scholar]
- 72.Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- 73.Skinner PJ, Koshy BT, Cummings CJ, Klement IA, Helin K, Servadio A, Zoghbi HY, Orr HT. Ataxin-1 with an expanded glutamine tract alters nuclear matrix- associated structures. Nature. 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 74.Kentsis A, Gordon RE, Borden KL. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc Natl Acad Sci U S A. 2002;99:15404–15409. doi: 10.1073/pnas.202608799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kentsis A, Gordon RE, Borden KL. From the Cover: Self-assembly properties of a model RING domain. Proc Natl Acad Sci U S A. 2002;99:667–672. doi: 10.1073/pnas.012317299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. Embo J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Culjkovic B, Topisirovic I, Borden KL. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle. 2007;6:65–69. doi: 10.4161/cc.6.1.3688. [DOI] [PubMed] [Google Scholar]
- 78.Woulfe JM, Prichett-Pejic W, Rippstein P, Munoz DG. Promyelocytic leukaemia-immunoreactive neuronal intranuclear rodlets in the human brain. Neuropathol Appl Neurobiol. 2007;33:56–66. doi: 10.1111/j.1365-2990.2006.00789.x. [DOI] [PubMed] [Google Scholar]
- 79.Scheper GC, Parra JL, Wilson M, Van Kollenburg B, Vertegaal AC, Han ZG, Proud CG. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol Cell Biol. 2003;23:5692–5705. doi: 10.1128/MCB.23.16.5692-5705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seker H, Rubbi C, Linke SP, Bowman ED, Garfield S, Hansen L, Borden KL, Milner J, Harris CC. UV-C-induced DNA damage leads to p53-dependent nuclear trafficking of PML. Oncogene. 2003;22:1620–1628. doi: 10.1038/sj.onc.1206140. [DOI] [PubMed] [Google Scholar]
- 81.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 82.Keene JD, Lager PJ. Post-transcriptional operons and regulons coordinating gene expression. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- 83.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 84.Sonenberg N, Gingras AC. The mRNA 5' cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 85.Borden KLB, Campbell Dwyer EJ, Salvato MS. The promyelocytic leukemia protein PML has a pro-apoptotic activity mediated through its RING domain. FEBS Lett. 1997;418:30–34. doi: 10.1016/s0014-5793(97)01344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen JC, de The H. PML induces a novel caspase-independent death process. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [see comments] [DOI] [PubMed] [Google Scholar]
- 87.Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi PP. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [see comments] [DOI] [PubMed] [Google Scholar]
- 88.Polunovsky VA, Rosenwald IB, Tan AT, White J, Chiang L, Sonenberg N, Bitterman PB. Translational control of programmed cell death: eukaryotic translation initiation factor 4E blocks apoptosis in growth-factor-restricted fibroblasts with physiologically expressed or deregulated Myc. Mol Cell Biol. 1996;16:6573–6581. doi: 10.1128/mcb.16.11.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tan A, Bitterman P, Sonenberg N, Peterson M, Polunovsky V. Inhibition of Myc-dependent apoptosis by eukaryotic translation initiation factor 4E requires cyclin D1. Oncogene. 2000;19:1437–1447. doi: 10.1038/sj.onc.1203446. [DOI] [PubMed] [Google Scholar]
- 90.Chen YC, Su YN, Chou PC, Chiang WC, Chang MC, Wang LS, Teng SC, Wu KJ. Overexpression of NBS1 contributes to transformation through the activation of phosphatidylinositol 3-kinase/Akt. J Biol Chem. 2005;280:32505–32511. doi: 10.1074/jbc.M501449200. [DOI] [PubMed] [Google Scholar]
- 91.Chen YC, Chiang HY, Yang MH, Chen PM, Chang SY, Teng SC, Vanhaesebroeck B, Wu KJ. Activation of phosphoinositide 3-kinase by the NBS1 DNA repair protein through a novel activation motif. J Mol Med. 2008 doi: 10.1007/s00109-008-0302-x. [DOI] [PubMed] [Google Scholar]
- 92.Le XF, Vallian S, Mu ZM, Hung MC, Chang KS. Recombinant PML adenovirus suppresses growth and tumorigenicity of human breast cancer cells by inducing G1 cell cycle arrest and apoptosis. Oncogene. 1998;16:1839–1849. doi: 10.1038/sj.onc.1201705. [DOI] [PubMed] [Google Scholar]
- 93.Naka K, Ikeda K, Motoyama N. Recruitment of NBS1 into PML oncogenic domains via interaction with SP100 protein. Biochem Biophys Res Commun. 2002;299:863–871. doi: 10.1016/s0006-291x(02)02755-9. [DOI] [PubMed] [Google Scholar]
- 94.Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Cordon-Cardo C, Pandolfi PP. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 2006;441:523–527. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, Piaggio G, Fanciulli M, Appella E, Soddu S. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 96.Yang S, Kuo C, Bisi JE, Kim MK. PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat Cell Biol. 2002;4:865–870. doi: 10.1038/ncb869. [DOI] [PubMed] [Google Scholar]
- 97.Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- 98.de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, Ferbeyre G, Lowe SW. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13:523–535. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 99.Louria-Hayon I, Grossman T, Sionov RV, Alsheich O, Pandolfi PP, Haupt Y. The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J Biol Chem. 2003;278:33134–33141. doi: 10.1074/jbc.M301264200. [DOI] [PubMed] [Google Scholar]
- 100.Ferbeyre G, de Stanchina E, Querido E, Baptiste N, Prives C, Lowe SW. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 2000;14:2015–2027. [PMC free article] [PubMed] [Google Scholar]
- 101.Phillips A, Blaydes JP. MNK1 and EIF4E are downstream effectors of MEKs in the regulation of the nuclear export of HDM2 mRNA. Oncogene. 2008;27:1645–1649. doi: 10.1038/sj.onc.1210785. [DOI] [PubMed] [Google Scholar]
- 102.Zhu N, Gu L, Findley HW, Zhou M. Transcriptional repression of the eukaryotic initiation factor 4E gene by wild type p53. Biochem Biophys Res Commun. 2005;335:1272–1279. doi: 10.1016/j.bbrc.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 103.Kurki S, Latonen L, Laiho M. Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization. J Cell Sci. 2003;116:3917–3925. doi: 10.1242/jcs.00714. [DOI] [PubMed] [Google Scholar]
- 104.Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–672. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- 105.Wei X, Yu ZK, Ramalingam A, Grossman SR, Yu JH, Bloch DB, Maki CG. Physical and functional interactions between PML and MDM2. J Biol Chem. 2003;278:29288–29297. doi: 10.1074/jbc.M212215200. [DOI] [PubMed] [Google Scholar]
- 106.Horton LE, Bushell M, Barth-Baus D, Tilleray VJ, Clemens MJ, Hensold JO. p53 activation results in rapid dephosphorylation of the eIF4E-binding protein 4E-BP1, inhibition of ribosomal protein S6 kinase and inhibition of translation initiation. Oncogene. 2002;21:5325–5334. doi: 10.1038/sj.onc.1205662. [DOI] [PubMed] [Google Scholar]
- 107.Mu ZM, Le XF, Vallian S, Glassman AB, Chang KS. Stable overexpression of PML alters regulation of cell cycle progression in HeLa cells. Carcinogenesis. 1997;18:2063–2069. doi: 10.1093/carcin/18.11.2063. [DOI] [PubMed] [Google Scholar]
- 108.Son SH, Yu E, Choi EK, Lee H, Choi J. Promyelocytic leukemia protein-induced growth suppression and cell death in liver cancer cells. Cancer Gene Ther. 2005;12:1–11. doi: 10.1038/sj.cgt.7700755. [DOI] [PubMed] [Google Scholar]
- 109.Cairo S, De Falco F, Pizzo M, Salomoni P, Pandolfi PP, Meroni G. PML interacts with Myc, and Myc target gene expression is altered in PML-null fibroblasts. Oncogene. 2005;24:2195–2203. doi: 10.1038/sj.onc.1208338. [DOI] [PubMed] [Google Scholar]
- 110.Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst. 2004;96:269–279. doi: 10.1093/jnci/djh043. [DOI] [PubMed] [Google Scholar]
- 111.Chan JY, Chin W, Liew CT, Chang KS, Johnson PJ. Altered expression of the growth and transformation suppressor PML gene in human hepatocellular carcinomas and in hepatitis tissues. Eur J Cancer. 1998;34:1015–1022. doi: 10.1016/s0959-8049(97)10138-1. [DOI] [PubMed] [Google Scholar]
- 112.Karkkainen MJ, Petrova TV. Vascular endothelial growth factor receptors in the regulation of angiogenesis and lymphangiogenesis. Oncogene. 2000;19:5598–5605. doi: 10.1038/sj.onc.1203855. [DOI] [PubMed] [Google Scholar]
- 113.Shinkaruk S, Bayle M, Lain G, Deleris G. Vascular endothelial cell growth factor (VEGF), an emerging target for cancer chemotherapy. Curr Med Chem Anticancer Agents. 2003;3:95–117. doi: 10.2174/1568011033353452. [DOI] [PubMed] [Google Scholar]
- 114.Topcu Z, Mack DL, Hromas RA, Borden KL. The promyelocytic leukemia protein PML interacts with the proline-rich homeodomain protein PRH: a RING may link hematopoiesis and growth control. Oncogene. 1999;18:7091–7100. doi: 10.1038/sj.onc.1203201. [In Process Citation] [DOI] [PubMed] [Google Scholar]
- 115.Topisirovic I, Borden KL. Homeodomain proteins and eukaryotic translation initiation factor 4E (eIF4E): an unexpected relationship. Histol Histopathol. 2005;20:1275–1284. doi: 10.14670/HH-20.1275. [DOI] [PubMed] [Google Scholar]
- 116.Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi PP. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 117.Guo A, Salomoni P, Luo J, Shih A, Zhong S, Gu W, Pandolfi PP. The function of PML in p53-dependent apoptosis. Nat Cell Biol. 2000;2:730–736. doi: 10.1038/35036365. [DOI] [PubMed] [Google Scholar]
- 118.Topisirovic I, Kentsis A, Perez JM, Guzman ML, Jordan CT, Borden KL. Eukaryotic translation initiation factor 4E activity is modulated by HOXA9 at multiple levels. Mol Cell Biol. 2005;25:1100–1112. doi: 10.1128/MCB.25.3.1100-1112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kentsis A, Volpon L, Topisirovic I, Soll CE, Culjkovic B, Shao L, Borden KL. Further evidence that ribavirin interacts with eIF4E. Rna. 2005;11:1762–1766. doi: 10.1261/rna.2238705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Phelps M, Phillips A, Darley M, Blaydes JP. MEK-ERK signaling controls Hdm2 oncoprotein expression by regulating hdm2 mRNA export to the cytoplasm. J Biol Chem. 2005;280:16651–16658. doi: 10.1074/jbc.M412334200. [DOI] [PubMed] [Google Scholar]