Abstract
Background
Sodium lithium countertransport (SLC) activity, an intermediate phenotype of essential hypertension, has been linked to a region of baboon chromosome 5, homologous to human chromosome 4p. Human SLC34A2, located at chromosome 4p15.1-p15.3, is a positional candidate gene for SLC. The specific aim of this study was to identify genetic variants of the SLC34A2 gene in both baboon and human, and to examine the relationship of these polymorphisms with SLC activity and blood pressure.
Methods
Single nucleotide polymorphism (SNP) was identified by sequencing the SLC34A2 gene in 24 baboon founders and 94 unrelated individuals. All tag SNPs in SLC34A2 were genotyped in 1856 individuals from 252 pedigrees of mixed European ancestry. Three SNPs in baboon were genotyped in 634 baboons comprising 11 pedigrees.
Results
In human, one SNP (rs12501856) was found to be significantly associated with SLC individually, though it did not pass multiple testing correction; however haplotype 2 containing allele C of SNP rs12501856 showed strong evidence of association with SLC (p=0.0037) after multiple comparison adjustment. This haplotype was also marginally associated with diastolic blood pressure and systolic blood pressure. This finding was confirmed in baboons, where a highly significant association was detected between SLC and baboon SNP Asn136Asn (p=0.0001). However, the associated SNP did not account for the linkage signal on baboon chromosome 5.
Conclusions
Consistent results in two different species imply that SLC34A2 is associated with SLC activity and blood pressure.
INTRODUCTION
Essential hypertension, a major risk factor for cardiovascular diseases, is a multifactorial disease with 30~50% genetic contribution. 1, 2 It is well documented that essential hypertension is highly heterogeneous, individuals with the same blood pressure levels may have mutations at completely different loci, and also hypertension may involve the same disease loci with different alleles.3–5 This characteristic may explain the often conflicting results from genetic studies in essential hypertension. To use an intermediate trait which is predictive of essential hypertension and regulated by fewer genes and environmental factors, such as sodium lithium countertransport (SLC), may provide substantial advantages to gene discovery studies.
Sodium-lithium countertransport (SLC), first described by Canessa et al. 6, is assessed by measuring the rate of lithium loss from lithium loaded erythrocytes incubated in sodium-free versus sodium-rich medium. Canessa et al. 6 further reported that SLC is elevated in individuals with hypertension. SLC is a stable, bimodally distributed trait 7 with high estimated heritability (55%–88%) in both humans and baboons 8–10. There is strong evidence of a major gene influencing the distribution of SLC in humans 11–13. Turner and Michels 14 showed that there was significant correlation of SLC with blood pressure in the general population of Rochester, MN, and that this correlation persisted after adjusting for body mass index, triglycerides and total cholesterol among both men and women. Turner et al. 15 showed that for each standard deviation increase in SLC, the risk of hypertension approximately doubled in men (OR 2.25, 95% CI 1.44–3.51) and women (OR=1.77, 95% 1.32–2.37). Weder et al. 16 reported that adults with elevated SLC exhibited higher blood pressure levels as children, and elevated SLC in normotensive offspring of hypertensive parents has been reported in U.S. Caucasians, African Americans and Africans.17, 18 These observations suggest that SLC is a premorbid marker of essential hypertension in humans. Kammerer et al. 10 have presented convincing evidence of a locus influencing SLC on baboon chromosome 5, although the measurement of blood pressure in the unrestrained baboon is not technically feasible.
The SLC34 family of solute carriers comprises three members: Type II Na/P(i)-cotransporters NaPi-IIa (SLC34A1), NaPi-IIb (SLC34A2) and NaPi-IIc (SLC34A3). SLC34A2 cotransported phosphate and sodium into cells in the presence of sodium.19 It plays a role in sodium and phosphate homeostasis. Human SLC34A2 is located at chromosome 4p15.1-p15.3 20 in a region of the genome homologous to the region of baboon chromosome 5, linked to SLC.10 Thus, SLC34A2 is a positional and biological candidate gene for SLC. To examine the relationship between SLC34A2 variation and SLC, we conducted a detailed sequence analysis of SLC34A2 in baboons of known phenotype and of its’ human ortholog, and conducted an association analysis between polymorphisms of SLC34A2 and variation in SLC activity in 634 baboons comprising 11 pedigrees and 1856 individuals from 252 pedigrees of mixed European ancestry.
MATERIAL AND METHODS
Subjects
Individuals in this study were participants in the Rochester Family Heart Study (RFHS) phase II, including 252 pedigrees containing 1856 individuals. Multi-generation pedigrees were ascertained through households having two or more children enrolled in the school of Rochester, MN, between 1984 and 1991. The samples are of mixed European ancestry. The details of the recruitment have been reported previously.21, 22 All procedures were approved by the Institutional Review Board at the Mayo Clinic, Rochester, MN and all subjects gave written informed consent.
634 noninbred baboons (Papio hamadryas) comprising 11 pedigrees ranging in size from 16 to 99 animals. These 2- and 3-generation pedigrees consisted of 202 founders (29 sires and 173 dams) that were not selected for blood pressure and their 432 offspring. All experimental protocols were approved by the Southwest Foundation for Biomedical Research Institutional Animal Care and Use Committee.
Phenotyping
For humans, blood pressure was taken with a random zero sphygmomanometer (Howkeley and Sons LTD., West Sussex, UK) while subjects were seated in a quiet room. Average of three systolic readings and three diastolic readings for each subject were used in all analyses and are referred to as systolic and diastolic blood pressure.22 The prevalence of hypertension in this sample was 15.9% and 87.4% of these were receiving antihypertensive medications. These individuals were included in this analysis to be representatives of the population of Rochester. Blood pressure in baboons could not be measured except under chemical restraint and are not considered reliable for analysis. The maximal velocity of the SLC was used as an indicator of SLC activity and was determined by measuring the external sodium-stimulated lithium efflux from lithium-loaded RBCs as previously reported.6, 23 The coefficient of variation for assays done with fresh cells from the same individual on different days was 8.9%, and for replicate assays done on the same day was 7.5%. Details of the baboon phenotyping are presented in Kammerer et al. 10
Sequencing
Initial SNP identification was performed by sequencing each exon, exon-intron boundaries, and the proximal 5′ region, containing the putative promoter region, of SLC34A2 in 94 unrelated individuals from RFHS phase II and 24 baboon founders. In order to maximize the potential genetic differences, those 94 subjects were selected from the highest and lowest deciles for SLC. The putative promoter region which was predicted by using Gene2Promotor program (Genomatix, Munich, Germany) is located around 500bp upstream of first coding exon (exon 2). Genomic DNA was isolated from peripheral blood leucocytes by standard procedures. DNA sequencing was performed by polymerase chain reaction (PCR) amplification (Supplement table 1 and 2) of the target fragment, purification of the product (ExoSAP-IT kit, USB corporation), and a sequencing using the ABI dRhodamine cycle sequencing kit (Applied Biosystems). Sequencing products were purified and applied to an ABI3700 capillary sequencer. Sequences were aligned and curated using the program SEQUENCHER (Gene Codes). All sequencing was carried out by the University of Pittsburgh, Genomics and Proteomics Core Laboratory (Pittsburgh, PA).
Genotyping
Human tag SNPs were selected using haploview 24 with data dumps from HapMap project, since the SNPs identified by resequencing are rare SNPs (MAF <0.05). All human tag SNPs with minor allele frequency (MAF) >0.05 were genotyped by Illumina Beadarray (Illumina Inc, San Diego, CA), genotypes of 4 SNPs rs12501856, rs6448389, rs3775909 and rs3796777 were further confirmed by fluorescence polarization (Supplement table 3) described by Chen et al.25, 26 using the L.J.L. Biosystems’ Analyst HT Assay Detection System. Baboon SNPs were chosen on the basis of the sequencing results and genotyped by sequencing and fluorescence polarization. All potentially functional sites, missense mutations and variants located in functional domains with minor allele frequency ≥ 10%, were genotyped. Functional domains in SLC34A2 were predicted using Bioinformatic Harvester.27 Since SNP Lys636Asn was in high Linkage Disequilibrium (LD) with Leu630Leu (r2=0.98) and Pro680Ala (r2=0.92), Asn136Asn was also in LD with Glu61Glu (r2=0.78), we eventually chose to genotype three SNPs: Lys636Asn, Asn136Asn and Lys645Glu across the entire baboon population. R2 was estimated by pair wise LD test in haploview software package.24
Data analysis
A basic X2 goodness-of-fit test was used to test the deviations from the Hardy-Weinberg Equilibrium (HWE) in all genotype data from unrelated founders. Mendelian error checking was performed using the INFER procedure in PEDSYS software version 2.0 (Southwest Foundation for Biomedical Research, San Antonio, TX).
Human SNP genotype association analyses were performed by SOLAR (Sequential Oligogenic Linkage Analysis Routines) Version 2.1.4.28 For each SLC34A2 polymorphism, we used maximum likelihood methods to estimate the possible linear effects of each genotype on SLC activity. Models were also adjusted for age, sex and weight.10 Using the likelihood ratio test, we compared this model to a nested model in which the effects of the SLC34A2 genotypes were set equal to zero. Associations were also confirmed by family-based association test (FBAT) version 1.5.1.29, 30 Option –e in FBAT, which could test for association in an area of known linkage, was used to complete the association analysis. 31 An additive model was tested. We used the Bonferroni correction to control for multiple testing. The haplotype version of FBAT (HBAT) was used to estimate the haplotype frequencies. Association of SLC and haplotypes having a ≥2% total frequency in the population (“major haplotypes”) were further analyzed by HBAT.32
For baboon, we tested for an association between SLC activity and the SLC34A2 polymorphisms using SOLAR. The adjusted means of SLC activity by SNP genotype were also estimated by SOLAR.33 Due to the complex family structure within the baboon pedigrees, we couldn’t use FBAT and HBAT in association analysis for baboons. We derived haplotypes from baboon SNP genotypes using PHASE version 2.1 34, 35 and then incorporated the effects of each haplotype into our analyses. After detecting a significant effect of the SLC34A2 SNP, variance component linkage analysis was used to evaluate whether this SNP account for the observed linkage signals on baboon chromosome 5.10 Variance component linkage analyses was performed by incorporating additive effects of the specific SNP genotype as a covariate, thus removing variation due to the SLC34A2 SNP genotype. For details of the variance component linkage analysis see Kammerer et al.10 Multipoint linkage results of the model containing SLC34A2 were compared to the original linkage results (that did not include SLC34A2). If the SLC34A2 SNP is the sole functional polymorphisms accounting for the heritable variation of the trait, the linkage signal will completely disappear and the LOD score should drop to 0. If the measured SNP is only one of the several functional polymorphisms or is in disequilibrium with true variant, the evidence for linkage should remain.36
RESULTS
Characteristics of subjects
In humans, the number of males and females was almost equal and the average SLC activity was around 298.1 umol/l RBC/hr. For baboons, the total 204 males and 430 females had a mean SLC activity 242 umol/l RBC/hr. Detailed subject characteristics were shown in table 1.
Table 1.
Characteristics of humans (RFHS II) and baboon used in this study
| Characteristics | Values* | |
|---|---|---|
| RFHS II | Age (year) | 41.1 ± 23.3 |
| Male (%) | 51.1 | |
| DBP(mmHg) | 69.5 ± 11.1 | |
| SBP(mmHg) | 116.9 ± 20.2 | |
| Height(cm) | 165.9 ± 12.4 | |
| Weight(kg) | 68.7 ± 19.3 | |
| BMI(kg/m2) | 24.6 ± 5.4 | |
| SLC activity(umol/l RBC/hr) | 298.1 ± 119.8 | |
|
| ||
| Baboon | Age (year) | 9.4 ± 6.0 |
| Male (%) | 32.2 | |
| Weight(kg) | 17.5±5.5 | |
| SLC activity(μmol/l RBC/hr) | 242±99 | |
Values are represented as mean ± SD (Standard Deviation)
Sequencing
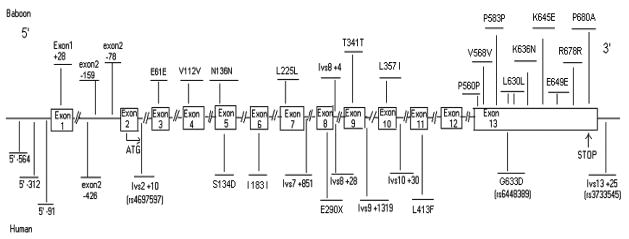
Figure 1 summarizes the sequence variation identified in 94 humans and 24 baboons. A total of 5 exonic single nucleotide polymorphisms (SNP) and 10 intronic SNPs were observed in the human compared 17 exonic SNPs and 3 intronic SNPs to in the baboon, despite the smaller sample size of baboons. This difference in occurrence of SNPs between humans and baboons has been observed for other loci 37 and may be due to the mixed nature of the founding population of baboons, which included both P.h. cynocephalus and P.h. anubis, or to the difference in population history between humans and baboons.
Figure 1. Baboon (top) and human (bottom) sequence variation in SLC34A2.
IVS: intervening sequence. For those SNPs previously reported for human, the rs# is given. All SNPs located in coding region were named in term of their amino acid locations. For SNPs located in non-coding region, the first base of any exon or IVS will be designed as “+1”, any base in the upstream (closer to 5′) of it will be denoted as “−”, and numbered from small to large according to their distance to “+1” from proximate to distal, e.g. the base next to it will be coded as “−1”; any base in the downstream (closer to 3′) of it will be denoted as “+”.
Comparison of human and baboon nucleotide and amino acid sequence for SLC34A2 is shown in Supplement Figure 1. Bioinformatic Harvester 27 predicted five domains, including two sodium-phosphate (Na_Pi) cotransport domains, two low compositional complexity domains and one transmembrane domain which are highlighted in different colors in Supplement Figure 1. Low complexity regions are not well understood but have been shown to be functionally important in some proteins.38 Several baboon SLC34A2 SNPs occur in regions important for SLC34A2 function, for example, Asn136Asn (exon5) is located in Na_Pi cotransport domain and Lys636Asn (exon 13) is located in low compositional complexity domain. These SNPs as well as another missense mutation Lys645Glu were further genotyped in pedigreed baboons.
Genotyping
Supplement table 4 summarizes genotype and allele frequencies (based on 48 chromosomes) for humans. Five exonic and 15 intronic SNPs were observed.
Supplement table 5 summarizes genotype and allele frequencies observed in baboon samples. Seventeen exonic single nucleotide polymorphisms (SNP) and 3 intronic SNP were identified. Only a small region flanking each exon was sequenced so the number of intronic SNPs detected is small. No SNPs were shared between the two species.
Association analysis in humans
Of the seven SLC34A2 SNPs genotyped in humans, SNP rs12501856 is associated with phenotypic variation in SLC (table 2). The p-values (0.037 for unadjusted and 0.024 for adjusted) by SOLAR are nominally significant, which was also confirmed by FBAT (P=0.03, data not shown), although it was not statistically significant after Bonferroni correction. However, haplotype association tests (table 3) show strong association (p=0.0027) between haplotype 2 in human SLC34A2 and SLC activity which was significant after correction for multiple testing. Haplotype 2 was marginally significant association with SBP (p=0.073) and DBP (p=0.049). Mean SLC activity for the CC genotype was 317.2±11.3 μmol Li/l RBC/hr whereas for CT and TT individuals, mean SLC activity was around 288.6±5.1 μmol Li/l RBC/hr (p<0.05) (supplement table 6).
Table 2.
Results of human SLC34A2 allelic association tests in Phase II
| SLC | SBP | DBP | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HWE | Unadjusted | Adjusted | Unadjusted | Adjusted | Unadjusted | Adjusted | |||
| SNP | Position | MAF | P | P | P* | P | P* | P | P* |
| rs12501856 | Intron 1 | 0.34 | 0.79 | 0.04 | 0.02 | 0.37 | 0.19 | 0.35 | 0.79 |
| rs3775909 | Intron 3 | 0.18 | 0.66 | 0.78 | 0.83 | 0.34 | 0.32 | 0.96 | 0.26 |
| rs3796777 | Intron 9 | 0.13 | 0.38 | 0.68 | 0.58 | 0.33 | 0.34 | 0.33 | 0.86 |
| rs2240995 | Intron 10 | 0.18 | 0.99 | 0.91 | 0.93 | 0.37 | 0.32 | 0.8 | 0.82 |
| rs2240996 | Intron 10 | 0.12 | 0.38 | 0.92 | 0.62 | 0.96 | 0.56 | 0.037 | 0.07 |
| rs12505556 | Intron 12 | 0.13 | 0.98 | 0.64 | 0.62 | 0.51 | 0.75 | 0.038 | 0.05 |
| rs6448389 | Exon13 | 0.13 | 0.27 | 0.12 | 0.39 | 0.98 | 0.44 | 0.52 | 0.74 |
SNP indicates single-nucleotide polymorphism; MAF, minor allele frequency; HWE, hardy-weinberg equilibrium; SLC, sodium lithium countertransport; SBP, systolic blood pressure; and DBP, diastolic blood pressure.
P value is adjusted by age, sex and body mass index.
The significance level for a single test is set as p=0.007 (α = 0.05/7; Seven SNPs).
Table 3.
Summary of haplotype association analysis results in human
| Hap | Haplotype | SLC | SBP | DBP | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | rs12501856 | rs3775909 | rs3796777 | rs2240995 | rs2240996 | rs12505556 | rs6448389 | Freq | P value | P value | P value |
| h1 | T | C | A | C | C | C | G | 0.346 | 0.233 | 0.621 | 0.563 |
| h2 | C | C | A | C | C | C | G | 0.159 | 0.004 | 0.073 | 0.049 |
| h3 | C | C | A | G | C | C | G | 0.051 | 0.455 | 0.385 | 0.692 |
| h4 | T | G | G | C | C | C | G | 0.043 | 0.659 | 0.431 | 0.428 |
| h5 | T | C | A | C | C | C | A | 0.034 | 0.269 | 0.462 | 0.501 |
| h6 | T | C | A | G | C | C | G | 0.03 | 0.786 | 0.68 | 0.6 |
| h7 | C | C | A | C | C | C | A | 0.03 | 0.023 | 0.027 | 0.029 |
| h8 | C | C | A | C | A | C | G | 0.03 | 0.717 | 0.324 | 0.378 |
| h9 | T | G | G | C | C | C | A | 0.027 | 0.751 | 0.921 | 0.993 |
| h10 | T | G | A | C | A | C | G | 0.022 | 0.558 | 0.652 | 0.622 |
Hap indicates haplotype; Freq, frequency.
The significance level for a single test is set as p=0.005 (α = 0.05/10; Ten haplotypes).
Linkage and association analysis in baboons
Among all genotyped sites in baboon, SNP Asn136Asn (exon5) shows (Table 4) strong evidence of association with SLC variation (p=0.0001) even after Bonferroni correction. The genotypes of this single SNP explained about 5% of total variance in SLC activity. The genotype specific effects on mean SLC activity for SNP Asn136Asn were similar as in human SNP rs12501856. The average SLC activity in individuals with CC genotype was much higher than ones in the other two groups (295.1±10.9 μmol Li/l RBC/hr, vs. 232.5±7.1 μmol Li/l RBC/hr, p<0.01). We also analyzed haplotypes at the SLC34A2 locus, and obtained a significant association with SLC activity (P= 0.0002). As expected, haplotypes containing the Asn136Asn C allele had increased SLC activity. (Data is not shown). In order to determine if Asn136Asn is the genetic variant that accounts for the baboon linkage signal identified by Kammerer et al,10 multipoint linkage analysis incorporating Asn136Asn genotypes was performed. The peak LOD score was only slightly reduced from 11.2 (in original linkage model that did not include SLC34A2) to 10.7 (in model containing SLC34A2) (data not shown), indicating this SNP did not account for the QTL (quantitative trait locus) effect.
Table 4.
Summary of results of baboon SLC34A2 allelic association analyses
| SNPs | Position | Minor Allele Frequency | HWE P value | SOLAR |
|---|---|---|---|---|
| Asn136Asn | exon 5 | 0.47 | 0.85 | 0.0001 |
| Lys 636 Asn | exon 13 | 0.46 | 0.17 | 0.28 |
| Lys 645 Glu | exon 13 | 0.1 | 0.14 | 0.27 |
The significance level for a single test is set as 0.0175 (α = 0.05/3; three SNPs).
DISCUSSION
Sodium lithium countertransport, a premorbid marker of essential hypertension, has been previously mapped to a region of baboon chromosome 5.10 Human SLC34A2 is located at chromosome 4p15.1-p15.3 20 in a region of the genome homologous to the linkage region of baboon chromosome 5, and near a suggestive linkage peak in humans making SLC34A2 is a positional candidate gene for SLC activity.39
Resequencing of SLC34A2 in the human and the baboon establishes the strong homology in exonic organization and sequence (96.7%) between the human and baboon SLC34A2 genes and extensive variation in both species (Supplement Figure 1). In humans, the SNP rs12501856, located in intron 1 was significantly associated with increased SLC activity in single locus analysis (p=0.02), and with a more profound effect in haplotype analysis (p=0.004). This SNP differentiates h2 from the most frequent haplotype h1 and suggests that it is making variation in the 5′ end of SLC34A2. Unfortunately, the other haplotypes, h3-h10, occur at frequencies too low, ≤ 5%, to be more specific. Genotyping of SNPs in baboon SLC34A2 revealed one variant of SNP Asn136Asn in exon 5 that is significantly (p=0.0001) associated with phenotypic variation in SLC activity. Baboons homozygous for the less common CC genotype had much higher levels of SLC activity than did carriers of the T allele. This SNP explained approximately 5% of the variation in SLC activity. It is worth noting that the baboon SNP which shows association with SLC is also in the 5′ region of the SLC34A2 gene. There are many possible explanations for a difference in allelic effect between humans and baboon: 1) The environments of baboons are more homogeneous than those of humans, for example, baboons were fed the same diet and raised in similar housing. 2) General genetic and physiological differences between the two species. 3) Specific genotype effects of different SNPs in the two species.
To address the question if SNP Asn136Asn accounts for the linkage signal identified by Kammerer et al.,10 variance component linkage analysis which incorporated the measured genotype effect of this SNP into model was conducted. After removing the SNP effect, evidence of linkage remained in the model. There are several possible explanations for why this polymorphism showed strong association with SLC but minimally influenced the linkage signal. Firstly, because association analysis is far more powerful than linkage analysis in detecting the common variants with modest effects, it’s possible that Asn136Asn is a surrogate for functional variation elsewhere in the gene. The most likely possibility is that another locus in linkage disequilibrium with gene SLC34A2 explains these findings. It is also possible that the linkage signal is explained by several genes, SLC34A2 being one of them but not the major gene influencing SLC activity. Lastly, the association is spurious; however, the consistent evidence of associations of SLC activity with SLC34A2 in two different species makes the last explanation less likely. In addition, since our study design was family-based, including ascertainment of relatively large family-based samples and application of family-based association tests (FBAT), the effect of major confounding factor-population stratification in the association study was avoided. Also, the very small p-values of association tests in both species make the likelihood of false positive result very small.
Comparison of results in the human and baboon are limited by the fact that blood pressure in the baboon can not be measured except under chemical restraint leaving the relationship between hypertension and sodium lithium countertransport in the baboon open to question.
CONCLUSIONS
This study provides strong evidence that variation in SLC34A2 is significantly associated with interindividual variation in sodium lithium countertransport in humans and baboons. In humans, a significantly higher SLC activity was associated with a haplotype marking the 5′ region of SLC34A2. In baboons, variation in SLC34A2 explains a small but significant proportion of the variation in sodium lithium countertransport (≤ 5%) but is not the locus responsible for the strong evidence of linkage to chromosome 5 in the baboon. Thus, SLC34A2 appears to be a relatively minor determinant of total sodium lithium countertransport, perhaps through linkage disequilibrium with major gene on baboon chromosome 5. The relationship of baboon chromosome 5 to the homologous region in the human genome is complicated by a complex inversion in this region in the baboon.
WEB RESOURCES
Accession numbers and URLs for data presented herein are as follows:
Bioinformatic Harvester, http://harvester.fzk.de/harvester/
Celera SNP Reference Database, http://www.celera.com/corporate/snpdata.html
CHIP Bioinformatics tools, http://snpper.chip.org/ (for the SNPper program)
GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for SLC34A2 cDNA sequence [accession number NM_006424], for genemic DNA sequence [accession number NC_000004]
Genomatix, http://www.genomatix.de/ (for the Gene2Promotor program)
Haploview, http://www.broad.mit.edu/mpg/haploview/
National Center for Health Statistics, http://www.cdc.gov/nchs/hus.htm
NCBI, http://www.ncbi.nlm.nih.gov/
University of Pittsburgh, Genomics and Proteomics core laboratory, http://www.genetics.pitt.edu
Supplementary Material
Acknowledgments
We would like to thank Shifra Birnbaum for his assistance in genotyping SNPs in baboon and Marget Kenney for her help with variance component linkage analysis in baboon. We thank the participants in the Rochester Family Heart Study.
SOURCES OF FUNDING
Support for this work was provided by National Heart, Lung, and Blood Institute Contract R01-HL-077491.
Footnotes
Disclosure: None
References
- 1.Ward R. Familial aggregation and genetic epidemiology of blood pressure. In: Laragh J, Brenner B, editors. Hypertension: Pathophysiology, Diagnosis, and Management. New York, NY: Raven Press; 1990. pp. 81–100. [Google Scholar]
- 2.Snieder H, Harshfield GA, Treiber FA. Heritability of Blood Pressure and Hemodynamics in African- and European-American Youth. Hypertension. 2003;41:1196–1201. doi: 10.1161/01.HYP.0000072269.19820.0D. [DOI] [PubMed] [Google Scholar]
- 3.Kurtz TW, Montano M, Chan L, Kabra P. Molecular evidence of genetic heterogeneity in Wistar-Kyoto rats: implications for research with the spontaneously hypertensive rat. Hypertension. 1989;13:188–192. doi: 10.1161/01.hyp.13.2.188. [DOI] [PubMed] [Google Scholar]
- 4.Laragh JH, Pecker MS. Dietary sodium and essential hypertension: some myths, hopes, and truths. Ann Intern Med. 1983;98:735–43. doi: 10.7326/0003-4819-98-5-735. [DOI] [PubMed] [Google Scholar]
- 5.Tournoy KG, Delanghe JR, Duprez DA, De Buyzere ML, Verbeeck RM, Vergauwe DA, Leroux-Roels GG, Clement DL. Genetic polymorphisms and erythrocyte sodium-lithium countertransport in essential hypertension. Clin Chim Acta. 1996;15(255):39–55. doi: 10.1016/0009-8981(96)06389-9. [DOI] [PubMed] [Google Scholar]
- 6.Canessa M, Adragna N, Solomon H, Connolly T, Tosteson D. Increased sodium-lithium countertransport in red cells of patients with essential hypertension. N Engl J Med. 1980;302:772–776. doi: 10.1056/NEJM198004033021403. [DOI] [PubMed] [Google Scholar]
- 7.Turner ST, Johnson M, Boerwinkle E, Richelson E, Taswell HF, Sing CF. Sodium-lithium countertransport and blood pressure in healthy blood donors. Hypertension. 1985;7:955–62. doi: 10.1161/01.hyp.7.6.955. [DOI] [PubMed] [Google Scholar]
- 8.Hasstedt SJ, Wu LL, Ash KO, Kuida H, Williams R. Hypertension and sodium-lithium countertransport in Utah pedigrees: evidence for major-locus inheritance. Am J Hum Genet. 1988;43:14–22. [PMC free article] [PubMed] [Google Scholar]
- 9.Dadone M, Hasstedt S, Hunt S, Smith J, Ash K, Williams R. Genetic analysis of sodium-lithium countertransport in 10 hypertension-prone kindreds. Am J Hum Genet. 1984;17:565–577. doi: 10.1002/ajmg.1320170304. [DOI] [PubMed] [Google Scholar]
- 10.Kammerer C, Cox L, Mahaney M, Rogers J, Shade R. Sodium-lithium countertransport activity is linked to chromosome 5 in baboons. Hypertension. 2001;37:398–402. doi: 10.1161/01.hyp.37.2.398. [DOI] [PubMed] [Google Scholar]
- 11.Boerwinkle E, Turner ST, Weinshilbaum R, Johnson M, Richelson E, Sing C. Analysis of the distribution of erythrocyte sodium lithium countertransport in a sample representative of the general population. Genet Epidemiol. 1986;3:365–378. doi: 10.1002/gepi.1370030509. [DOI] [PubMed] [Google Scholar]
- 12.Rebbeck TR, Turner ST, Sing CF. Sodium-lithium countertransport genotype and the probability of hypertension in adults. Hypertension. 1993;22:560–568. doi: 10.1161/01.hyp.22.4.560. [DOI] [PubMed] [Google Scholar]
- 13.Hunt SC, Stephenson SH, Hopkins PN, Hasstedt SJ, Williams RR. A prospective study of sodium-lithium countertransport and hypertension in Utah. Hypertension. 1991;17:1–7. doi: 10.1161/01.hyp.17.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Turner ST, Michels VV. Sodium-lithium countertransport and hypertension in Rochester, Minnesota. Hypertension. 1991;18:183–190. doi: 10.1161/01.hyp.18.2.183. [DOI] [PubMed] [Google Scholar]
- 15.Turner ST, Rebbeck TR, Sing CF. Sodium-lithium countertransport and probability of hypertension in Caucasians 47–89 years old. Hypertension. 1992;20:841–850. doi: 10.1161/01.hyp.20.6.841. [DOI] [PubMed] [Google Scholar]
- 16.Weder AB, Schork NJ, Krause L, Julium S. Red blood cell lithium-sodium countertransport in the Tecumseh blood pressure study. Hypertension. 1991;17:652–660. doi: 10.1161/01.hyp.17.5.652. [DOI] [PubMed] [Google Scholar]
- 17.Woods JW, Falk RJ, Pittman AW, Klemmer PJ, Watson BS, Namboodiri K. Increased red-cell sodium-lithium countertransport in normotensive sons of hypertensive patients. N Engl J Med. 1982;306:593–595. doi: 10.1056/NEJM198203113061007. [DOI] [PubMed] [Google Scholar]
- 18.Obasohan AO, Osuji CO, Oforofuo IAO. Sodium-lithium countertransport activity in normotensive offspring of hypertensive black Africans. J Hum Hypertens. 1998;12:373–377. doi: 10.1038/sj.jhh.1000597. [DOI] [PubMed] [Google Scholar]
- 19.Feild JA, Zhang L, Brun KA, Brooks DP, Edwards RM. Cloning and functional characterization of a sodium-dependent prostate transporter expressed in human lung and small intestine. Biochem Biophys Res Commun. 1999;258:578–582. doi: 10.1006/bbrc.1999.0666. [DOI] [PubMed] [Google Scholar]
- 20.Xu H, Bai L, Collins JF, Ghishan FK. Molecular cloning, functional characterization, tissue distribution, and chromosomal localization of a human, small intestinal sodium-phosphate (Na+-Pi) transporter (SLC34A2) Genomics. 1999;62:281–284. doi: 10.1006/geno.1999.6009. [DOI] [PubMed] [Google Scholar]
- 21.Moll P, Sing C, Weidman W, Gordon H, Ellefson R, Hogdson P, Kottke B. Total cholesterol and lipoproteins in school children prediction of coronary heart disease in adult relatives. Circulation. 1983;67:127–134. doi: 10.1161/01.cir.67.1.127. [DOI] [PubMed] [Google Scholar]
- 22.Turner ST, Weidman WH, Michels VV, Reed TJ, Ormson CL, Fuller T, Sing CF. Distribution of sodium-lithium countertransport and blood pressure in Caucasians five to eighty-nine years of age. Hypertension. 1989;13:378–391. doi: 10.1161/01.hyp.13.4.378. [DOI] [PubMed] [Google Scholar]
- 23.Smith JB, Ash KO, Hentschel WM, Sprowell WL, Williams R. A simplified method for simultaneously determining countertransport and cotransport in human erythrocytes. Clin Chim Acta. 1984;137:168–177. doi: 10.1016/0009-8981(84)90177-3. [DOI] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Chen X, Levine L, Kwok PY. Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res. 1999;9:492–8. [PMC free article] [PubMed] [Google Scholar]
- 26.Kwok PY. SNP genotyping with fluorescence polarization detection. Hum Mutat. 2002;19:315–323. doi: 10.1002/humu.10058. [DOI] [PubMed] [Google Scholar]
- 27.Liebel U, Kindler B, Pepperkok R. Bioinformatic “Harvester”: a search engine for genome-wide human, mouse, and rat protein resources. Methods Enzymol. 2005;404:19–26. doi: 10.1016/S0076-6879(05)04003-6. [DOI] [PubMed] [Google Scholar]
- 28.Almasy L, Blangero J. Multipoint Quantitative-Trait Linkage Analysis in General Pedigrees. Am J Hum Gbnet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones KA, Porjesz B, Almasy L, Bierut L, Goate A, Wang JC, Dick DM, Hinrichs A, Kwon J, Rice JP, Rohrbaugh J, Stock H, Wu W, Bauer LO, Chorlian DB, Crowe RR, Edenberg HJ, Foroud T, Hesselbrock V, Kuperman S, Nurnberger J, Jr, O’Connor SJ, Schuckit MA, Stimus AT, Tischfield JA, Reich T, Begleiter H. Linkage and linkage disequilibrium of evoked EEG oscillations with CHRM2 receptor gene polymorphisms: implications for human brain dynamics and cognition. Int J Psychophysiol. 2004;53:75–90. doi: 10.1016/j.ijpsycho.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotyp reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almasy L, Blangero J. Exploring positional candidate genes: linkage conditional on measured genotype. Behav Genet. 2004;34:173–177. doi: 10.1023/B:BEGE.0000013731.03827.69. [DOI] [PubMed] [Google Scholar]
- 33.Laird NM, Horvath S, Xu X. Implementing a unified approach to family based tests of association. Genetic Epi. 2000;(supp 1):36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 34.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 35.Lake SL, Blacker D, Laird NM. Family-Based Tests of Association in the Presence of Linkage. Am J Hum Genet. 2000;67:1515–1525. doi: 10.1086/316895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: Application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 37.Wang QF, Liu X, O’Connell J, Peng Z, Krauss RM, Rainwater DL, VandeBerg JL, Rubin EM, Cheng JF, Pennacchio LA. Haplotypes in the APOA1-C3-A4-A5 gene cluster affect plasma lipids in both humans and baboons. Hum Mol Genet. 2004;13:1049–1056. doi: 10.1093/hmg/ddh121. [DOI] [PubMed] [Google Scholar]
- 38.Wan H, Wootton JC. A global compositional complexity measure for biological sequences: AT-rich and GC-rich genomes encode less complex proteins. Comput Chem. 2000;24:71–94. doi: 10.1016/s0097-8485(99)00048-0. [DOI] [PubMed] [Google Scholar]
- 39.Morrison AC, Boerwinkle E, Turner ST, Ferrell RE. Genome-wide linkage study of erythrocyte sodium-lithium countertransport. Am J Hypertens. 2005;18:653–656. doi: 10.1016/j.amjhyper.2004.11.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.