SUMMARY
Animal cells control their membrane lipid composition within narrow limits, but the sensing mechanisms underlying this control are largely unknown. Recent studies disclosed a protein network that controls the level of one lipid – cholesterol. This network resides in the endoplasmic reticulum (ER). A key component is Scap, a tetrameric ER membrane protein that binds cholesterol. Cholesterol binding prevents Scap from transporting SREBPs to the Golgi for activation. Using a new method to purify ER membranes from cultured cells, we here show that Scap responds cooperatively to ER cholesterol levels. When ER cholesterol exceeds 5% of total ER lipids, SREBP-2 transport/activation are abruptly blocked. They resume when ER cholesterol falls below the 5% threshold. The 5% threshold is lowered to 3% when cells overexpress Insig-1, a Scap-binding protein. Cooperative interactions between cholesterol, Scap, and Insig create a sensitive switch that controls the cholesterol composition of cell membranes with remarkable precision.
INTRODUCTION
The endoplasmic reticulum (ER) of mammalian cells contains an elaborate feedback system that senses the level of membrane cholesterol and modulates the transcription of genes that mediate cholesterol synthesis and uptake. This system maintains cholesterol homeostasis by increasing cholesterol synthesis and uptake when membrane cholesterol falls and by down-regulating these processes when excess cholesterol accumulates. The regulatory system revolves around a family of membrane bound transcription factors called Sterol Regulatory Element-binding Proteins (SREBPs), and it has been called the SREBP pathway (Brown and Goldstein, 1997).
Central to the SREBP pathway is a polytopic membrane protein called Scap. Scap forms a complex with SREBPs, and it also binds CopII proteins. The latter cluster the Scap/SREBP complex into coated vesicles that bud from the ER and travel to the Golgi complex (Sun et al., 2007). There the SREBP is processed by two proteases, liberating the active transcription factor domain which enters the nucleus and activates the genes for cholesterol synthesis and uptake. When excess cholesterol accumulates, the Scap/SREBP complex binds to a resident ER protein, designated Insig. This binding precludes the binding of CopII proteins. As a result, the Scap/SREBP complex remains in the ER, transcription of the target genes declines, and cholesterol synthesis and uptake fall (Goldstein et al., 2006).
The mechanism by which Scap senses cholesterol has been elucidated through cholesterol-binding studies conducted with the purified octahelical membrane domain of Scap which forms tetramers in detergent solution (Radhakrishnan et al., 2004). This domain of Scap binds cholesterol with saturation kinetics and with stereospecificity. When studied in membrane vesicles, cholesterol binding leads to a conformational change in the membrane domain of Scap that can be monitored by the appearance of a new cleavage site that is accessible to trypsin (Brown et al., 2002; Adams et al., 2004). This conformational change promotes the binding of Scap to Insig. The Scap:Insig ratio is crucial for determining the sensitivity of the cellular response to cholesterol. Overexpression of Insig lowers the threshold for suppression of SREBP cleavage by cholesterol in intact cells (Yang et al., 2002).
A major unanswered question relates to the level of cholesterol in ER membranes that is necessary in order to inhibit SREBP processing. It has long been known that the concentration of cholesterol in ER membranes is much lower than the concentration of cholesterol in plasma membranes when expressed as the molar ratio of cholesterol to phospholipids (Bergstrand and Dallner, 1969; Zambrano et al., 1975). However, there have been no measurements of the changes in ER cholesterol that are necessary to regulate SREBP processing. In part, this lack of information is attributable to the difficulty in attempting to obtain a pure population of ER membranes that would permit direct measurements of cholesterol levels. Lange and Steck (Lange and Steck, 1997; Lange et al., 1999) approached the problem indirectly by measuring the percentage of total cholesterol in human fibroblasts that could be esterified in vitro by the resident ER enzyme acyl-CoA:cholesterol acyltransferase (ACAT). From these data, they concluded that the ER contained 0.5% of total cholesterol when cells were depleted of sterols. This value rose to 5% when the total cellular cholesterol content was increased 2-fold by addition of cholesterol complexed to hydroxypropyl-cyclodextrin (Lange et al., 1999).
In the current studies, we have developed a procedure for purification of ER membranes from cultured Chinese hamster ovary (CHO) cells, which allows direct measurement of the ER cholesterol content by mass spectrometry. With this assay, we demonstrate that the suppression of SREBP cleavage by cholesterol exhibits a cooperative threshold response. Suppression occurs abruptly when the molar cholesterol level in ER membranes reaches 4–5% of total lipids on a molar basis. Through such cooperativity, cells are able to respond with great precision to changes in membrane cholesterol, and thereby to fine-tune their cholesterol content.
RESULTS
Isolation of Pure ER Membranes from CHO Cells
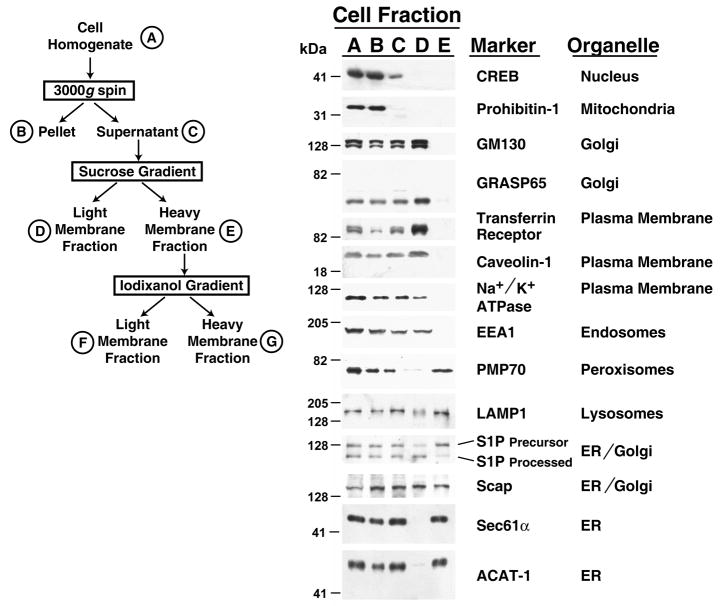
We developed a fractionation scheme to isolate pure ER membranes from CHO-K1 cells as outlined in Figure 1 (left panel). Efficient and reproducible cell homogenization was achieved through the use of a ball-bearing homogenizer (Balch and Rothman, 1985). The cell homogenate (designated Fraction A in Figure 1) was subjected to three centrifugation steps, which progressively separated ER membranes from other organelles. The results of the first two centrifugation steps are summarized in Figure 1 (right panel). Centrifugation of the homogenate at 3000g eliminates unbroken cells and the heaviest membranes. As assessed by immunoblot analysis of the fractions, a soluble nuclear protein marker (CREB) and a mitochondrial membrane protein marker (Prohibitin-1) are mostly retained in the 3000g pellet (Fraction B), whereas significant amounts of all other membrane markers escape into the 3000g supernatant (Fraction C). Fraction C was then subjected to centrifugation through a discontinuous sucrose gradient and yielded two distinct bands of membranes (Fractions D and E). The small amount of CREB in Fraction C is absent in Fractions D or E and likely represents a portion that was released into the cytosol, owing to rupture of the nuclear membranes during the fractionation procedure. The Golgi membrane proteins (GM130 and GRASP65), plasma membrane proteins (transferrin receptor, caveolin-1, and Na+/K+ ATPase), and the early endosomal membrane protein (EEA1) are found in the light membrane Fraction D and are almost completely absent from the heavy membrane Fraction E. The peroxisomal membrane protein (PMP70) is localized predominantly in Fraction E, and a lysosomal membrane protein (LAMP1) is present in both Fractions D and E. ER-resident membrane proteins (Sec61α and ACAT-1) are localized almost completely to Fraction E. Scap, a membrane protein that cycles between ER and Golgi membranes (Goldstein et al., 2006), is found in both Fractions D and E. The two forms of Site-1 protease, the inactive precursor form and the processed active form (Espenshade et al., 1999), are separated into Fraction E and Fraction D, respectively.
Figure 1. Step 1 in Purification of ER Membranes from CHO-K1 Cells: Elimination of Nuclear Mitochondrial, Plasma, Golgi, and Early Endosomal Membranes.
(Left Panel) Diagram of ER membrane fractionation scheme. A-G denote major fractions recovered and analyzed by immunoblot analysis.
(Right Panel) CHO-K1 cells (~2×108) were treated according to the fractionation scheme as described in Experimental Procedures and shown at the left. Cells were disrupted using a ball-bearing homogenizer and centrifuged at 3000g. The supernatant was then loaded at the top of a discontinuous sucrose gradient and centrifuged at 100,000g for 1 hr, yielding two distinct membrane layers. After this step, aliquots representing equal volumes of each fraction (A–E) were subjected to immunoblot analysis for the indicated organelle markers.
The aforementioned data suggest that Fraction E contains partially purified ER membranes devoid of membranes from the nucleus, mitochondria, plasma membrane, Golgi and early endosomes, but still contaminated with lysosomal and peroxisomal membranes.
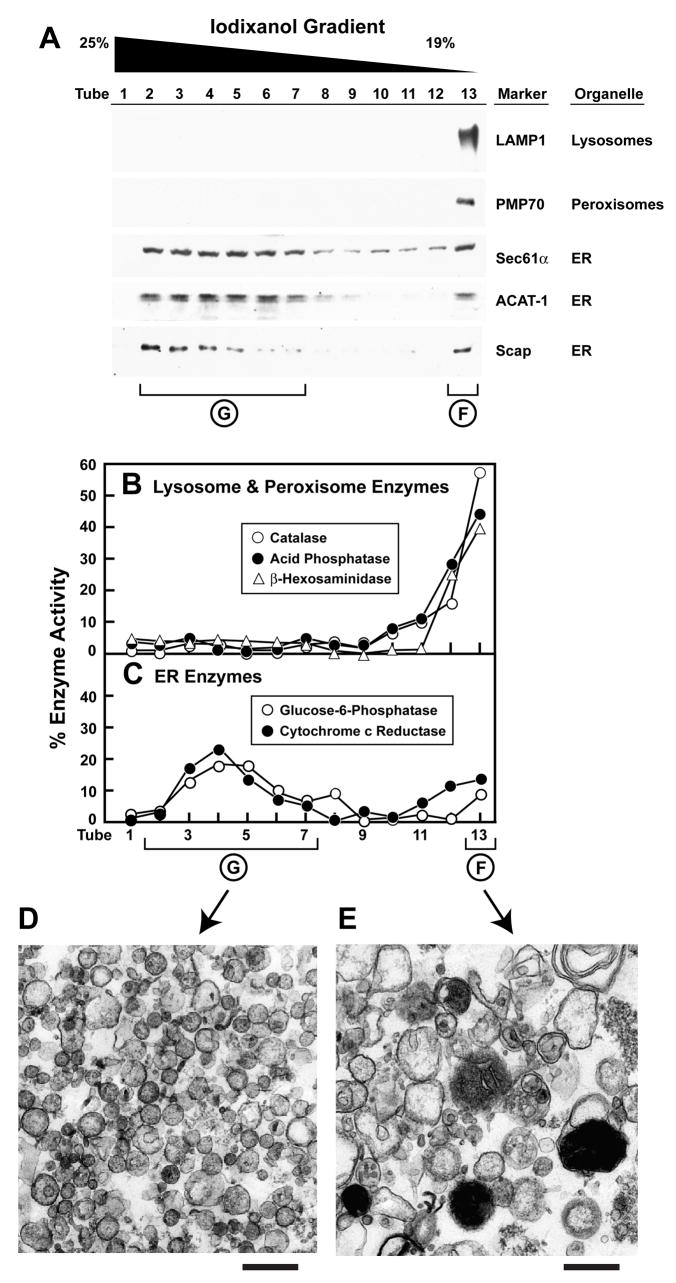
In order to further purify ER membranes, we subjected Fraction E to an additional centrifugation step through a continuous iodixanol gradient (van Veldhoven et al., 1996). As shown in Figure 2A, the lysosomal and peroxisomal membrane proteins, LAMP1 and PMP70, are exclusively localized to a floating membrane layer (tube 13, which is designated as Fraction F). On the other hand, ER membrane proteins (Sec61α, ACAT-1, and Scap) divide into two portions. A portion of all three of these proteins float (tube 13, Fraction F), but the majority of all three are found in denser regions of the gradient (tubes 2–7), which were pooled and designated Fraction G. Fraction G is devoid of the lysosomal protein LAMP1 and the peroxisomal protein PMP70. Moreover, fraction G did not show any detectable signal for plasma membrane markers (Na+/K+ ATPase and transferrin receptor) even when we analyzed aliquots that were 10-fold higher than those shown in Figure 2.
Figure 2. Step 2 in Purification of ER Membranes from CHO-K1 Cells: Elimination of Lysosomal and Peroxisomal Membranes.
CHO-K1 cells (~2×108) were fractionated as described in Experimental Procedures. The heavy membrane fraction obtained after the discontinuous sucrose gradient step (Fraction E, see Figure 1) was loaded below a continuous 19%–25% iodixanol gradient, centrifuged for 2 hr at 110,000g, after which fractions were collected from the bottom. Equal volumes of each fraction were subjected to immunoblot analysis for the indicated organelle markers (A) or assayed for the indicated enzyme activity (B, C). Tubes 2–7 correspond to (Fraction G) and Tube 13 corresponds to Fraction F (see Figure 1). Membranes from Fractions F and G (obtained after fractionation of ~4×108 CHO-K1 cells) were processed and visualized by election microscopy as described in Experimental Procedures. Representative images of Fraction G (purified ER) and Fraction F (mixture of lysosomes, peroxisomes, and ER) are shown in panels D and E, respectively. Scale bar = 500 nm.
We further characterized the iodixanol gradient fractions with enzyme assays as shown in Figures 2B and 2C. Consistent with the immunoblot results of Figure 2A, the activities of two lysosomal enzymes (acid phosphatase and β-hexosaminidase) and a peroxisomal enzyme (catalase) were localized exclusively to Fraction F (Figure 2B), whereas the activities of ER enzymes (glucose-6-phosphatase and cytochrome c reductase) were present in both Fractions F and G (Figure 2C). The activities of a Golgi enzyme (mannosidase II) and a plasma membrane enzyme (alkaline phosphodiesterase I) were localized to Fraction D. Neither of these enzyme activities were detected in any fractions from the iodixanol gradient (data not shown). Based on both the immunoblot and enzymatic analysis, we conclude that Fraction G contains ER membranes and contains less than 10% contamination with lysosomal, peroxisomal, plasma, or other membranes. Fraction G is hereafter designated as “purified ER membranes.”
We examined the morphology of membranes from Fractions F and G by electron microscopy. Figures 2D and 2E show images representative of more than 50 different sections examined. Fraction F is a heterogeneous mixture, containing vesicles 50–200 nm in diameter, large structures with intense electron staining (characteristic of lysosomes), and other irregularly shaped structures (Figure 2E). Fraction G is more homogeneous, containing primarily spherical vesicles 50–200 nm in diameter (Figure 2D). Since we did not take measures to preserve ribosome attachment, the ER vesicles are not decorated by ribosomes. Immunoblot analysis of ribosomal subunit S6 showed that it did not localize with the membranes of Fractions D or E and was likely dissociated into the cytosol (data not shown).
Relating Cholesterol Levels of Purified ER to SREBP Activation
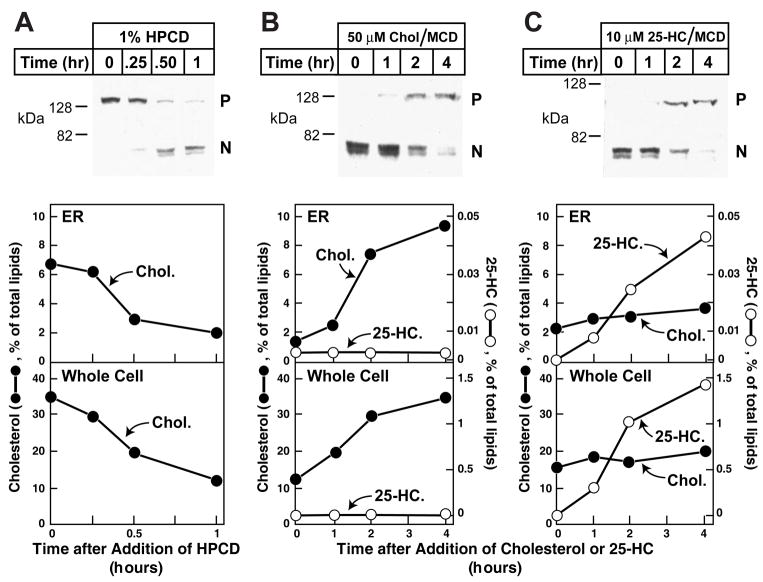
The availability of purified ER membranes allowed us to study how changes in cell lipids alter the ER cholesterol content and how these alterations regulate the exit of the Scap/SREBP complex. We first sought to measure the effects of cyclodextrin-induced cholesterol depletion on cholesterol concentrations in both the whole cell and ER and on proteolytic processing of SREBP-2. Cholesterol percentages are expressed as moles of cholesterol divided by moles of total lipid × 100. The lipids are defined as the measured moles of sterols plus phospholipids. As shown in Figure 3A, switching cells from medium containing 5% FCS to a lipid-poor medium containing 1% hydroxypropyl cyclodextrin (HPCD) triggered the conversion of SREBP-2 from an inactive precursor form to an active cleaved nuclear form in 0.5 to 1 hr (top panel). This increase in SREBP-2 processing was accompanied by a decrease in the ER cholesterol content from ~7% to ~2% of the total lipid in the ER (middle panel) and a decrease in whole cell cholesterol from ~35% to ~10% of whole cell total lipids (bottom panel). We then analyzed the consequences of adding back either cholesterol or 25-hydroxycholesterol (25-HC). Both sterols were added in complexes with methyl-β-cyclodextrin (MCD). After the initial cholesterol depletion, all of the visible SREBP-2 was found in the cleaved nuclear form (zero time in Figures 3B and 3C, top panels). After addition of 50 μM cholesterol/MCD, the nuclear form declined and the uncleaved membrane precursor form of SREBP-2 became visible at 2 hr (Figure 3B). By 4 hr the nuclear form was barely visible, indicating that SREBP-2 processing had been inhibited nearly completely. Over the 4-hr time period, the addition of 50 μM cholesterol/MCD increased ER cholesterol content from ~0.13 to ~1.3 nmol/dish (representing 1% and 10% of total ER lipids, respectively) and whole cell cholesterol content from ~40 to ~90 nmol/dish (representing 13% and 35% of total cellular lipids, respectively) (Figure 3B, middle and bottom panels). In the sterol-depleted cells, the content of 25-HC was below the detectable limit (~25 fmol) in both the ER and whole cell fractions (Figure 3C). Four hr after addition of 10 μM 25-HC to the culture medium, SREBP-2 cleavage was suppressed to the same degree as seen with cholesterol addition. This treatment did not alter cholesterol levels significantly in either the whole cell or the ER (Figure 3C, middle and bottom panels). On the other hand, the level of 25-HC increased from below the detectable limit to ~5 pmol/dish in the ER (representing 0.042% of total lipids) and to ~4 nmol/dish in the whole cell (representing ~1.5% of total lipids). Thus, a relatively small amount of 25-HC in the ER was able to inhibit SREBP-2 cleavage without causing a measurable change in ER cholesterol. In experiments not shown, we verified that the ER purification procedure is not altered at the extreme conditions of lipid depletion or addition used in these studies.
Figure 3. Analysis of SREBP-2 Processing and Its Relation to Sterol Content of Purified ER Membranes in CHO-K1 Cells after Various Treatments.
On day 0, CHO-K1 cells were set up as described in Experimental Procedures in medium B (5% FCS). On day 3, cells were washed with phosphate-buffered saline and each dish received one of the following types of medium: (A) Medium C (lipoprotein-deficient serum) containing 1% HPCD for the indicated times; (B) Medium C containing 1% HPCD for 1 hr, then switched to Medium C containing 50 μM cholesterol complexed to MCD for the indicated time; or (C) Medium C containing 1% HPCD for 1 hr, then switched to Medium C containing 10 μM 25-HC complexed to MCD for the indicated time. At the indicated time, cells were harvested and disrupted by a ball-bearing homogenizer. A portion of the homogenate (5% of total) was saved for lipid analysis and immunoblot analysis of SREBP-2 (30 μg/lane) (P, precursor form; N, cleaved nuclear form). The remainder (95%) of the homogenized cells was used to purify ER membranes. Lipids were extracted from both the homogenate and the purified ER, and the amount of cholesterol, 25-HC, and phospholipids was quantified as described in Experimental Procedures. Approximately 5×107 cells (from 5 10-cm dishes) were used for each data point. The amount of sterol in each fraction was expressed as the percent of total amount of phospholipid, cholesterol, and 25-HC.
Cooperative Threshold Response Fine-tunes SREBP Activation
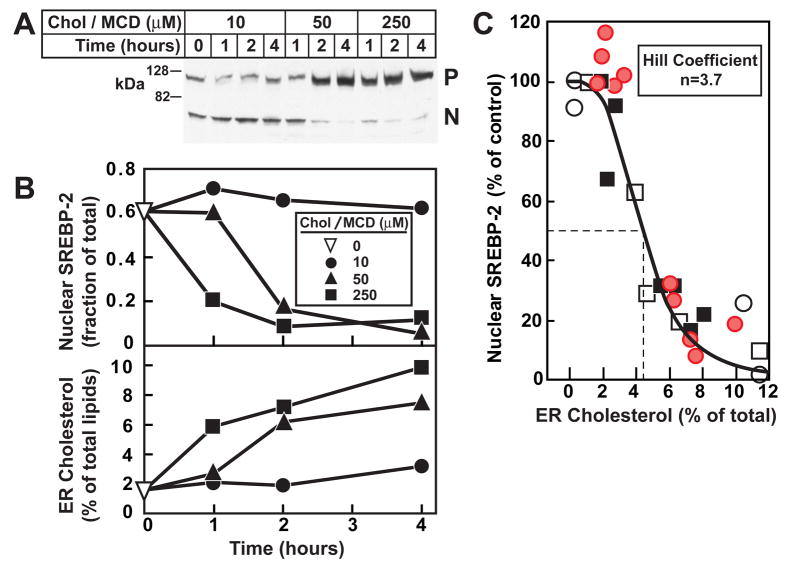
We next sought to determine whether a threshold exists between ER cholesterol levels and SREBP-2 processing. We again depleted cellular cholesterol by treament with 1% HPCD for 1 hr. We then added varying levels of cholesterol in MCD for various times. As shown in Figure 4A, addition of 10 μM cholesterol/MCD had no effect on SREBP-2 processing even after 4 hr, whereas 50 μM cholesterol/MCD suppressed SREBP-2 processing after 2 hr and 250 μM cholesterol/MCD suppressed SREBP-2 processing even earlier than 1 hr. Figure 4B (top panel) shows a quantification of the immunoblot from Figure 4A by densitometry, where the active nuclear form of SREBP-2 is expressed as a fraction of the total amount of SREBP-2 (precursor form plus nuclear cleaved form). The bottom panel of Figure 4B shows the cholesterol content of purified ER membranes at each level of added cholesterol/MCD. To gain additional data, we carried out two other similar experiments where cells were first depleted of cholesterol by treatment with 1% HPCD for 1 hr and then with different concentrations of cholesterol (in MCD) for various times. In a fourth experiment, we varied membrane cholesterol in the other direction, i.e., by first incubating cells with 50 μM cholesterol/MCD for 4 hr and then depleting cholesterol with 1% HPCD for various times (open squares in Figure 4C). We then plotted nuclear SREBP-2 as a function of ER cholesterol using the data from all four experiments. This analysis revealed a threshold effect. When the mole fraction of cholesterol in the ER was less than 2%, nuclear SREBP-2 levels were maximal. As the ER cholesterol rose above 2%, there was a sharp drop in SREBP-2 processing with 50% inhibition occurring at about 4.5% cholesterol. The sigmoidal nature of this curve was confirmed by subjecting the data to analysis by the Hill equation, which revealed a high degree of positive cooperativity with a Hill coefficient of 3.7.
Figure 4. Relation of SREBP-2 Processing to Cholesterol Content of Purified ER Membranes after Treatment of CHO-K1 Cells with Increasing Amounts of Cholesterol Complexed to MCD for Various Times.
(A) On day 0, CHO-K1 cells were set up in Medium B (5% FCS). On day 3, cells were switched to Medium C (lipoprotein-deficient serum) containing 1% HPCD for 1 hr, then switched to Medium C containing the indicated concentration of cholesterol complexed to MCD. At the indicated time, cells were harvested and disrupted by a ball-bearing homogenizer. A portion of the homogenate (5% of total) was saved for immunoblot analysis of SREBP-2 (30 μg/lane). P, precursor form of SREBP-2; N, cleaved nuclear form of SREBP-2.
(B, top panel) Densitometric quantification of SREBP-2 in (A) expressed as the amount of the nuclear form relative to the total (nuclear plus precursor). ▽, zero-time value.
(B, bottom panel) The remainder (95%) of the homogenate from (A) were used to purify ER membranes, after which the lipids were extracted and the amount of cholesterol and phospholipids was quantified as described in Experimental Procedures.
(C) This graph represents the data from panels A and B (red circles) and three other similar experiments (●, ○, □). For each of the four experimemts, the amount of nuclear SREBP-2 at zero time was normalized to 100%. Approximately 5×107 cells (from 5 10-cm dishes) were used for each data point. The solid line represents a best-fit of the experimental data to the Hill equation as described in Experimental Procedures. The best-fit values with 95% confidence intervals for cholesterol concentration (mol%) corresponding to 50% inhibition of SREBP-2 processing and the Hill coefficient were 4.5% ± 0.45% amd 3.7 ± 0.23, respectively.
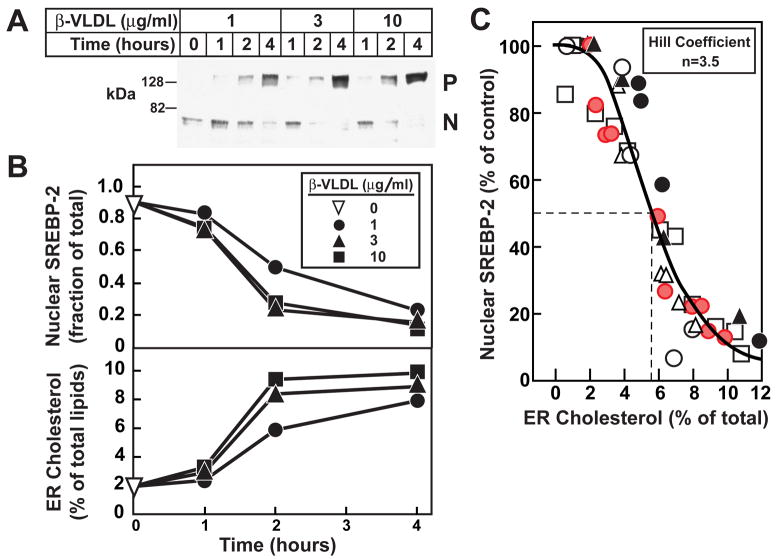
In addition to delivery in MCD complexes, cholesterol can be delivered to cells by receptor-mediated endocytosis of lipoprotein particles that bind to LDL receptors. One of these lipoproteins, β-VLDL, is isolated from plasma of cholesterol-fed rabbits (Kovanen et al., 1981) and is highly enriched in cholesteryl esters. These esters are hydrolyzed in lysosomes and the free cholesterol is transported to the ER. We used β-VLDL to deliver cholesterol to lysosomes in CHO-K1 cells and monitored SREBP-2 processing and ER cholesterol as in Figure 4. We first treated the cells with 1% HPCD for 1 hr to deplete cholesterol and then incubated them with increasing levels of β-VLDL for various times. As shown in Figure 5A, β-VLDL at 3 μg protein/ml showed maximal suppression of SREBP-2 cleavage at 2 hr. Increasing the β-VLDL concentration to 10 μg/ml did not shorten this time. The top panel of Figure 5B shows a densitometric quantification of the immunoblot from Figure 5A, and the bottom panel of Figure 5B shows the cholesterol content from purified ER membranes after each treatment. The data suggest that cholesterol from β-VLDL requires more than 1 hr to reach the ER, whereas cyclodextrin-delivered cholesterol reaches the ER in less than 1 hr (compare Figures 5 and 4).
Figure 5. Relation of SREBP-2 Processing to Cholesterol Content of Purified ER Membranes in CHO-K1 Cells after Incubation with Increasing Amounts of β-VLDL for Various Times.
(A) On day 0, CHO-K1 cells were set up in medium B (5% FCS). On day 3, cells were switched to medium C (lipoprotein-deficient serum) containing 1% HPCD for 1 hr, then switched to medium C containing the indicated concentration of β-VLDL. At the indicated time, cells were harvested and disrupted by a ball-bearing homogenizer. A portion of the homogenate (5% of total) was saved for immunoblot analysis of SREBP-2 (30 μg/lane). P, precursor form of SREBP-2; N, cleaved nuclear from of SREBP-2.
(B, top panel) Densitometric quantification of SREBP-2 in (A) expressed as the amount of the nuclear form relative to the total (nuclear plus precursor). ▽, zero-time value.
(B, bottom panel) The remainder (95%) of the homogenized cells from (A) were used to purify ER membranes, after which the lipids were extracted and the amount of cholesterol and phospholipids was quantified
(C) This graph represents the data from panels A and B (red circles) and five other similar experiments (○, ●, □, △, ▲). For each of the six experiments, the amount of nuclear SREBP-2 at zero time was normalized to 100%. Approximately 5×107 cells (from 5 10-cm dishes) were used for each data point. The solid line represents a best-fit of the experimental data to the Hill equation as described in Experimental Procedures. The best-fit values with 95% confidence intervals for cholesterol concentration (mol%) corresponding to 50% inhibition of SREBP-2 processing and the Hill coefficient were 5.7% ± 0.28% and 3.5 ± 0.13, respectively.
To quantify the correlation between ER cholesterol and SREBP-2 processing after β-VLDL treatment, we carried out five additional experiments using different concentrations of β-VLDL for various times. The combined analysis is shown in Figure 5C and again there is a clear threshold effect. We observed 50% inhibition of SREBP-2 cleavage at mole fraction of ER cholesterol of about 5.7%. This is slightly higher than the 4.5% value obtained with cholesterol/MCD (Figure 4). The significance of this difference is uncertain. Once again, analyzing data using the Hill equation reveals a high degree of positive cooperativity with a Hill coefficient of 3.5, similar to that observed with cholesterol/MCD.
Lowering the Threshold with Increased Insig
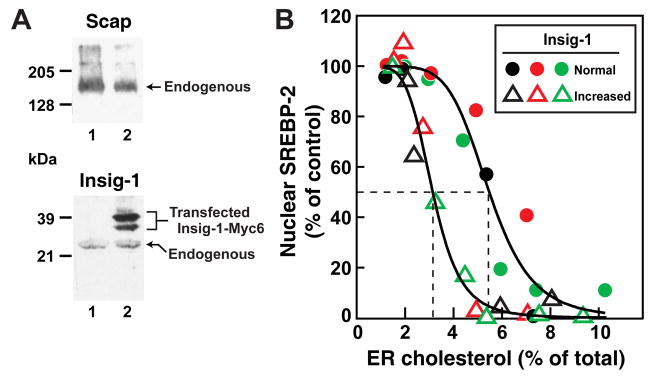
We next sought to determine whether an increase in Insig-1 in the ER membrane would lower the threshold for inhibition of SREBP-2 processing. In order to test this hypothesis, we took advantage of a CHO-7 cell line that stably overexpresses human Insig-1 (designated CHO/pInsig1-Myc cells) (Yang et al., 2002). Immunoblotting and densitometry show that the total amount of Insig-1 is elevated ~6.2-fold in CHO/pInsig1-Myc cells compared to the parental CHO-7 cells (Figure 6A). (The two bands of Insig-1-Myc6 in the immunoblots of the transfected cells represent versions with alternative initiator methionines, both of which are equally active (Yang et al., 2002)). Cholesterol levels in both cell lines were manipulated by first treating them with 1% HPCD for 1 hr to deplete cholesterol and then with increasing levels of cholesterol/MCD or β-VLDL for various times. We then assayed SREBP-2 processing and measured cholesterol levels in purified ER membranes as described for Figures 4 and 5. In experiments not shown, we verified that the ER purification procedure developed for CHO-K1 cells applies equally well to these two cell lines. The combined analysis from three different experiments (Figure 6B) shows clear threshold effects, just as in Figures 4 and 5. The mole fraction of cholesterol in the ER at which the processing of SREBP-2 is blocked by more than 50% is shifted to 1.8-fold lower values in the cells expressing increased vs. normal Insig-1 levels (i.e., from 5.5% to 3.1%). Analysis of the data by the Hill equation shows that the additional Insig increased the Hill coefficient from 5.1 to 6.9, sharpening the threshold for inhibition of SREBP-2 processing.
Figure 6. Relation of SREBP-2 Processing to Cholesterol Content of Purified ER Membranes in CHO-7 Cells Expressing Normal or Increased Ratio of Insig to Scap.
On day 0, CHO-7 or CHO/pInsig1-Myc cells were set up in Medium B (5% FCS). On day 3, cells were switched to Medium C (lipoprotein-deficient serum) containing 1% HPCD for 1 hr, then switched to Medium C containing various concentrations of cholesterol complexed to MCD or β-VLDL. After various times of incubation, cells were harvested and disrupted by a ball-bearing homogenizer. A portion of the homogenate (5% of total) was saved for immunoblot analysis of SREBP-2 (30 μg/lane). The extent of SREBP-2 processing to its cleaved nuclear form was quantified as described in the legend to Figure 4. The remainder (95%) of the homogenate was used to purify ER membranes, after which lipids were extracted, and the amount of cholesterol and phospholipids was quantified as described in Experimental Procedures.
(A) Immunoblot analysis of Scap and Insig-1 levels in the whole cell lysate (400 μg/lane) from CHO-7 cells (lane 1) and CHO/pInsig1-Myc cells (lane 2).
(B) The graph represents the combined data from two experiments (red and green symbols) where cholesterol was delivered by cyclodextrin complexes (cholesterol/MCD, see Figure 4) and one experiment (black symbols) where cholesterol was delivered by β-VLDL (see Figure 5). For each experiment, the amount of nuclear SREBP-2 at zero time was normalized to 100%. Approximately 108 cells (from ten 10-cm dishes) were used for each data point. Circles denote data from CHO-7 cells (normal Insig-1 content); triangles denote data from CHO/pInsig1-Myc cells (increased Insig-1 content). The solid lines represent a best-fit of the experimental data to the Hill equation as described in Experimental Procedures. The best-fit values with 95% confidence intervals for cholesterol concentration (mol%) corresponding to 50% inhibition of SREBP-2 processing and the Hill coefficient were 5.5% ± 0.74% and 5.1 ± 0.36, respectively, for CHO-7 cells, and 3.1% ± 0.24% and 6.9 ± 0.48, respectively, for CHO/pInsig1-Myc cells.
DISCUSSION
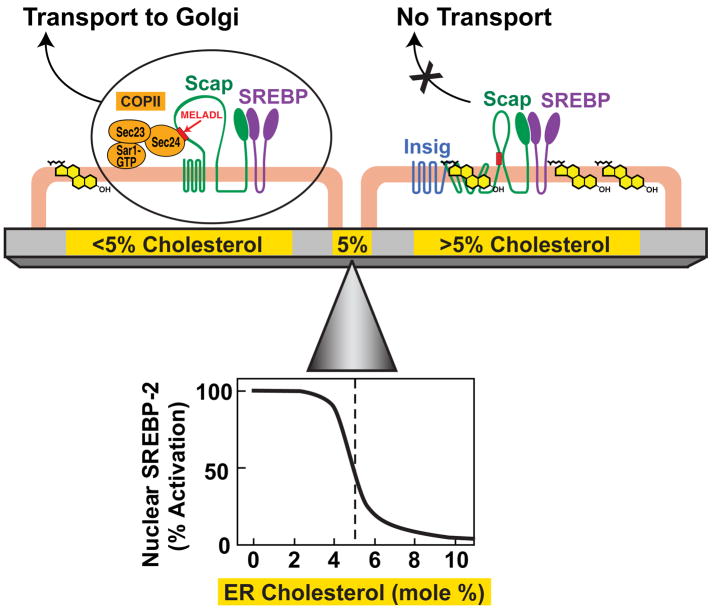
Together with previous data, the current studies reveal a delicate balance that controls the level of membrane cholesterol in mammalian cells (Figure 7). When cultured cells are depleted of cholesterol, the cholesterol content of ER membranes falls below 5 mol%. Under these conditions, the MELADL sequence in Scap binds CopII proteins which cluster the Scap/SREBP complex into transport vesicles that move to the Golgi where SREBPs are processed (Sun et al., 2007). When ER cholesterol rises above 5 mol%, the cholesterol binds to Scap, inducing Scap to bind Insig. As a result, CopII proteins no longer bind to Scap, and the Scap/SREBP complex is not transported to the Golgi. Inasmuch as Golgi processing is essential for SREBP activation, the net effect of cholesterol accumulation is to reduce transcription of genes required for cholesterol biosynthesis and uptake, thereby lowering membrane cholesterol and restoring cholesterol balance (Sun et al., 2007; Goldstein et al., 2006). Previous studies have shown that sterols regulate the SREBP pathway at the ER-to-Golgi transport step (which produces the active nuclear form of SREBPs) and not at other points such as degradation of the nuclear form (Yang et al., 1994; Yang et al., 1995).
Figure 7. Cholesterol Homeostasis – A Delicate Balance.
ER cholesterol levels are tuned to the overall cholesterol content of cells. When cellular cholesterol levels are low, ER cholesterol concentration is below a threshold value (<5 mol%). Under these conditions, Scap escorts SREBPs from ER to Golgi by binding to Sec24, a component of the Sar1/Sec23/Sec24 complex of the CopII protein coat. Once in the Golgi, the SREBPs are proteolytically processed to generate their nuclear forms that activate genes for cholesterol synthesis and uptake. When cellular cholesterol levels are no longer limiting, ER cholesterol concentration rises above a threshold value (>5 mol%). Under these conditions, cholesterol binds to Scap, initiating its binding to Insig, an ER retention protein. This interaction prevents the hexapeptide sorting signal (MELADL) in Scap from binding to CopII proteins, blocking transport of SREBPs to Golgi and thus preventing its subsequent proteolytic cleavage and transcriptional activation. Cholesterol levels are critically balanced by this sharp switch. The concentration of ER cholesterol that defines the tip of the fulcrum (~5 mol%) is set by the ratio of Scap to Insig. Upper half of schematic diagram adapted from Sun, et al. (2007); lower half from data in current paper.
The current study was made possible by obtaining highly purified ER membranes, which allowed us to accurately measure changes in ER cholesterol using mass spectrometry. We established a three-step protocol, beginning with gentle but efficient disruption of cells with a ball-bearing homogenizer followed by two gradient centrifugation steps that efficiently separate ER membranes from those of other organelles. The method was shown to work successfully in three CHO cell lines – CHO-K1, CHO-7, and a line of CHO-7 cells that are stably transfected with Insig-1. In studies to be reported elsewhere, we found that the method also works for cultured human fibroblasts (L. Abi-Mosleh, A. Radhakrishnan, M. S. Brown, and J. L. Goldstein, unpublished). We were especially careful to eliminate plasma membranes, lysosomes, and other membranes of the endocytic pathway as these have the highest concentrations of cholesterol (Figures 1 and 2).
Despite its virtual insolubility, cholesterol can be transported between various organelle membranes. Cholesterol depletion from the plasma membrane results in a drop in ER cholesterol from a resting value of ~7 mol% to ~2 mol% in 0.5 hr, triggering the processing of SREBP-2 to its active nuclear form (Figure 3A). This pathway is reversible: replenishing the plasma membrane cholesterol pool by the delivery of cholesterol from MCD complexes or from β-VLDL particles returns ER cholesterol to its original resting value, stopping SREBP-2 processing (Figures 3B, 4, and 5). When cholesterol is delivered in MCD, the kinetics are dominated by mass action as higher concentrations of cholesterol/MCD increase the speed with which cholesterol accumulates in the ER (Figures 4A and 4B). The kinetics differ when cholesterol is delivered in β-VLDL. Under these conditions, ER cholesterol does not increase until 1 hr even at the highest level of β-VLDL tested (Figure 5). This difference in kinetics is likely attributable to the distinct pathways taken by MCD- or β-VLDL-derived cholesterol to reach the ER. MCD-cholesterol likely partitions into the plasma membrane, from which it is transferred rapidly to the ER by vesicular or non-vesicular carriers (Prinz, 2007). On the other hand, β-VLDL must be taken up by endocytosis, its cholesteryl esters must be hydrolyzed, and the resultant cholesterol must be exported from lysosomes. One or more of these steps constitutes a rate-limiting barrier.
The pathways that transport cholesterol from one organelle to another allowing the ER to efficiently monitor cellular cholesterol levels are largely unknown. In studies by Xu and Tabas (1991) and Lange, et al. (Lange et al., 1999), ER cholesterol levels (as estimated by ACAT activity) rose only after total cell cholesterol crossed a threshold value, suggesting that the step(s) mediating transport between different cellular organelles may also be subject to regulation. Moreover, studies in artificial membranes have suggested a model whereby the chemical activity of cholesterol in the plasma membrane, by being poised near a threshold point, could serve as a regulatory sensor (Radhakrishnan and McConnell, 2000).
It has long been known that oxysterols like 25-HC can block cholesterol synthesis when added to cultured cells (Kandutsch and Chen, 1974; Brown and Goldstein, 1974). Once the SREBP pathway was discovered, it became clear that 25-HC acted through the same mechanism as cholesterol, i.e., by sequestering the Scap/SREBP complex in the ER (Brown and Goldstein, 1997). Yet, 25-HC was shown not to bind to Scap (Radhakrishnan et al., 2004). Rather, it binds to the other member of the Scap/Insig complex, i.e., Insig (Radhakrishnan et al., 2007). 25-HC binding to Insig promotes the same reaction as cholesterol binding to Scap, i.e., formation of the Scap/Insig complex (Sun et al., 2007). The current results support this explanation. Thus, addition of 10 μM 25-HC/MCD to cells caused a reduction in SREBP-2 processing at a time when the ER cholesterol level had not risen measurably (Figure 3C). When added at higher concentrations for longer periods of time, 25-HC can increase ER cholesterol as evidenced by an increase in the rate at which endogenous cholesterol is esterified by the ER resident enzyme ACAT (Brown et al., 1975). Consistent with this conclusion, Lange and Steck (1997) used their indirect method to demonstrate an increase in ER cholesterol 18–48 hr after adding 50 μM 25-HC to cells. The current results show that SREBP-2 cleavage is suppressed as early as 2 hr when no increase in ER cholesterol occurs.
Detailed analysis of the ER cholesterol concentration required to block SREBP-2 processing revealed a sharp, switch-like response at 4.5–5.7 mol% cholesterol in both CHO-K1 and CHO-7 cells (Figures 4–6). The threshold could be reached in either direction, i.e., whether cholesterol was being removed or added. In an initial attempt to model this response curve, we performed a Hill analysis, which assumes that a single reaction governs the inhibition of SREBP-2 cleavage. This is an oversimplification of the complexity of the SREBP pathway, but it provides us with a convenient starting point to quantify the sigmoidal behavior. The analysis suggested a high degree of positive cooperativity (best-fit Hill coefficient of 3.5–5.1 in Figures 4–6). Since the Hill coefficient is close to 4 and since Scap is a tetramer (Radhakrishnan et al., 2004), it is tempting to speculate that, analogous to the binding of oxygen to tetrameric hemoglobin (Koshland et al., 1966), the binding of cholesterol to one of the four binding sites in the Scap tetramer increases the affinity of the other sites for cholesterol, resulting in sharp cooperativity. Unfortunately, this hypothesis is difficult to test in our in vitro binding assay which contains purified Scap and cholesterol in detergent solutions (Radhakrishnan et al., 2004). In this system the rate-limiting step is transfer of cholesterol from detergent micelles to micelles that contain Scap. Because of this detergent effect, we are unable to measure cooperativity in the binding reaction (Radhakrishnan et al., 2004).
In CHO-7 cells with increased Insig-1 levels, the ER cholesterol that caused 50% inhibition of SREBP-2 processing was shifted from 5.5 mol% to 3.1 mol% (Figure 6), and the Hill coefficient increased from 5.1 to 6.9. The mechanism for this enhanced sensitivity is currently not known. It is possible that the interaction of Insig with Scap enhances the affinity of Scap for cholesterol, providing yet another source of cooperativity. The dependence of the cholesterol threshold on the Insig/Scap ratio increases the importance of the relative levels of these two proteins in controlling cholesterol metabolism in cells and in the body. In the liver, Insig levels are under exquisite control by insulin, a phenomenon that may play a role in regulating lipid synthesis in that crucial organ (Engelking et al., 2005; Yabe et al., 2003).
EXPERIMENTAL PROCEDURES
Materials
We obtained hydroxypropyl-β-cyclodextrin (HPCD) and methyl-β-cyclodextrin (MCD) from Cyclodextrin Technologies (High Springs, FL); 25-hydroxycholesterol (25-HC) from Steraloids, Inc.; cholesterol and iodixanol (Optiprep density gradient medium) from Sigma; monoclonal anti-CREB and anti-transferrin receptor antibodies from Invitrogen; monoclonal anti-caveolin-1 antibody from BD Biosciences; polyclonal anti-prohibitin-1 antibody from Abcam (Cambridge, MA); polyclonal anti-Na+/K+ ATPase antibody from Cell Signaling Technology (Danvers, MA); polyclonal anti-EEA1 and anti-PMP70 antibodies from Affinity BioReagents (Golden, CO); polyclonal anti-Sec61α from Millipore; and horseradish peroxidase-conjugated, affinity-purified donkey anti-mouse and anti-rabbit IgGs from Jackson ImmunoResearch Laboratories. Rabbit polyclonal antibody IgG-R139 raised against hamster Scap (amino acids 54–277 and 540–707) (Sakai et al., 1997); rabbit polyclonal antibody U1683 against hamster S1P (amino acids 1023–1052) (Espenshade et al., 1999); rabbit polyclonal antibody 81-2 against hamster ACAT-1 (amino acids 1–140) (Cao et al., 1996); and monoclonal antibody IgG-7D4 against hamster SREBP-2 (amino acids 32–350) (Yang et al., 1995) have been described in the indicated references. Other antibodies used in this study were generous gifts from J. Seemann, (polyclonal anti-GM130, anti-GRASP65); W. Dunn, (polyclonal anti-LAMP1); and from Y. K. Ho and Linda Donnelly (anti-Insig-1, hybridoma culture medium). Solutions of compactin and sodium mevalonate were prepared as described (Brown et al., 1978). Rabbit β-migrating very low density lipoproteins (β-VLDL, d <1.006 g/ml) (Kovanen et al., 1981) and newborn calf lipoprotein-deficient serum (Goldstein et al., 1983) were prepared by ultracentrifugation as described in the indicated reference. Stock solutions of complexes of cholesterol/MCD or 25-HC/MCD were prepared at a final sterol concentration of 2.5 mM and a sterol/MCD ratio of 1:10 as described (Brown et al., 2002). From many experiments in our laboratory, we have consistently found that HPCD is more effective than MCD in removing cholesterol from cells and MCD is more effective in delivering sterols in cells.
Buffers and Media
Buffer A contains 50 mM Tris-HCl (pH 7.5) and 150 mM NaCl. Buffer B is Buffer A supplemented with a protease inhibitor mixture (50 μg/ml leupeptin, 25 μg/ml pepstatin A, 10 μg/ml aprotinin, 25 μg/ml phenylmethylsulfonylfluoride, and 1/100 tablet/ml of complete EDTA-free protease inhibitor cocktail (Roche)). Medium A is a 1:1 mixture of Ham’s F-12 medium and Dulbecco’s modified Eagle’s medium containing 100 units/ml penicillin and 100 μg/ml streptomycin sulfate. Medium B is Medium A supplemented with 5% (v/v) fetal calf serum. Medium C is Medium A supplemented with 5% (v/v) newborn calf lipoprotein-deficient serum, 50 μM sodium compactin, and 50μM sodium mevalonate.
Cell Culture
Cells were maintained in monolayer culture at 37°C in 8–9% CO2. CHO-7 cells are a clone of CHO-K1 cells selected for growth in lipoprotein-deficient serum (Metherall et al., 1989). CHO/pInsig1-Myc cells are a clone of CHO-7 cells stably transfected with pCMV-Insig-1-Myc6 (Yang et al., 2002).
Cell Fractionation
On day 0, cells were set up in medium B at 700,000 cells/10-cm dish. On day 3, the cells were subjected to the indicated treatments, then washed twice with phosphate-buffered saline, and then scraped into 50-ml tubes. All further operations were carried out at 4°C. The cell pellets were collected after centrifugation at 500g for 10 min and resuspended in 1 ml of ice-cold Buffer B containing 15% (w/v) sucrose. Cells were then disrupted by passage 13 times through a ball bearing-homogenizer with a 10 μm clearance (Balch and Rothman, 1985). The homogenized cells (Fraction A, see fractionation scheme in Figure 1) were centrifuged at 3000g for 10 min to yield a pellet (Fraction B) and a supernatant (Fraction C). The supernatant was diluted to a total volume of 3 ml using Buffer A containing 15% sucrose. A discontinuous sucrose gradient was generated in a SW41 tube (Beckman Instruments) by overlaying the following sucrose solutions all in Buffer A: 2 ml 45%, 4 ml 30%, 3 ml of the diluted supernatant in 15% sucrose, and 1 ml 7.5%. The gradient was centrifuged at 100,000g in a SW40Ti rotor (Beckman) for 1 hr and allowed to slow down without the application of a brake, after which two bands of membranes were clearly visible and collected using a Pasteur pipet. The sucrose concentration in the light membrane (Fraction D) ranged from 14–18% and that in the heavy membrane fraction (Fraction E) ranged from 34–38%, as measured using a refractometer (model ABBE-3L, Milton Roy Co., Rochester, NY). A discontinuous iodixanol gradient was generated by underlaying, in succession, 2.25 ml of 19%, 21%, 23%, and 25% (v/v) iodixanol, all in buffer A. This discontinuous gradient was allowed to stand for 1–2 hr, allowing for diffusion across the interfaces to form a continuous gradient. The heavy membrane fraction from above (E) was loaded at the bottom of this gradient, which was then centrifuged for 2 hr at 110,000g in a SW40Ti rotor and allowed to slow down without the application of a brake. Fractions (0.8 ml each) were collected from the bottom. The entire fractionation process was completed in ~ 5 hr. Aliquots of Fraction (A) containing equal protein amounts were subjected to immunoblot analysis to monitor SREBP-2 processing. Protein concentrations were measured with a BCA kit (Pierce). Organelle enrichment was assessed by assaying equal volumes of all fractions for either enzyme activity or presence of organelle markers by immunoblot analysis.
Enzyme Assays
Enzyme assays for acid phosphatase, catalase, and cytochrome c reductase were carried out with assay kits (Sigma). Glucose-6-phosphatase activity was measured as described (Nordlie and Arion, 1966). β-Hexosaminidase activity was measured as described (Green et al., 1987).
Immunoblot Analysis
Samples were mixed with 5x SDS loading buffer, heated for 10 min at 95°C, and then subjected to 10% SDS-PAGE. After SDS-PAGE, the proteins were transferred to Hybond-C extra nitrocellulose filters (Amersham) and then incubated for 1 hr at room temperature with the indicated antibodies at the following concentrations: anti-CREB, 0.5 μg/ml; anti-transferrin receptor, 0.5 μg/ml; anti-caveolin-1, 0.25 μg/ml; anti-prohibitin-1, 1 μg/ml; anti-Na+/K+ ATPase, 1:1000 dilution; anti-EEA1, 2 μg/ml; anti-PMP70, 2 μg/ml; anti-Sec61α, 1:1000 dilution; anti-GM130, 1:1000 dilution; anti-GRASP65, 1:1000 dilution; anti-LAMP1, 1:5000 dilution; anti-SCAP IgG-R139, 10 μg/ml; anti-S1P U1683, 1:1000 dilution; anti-ACAT-1 81-2, 1:500 dilution; anti-SREBP-2 IgG-7D4, 3 μg/ml; anti-mouse IgG, 1:5000 dilution; anti-Insig-1, undiluted hybridoma culture medium; anti-rabbit IgG, 1:5000 dilution. Bound antibodies were visualized by chemiluminescence (Super Signal Substrate, Pierce). Filters were exposed to Kodak X-Omat Blue XB-1 film at room temperature for 1–120 sec.
Electron Microscopy
Membranes were fixed with 2% (v/v) gluteraldehyde in 100 mM cacodylate (pH 7.4) containing 200 mM sucrose at room temperature for 30 min, washed with 100 mM cacodylate (pH 7.4), embedded in 2.5% low melt agarose, and postfixed for 1 hr with 1% (w/v) osmium tetroxide, 1.5% (w/v) potassium cyanoferrate in 100 mM cacodylate (pH 7.4). The membranes were then washed with water, stained with 2% (w/v) aqueous uranyl acetate, dehydrated by washing with increasing concentrations of ethanol, and embedded in EPON 812 according to the manufacturer’s instructions (Electron Microscopy Sciences, Hatfield, PA). Ultrathin sections (70–90 nm) were cut by using a Leica Ultracut E ultramicrotome, placed on Formvar coated grids, and post-stained with 2% aqueous uranyl acetate (10 min) and lead citrate (5 min). Images were taken on a JEOL 1200EXII transmission electron microscope operated at 120 kV, with a Sis Morada 11 Mpixel CCD camera.
Lipid Extraction and Quantification
Lipids were extracted from cellular fractions using a chloroform/methanol (1:1, v/v) mixture (Bligh and Dyer, 1959). The organic solvent was evaporated under a gentle stream of nitrogen, and the dried lipid extracts were reconstituted in 95% methanol. Free unesterified cholesterol levels were measured with isotope dilution mass spectrometry using a deuterated analog of cholesterol (d7) (Avanti Polar Lipids) added to the sample prior to extraction as a standard for quantification. Lipid extracts were resolved and detected using high-performance liquid chromatography (HPLC) coupled to a triple quadrupole mass spectrometer (MS) through an electrospray ionization interface (McDonald et al., 2007). Total phospholipid levels were determined by a colorimetric assay that measures inorganic phosphate released after acidic digestion (detection limit ~ 1 μg) (Chalvardjian and Rudnicki, 1970). In estimating cholesterol:phospholipid molar ratios, we assumed that the mean molecular mass of the phospholipids (~800 Da) was twice that of cholesterol (388 Da).
Hill Analysis
The dependence of SREBP-2 processing on cholesterol levels in the ER was analyzed by a least squares fit of the Hill equation: Y = 100 (1 − Xn/(C + Xn)), where Y is the fraction of SREBP-2 in the processed nuclear form (expressed as % of control as described in the legends to Figures 4–6) and X is the mol% of free cholesterol in ER. The quantity 1/C can be considered as a cumulative binding constant for the species containing n ligands, assuming that this species dominates over all other liganded forms; n is regarded as a measure of positive cooperativity and is usually referred to as the Hill coefficient (Dahlquist, 1978; Wyman, Jr., 1964). Curve fitting and statistical analysis were performed with Mathematica (Wolfram Research, Inc.).
Acknowledgments
We thank our colleague Joachim Seemann for helpful discussion; Lisa Beatty, Shomanike Head, and Ly Le for invaluable help with tissue culture; Oliver Ritim and Matthew Francis for excellent technical assistance; and the UT Southwestern Live Cell Imaging Facility for electron microscopy. This research was supported by grants from the NIH (HL20948 and GM069338) and the Perot Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams CM, Reitz J, DeBrabander JK, Feramisco JD, Brown MS, Goldstein JL. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- Balch WE, Rothman JE. Characterization of protein transport between successive compartments of the Golgi apparatus: Asymmetric properties of donor and acceptor activities in a cell-free system. Arch Biochem Biophysics. 1985;240:413–425. doi: 10.1016/0003-9861(85)90046-3. [DOI] [PubMed] [Google Scholar]
- Bergstrand A, Dallner G. Isolation of rough and smooth microsomes from rat liver by means of a commercially available centrifuge. Analytical Biochem. 1969;29:351–356. doi: 10.1016/0003-2697(69)90321-2. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brown AJ, Sun L, Feramisco JD, Brown MS, Goldstein JL. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- Brown MS, Dana SE, Goldstein JL. Cholesteryl ester formation in cultured human fibroblasts: Stimulation by oxygenated sterols. J Biol Chem. 1975;250:4025–4027. [PubMed] [Google Scholar]
- Brown MS, Faust JR, Goldstein JL, Kaneko I, Endo A. Induction of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts incubated with compactin (ML-236B), a competitive inhibitor of the reductase. J Biol Chem. 1978;253:1121–1128. [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Suppression of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity and inhibition of growth of human fibroblasts by 7-ketocholesterol. J Biol Chem. 1974;249:7306–7314. [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Cao G, Goldstein JL, Brown MS. Complementation of mutation in acyl-CoA: cholesterol acyltransferase (ACAT) fails to restore sterol regulation in ACAT-defective sterol-resistant hamster cells. J Biol Chem. 1996;271:14642–14648. doi: 10.1074/jbc.271.24.14642. [DOI] [PubMed] [Google Scholar]
- Chalvardjian A, Rudnicki E. Determination of lipid phosphorus in the nanomolar range. Anal Biochem. 1970;36:225–226. doi: 10.1016/0003-2697(70)90352-0. [DOI] [PubMed] [Google Scholar]
- Dahlquist FW. The meaning of scatchard and hill plots. Methods in Enzymology. 1978;48:270–299. doi: 10.1016/s0076-6879(78)48015-2. [DOI] [PubMed] [Google Scholar]
- Engelking LJ, Liang G, Hammer RE, Takaishi K, Kuriyama H, Evers BM, Li WP, Horton JD, Goldstein JL, Brown MS. Schoenheimer Effect explained -Feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J Clin Invest. 2005;115:2489–2498. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade PJ, Cheng D, Goldstein JL, Brown MS. Autocatalytic processing of Site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J Biol Chem. 1999;274:22795–22804. doi: 10.1074/jbc.274.32.22795. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Basu SK, Brown MS. Receptor-mediated endocytosis of low density lipoprotein in cultured cells. Meth Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Green SA, Zimmer KP, Griffiths G, Mellman I. Kinetics of intracellular transport and sorting of lysosomal membrane and plasma membrane proteins. J Cell Biol. 1987;105:1227–1240. doi: 10.1083/jcb.105.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandutsch AA, Chen HW. Inhibition of sterol synthesis in cultured mouse cells by cholesterol derivatives oxygenated in the side chain. J Biol Chem. 1974;249:6057–606l. [PubMed] [Google Scholar]
- Koshland DE, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochem. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- Kovanen PT, Brown MS, Basu SK, Bilheimer DW, Goldstein JL. Saturation and suppression of hepatic lipoprotein receptors: A mechanism for the hypercholesterolemia of cholesterol-fed rabbits. Proc Natl Acad Sci USA. 1981;78:l396–l400. doi: 10.1073/pnas.78.3.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange Y, Steck TL. Quantitation of the pool of cholesterol associated with acyl-CoA: Cholesterol acyltransferase in human fibroblasts. J Biol Chem. 1997;272:13103–13108. doi: 10.1074/jbc.272.20.13103. [DOI] [PubMed] [Google Scholar]
- Lange Y, Ye J, Rigney M, Steck TL. Regulation of endoplasmic reticulum cholesterol by plasma membrane cholesterol. J Lipid Res. 1999;40:2264–2270. [PubMed] [Google Scholar]
- McDonald JG, Thompson BM, McCrum EC, Russell DW. Extraction and analysis of sterols in biological matrices by high performance liquid chromatography electrospray ionization mass spectrometry. Methods Enzymol. 2007;432:145–170. doi: 10.1016/S0076-6879(07)32006-5. [DOI] [PubMed] [Google Scholar]
- Metherall JE, Goldstein JL, Luskey KL, Brown MS. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J Biol Chem. 1989;264:15634–15641. [PubMed] [Google Scholar]
- Nordlie RC, Arion WJ. Glucose-6-phosphatase. Methods in Enzymology. 1966;9:619–625. [Google Scholar]
- Prinz WA. Non-vesicular sterol transport in cells. Prog Lipid Res. 2007;46:297–314. doi: 10.1016/j.plipres.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Ikeda Y, Kwon HJ, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Oxysterols block transport by binding to Insig. Proc Natl Acad Sci USA. 2007;104:6511–6518. doi: 10.1073/pnas.0700899104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, McConnell HM. Chemical activity of cholesterol in membranes. Biochemistry. 2000;39:8119–8124. doi: 10.1021/bi0005097. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell. 2004;15:259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Sakai J, Nohturfft A, Cheng D, Ho YK, Brown MS, Goldstein JL. Identification of complexes between the COOH-terminal domains of sterol regulatory element binding proteins (SREBPs) and SREBP cleavage-activating protein (SCAP) J Biol Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- Sun LP, Seemann J, Brown MS, Goldstein JL. Sterol-regulated transport of SREBPs from endoplasmic reticulum to Golgi: Insig renders sorting signal in Scap inaccessible to COPII proteins. Proc Natl Acad Sci USA. 2007;104:6519–6526. doi: 10.1073/pnas.0700907104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veldhoven PP, Baumgart E, Mannaerts GP. Iodixanol (Optiprep), an improved density gradient medium for the Iso-osmotic isolation of rat liver peroxisomes. Analytical Biochem. 1996;237:17–23. doi: 10.1006/abio.1996.0194. [DOI] [PubMed] [Google Scholar]
- Wyman J., Jr Linked functions and reciprocal effects in hemoglobin: A second look. Advan Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- Xu XX, Tabas I. Lipoproteins activate acyl-coenzyme A: cholesterol acyltransferase in macrophages only after cellular cholesterol pools are expanded to a critical threshold level. J Biol Chem. 1991;266:17040–17048. [PubMed] [Google Scholar]
- Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: Implications for fatty acid synthesis. Proc Natl Acad Sci USA. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Brown MS, Ho YK, Goldstein JL. Three different rearrangements in a single intron truncate SREBP-2 and produce sterol-resistant phenotype in three cell lines. J Biol Chem. 1995;270:12152–12161. doi: 10.1074/jbc.270.20.12152. [DOI] [PubMed] [Google Scholar]
- Yang J, Sato R, Goldstein JL, Brown MS. Sterol-resistant transcription in CHO cells caused by gene rearrangement that truncates SREBP-2. Genes Dev. 1994;8:1910–1919. doi: 10.1101/gad.8.16.1910. [DOI] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Zambrano F, Fleischer S, Fleischer B. Lipid composition of the Golgi apparatus of rat kidney and liver in comparison with other subcellular organelles. Biochimica et Biophysica Acta. 1975;380:357–369. doi: 10.1016/0005-2760(75)90104-6. [DOI] [PubMed] [Google Scholar]