Summary
Kidney development is based on differential cell type specific expression of a vast number of genes. While multiple critical genes and pathways have been elucidated, a genomewide analysis of gene expression within individual cellular and anatomic structures is lacking. Accomplishing this could provide significant new insights into fundamental developmental mechanisms such as mesenchymal-epithelial transition, inductive signaling, branching morphogenesis and segmentation. We describe here a comprehensive gene expression atlas of the developing mouse kidney based on the isolation of each major compartment by either laser capture microdissection or fluorescent activated cell sorting, followed by microarray profiling. The resulting data agrees with known expression patterns and additional in situ hybridizations. This kidney atlas allows a comprehensive analysis of the progression of gene expression states during nephrogenesis, as well as discovery of novel growth factor-receptor interactions. In addition, the results provide deeper insight into the genetic regulatory mechanisms of kidney development.
Introduction
We describe here the first exhaustive atlas of gene expression driving the formation of an organ, the kidney. Organogenesis is a complex process that we are only beginning to understand. While single gene based studies have provided key insights, the resulting picture remains quite incomplete. A more global analysis can create an overview, discover new developmental pathways, identify novel molecular markers of specific components, define the changing patterns of gene utilization as a function of developmental time, and provide insight into the genetic regulatory mechanisms of nephrogenesis. To generate a development gene expression resource the NIH has created an international consortium, termed GUDMAP (GenitoUrinary Development Molecular Anatomy Project), with kidney microarray results reported here.
The kidney is an excellent model system for studying the principles of organogenesis as it employs many common developmental mechanisms, including reciprocal inductive interactions, stem cell growth and differentiation, mesenchyme to epithelia conversion, branching morphogenesis, and proximal-distal segmentation along the length of the nephron (Dressler, 2006). In this study we used either laser capture microdissection (LCM) or fluorescent activated cell sorting (FACS) combined with component specific-GFP transgenic mice to purify the discrete elements of the developing kidney, which were then transcriptionally profiled with microarrays. The gene expression states of the kidney progenitor cells and multiple components of the developing nephrons and collecting ducts were characterized, thus creating a comprehensive data set of changing gene expression programs used during the progression of nephrogenesis.
The kidney is well-suited for a comprehensive gene expression analysis of organogenesis. It is intermediate in complexity among organs, far simpler than for example the brain, yet sufficiently complex to provide an instructive model. The adult human kidney contains approximately one million nephrons. At one end of the nephron is the renal corpuscle (glomerulus), the filtration unit, followed by a segmented tubule devoted to the recapture of essential filtrate elements. Nephrogenesis is induced at the periphery of the developing kidney by the branching ureteric bud. As the kidney grows outward newly initiated nephrons are near the surface and more mature nephrons are located deeper within the kidney. A single developmental time point, such as E15.5, can therefore be used to examine multiple stages of nephron formation. We present here comprehensive gene expression profiles of the major elements of kidney development. This dataset represents the first genomics level analysis of organogenesis, with each key developmental component examined. This resource allows one to choose a gene of interest and to define quantitative expression levels in the many different parts of the developing kidney. It also allows one to choose a developmental component of interest, such as the renal vesicle, and to define its gene expression state. Moreover, gene expression profiles of different compartments can be compared, to determine changing patterns of gene utilization as a function of nephrogenesis. The data can be used, for example, to identify previously unrecognized growth factor-receptor signaling pathways active during kidney development. In addition, the dataset provides novel sets of genes expressed in a component specific manner; a compendium of useful molecular markers for the analysis of mutants, and for the production of additional useful transgenic tools. The universal gene expression patterns generated also facilitate analysis of the genetic regulatory network of kidney development. During nephrogenesis new sets of expressed genes show highly significant shared transcription factor binding sites within their evolutionarily conserved promoter regions, implicating specific regulatory pathways. In general the data yield a global view of the gene expression blueprint of kidney development.
Results
The nephron, the functional unit of the kidney, develops through an intricate progression of morphological structures as shown in Fig. 1. In the E 15.5 kidney, the formation of the nephron is initiated when signals derived from the ureteric bud induce the overlaying capping mesenchymal cells to aggregate and undergo a mesenchymal-to-epithelial transition to form the renal vesicle. The cells of the renal vesicle, in turn, differentiate, elongate and convolute to form an S-shaped body, which is patterned along the proximal-distal axis, and is the structure from which the glomerulus, proximal tubule, loop of Henle and distal tubule are derived. In addition to the nephron, the kidney is also composed of a network of collecting ducts, which is formed through signals from the capping mesenchyme that induces the ureteric epithelium to undergo a complex series of differentiation, growth and branching events to eventually form medullary, cortical and tip regions, all of which have been shown to have distinct properties. Where the ureteric bud exits the kidney it forms the urothelium of the ureter, which is surrounded by a smooth muscle forming region. Finally, there are interstitial elements, the stromal cells, which provide important signaling function, and can be divided into the renal cortical and medullary interstitium. In this report we present the comprehensive gene expression profiles of each of these major components of kidney development.
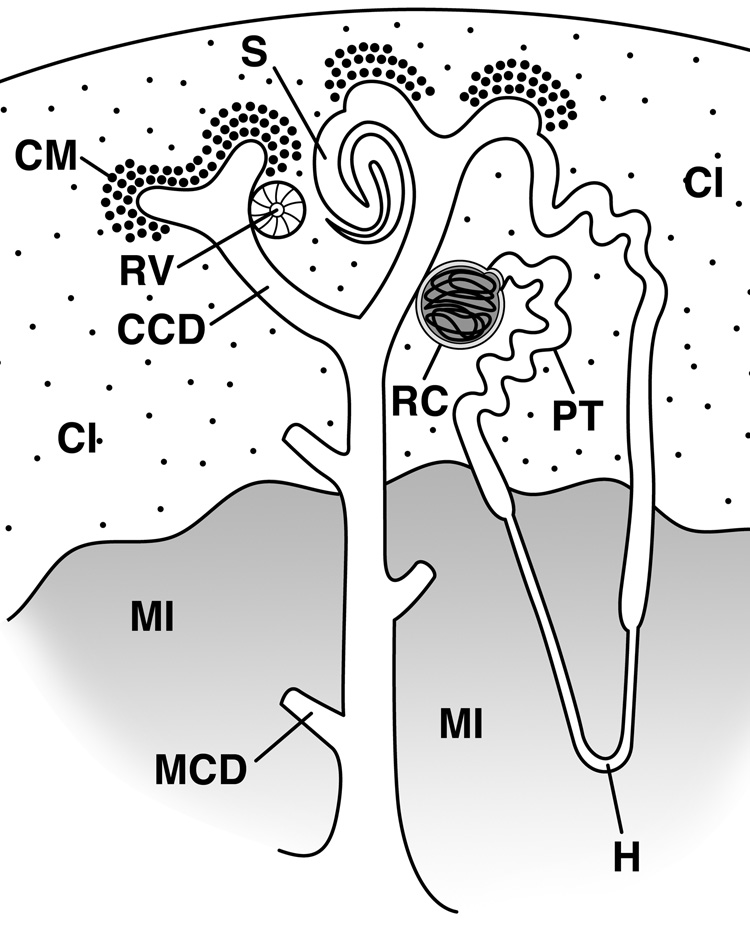
Fig. 1. Kidney development.
The branching ureteric bud induces the cap mesenchyme (CM) to give rise to the renal vesicle (RV), which expands, folds, and fuses to the developing collecting duct to form the S-shaped body (S), which in turn gives rise to the renal corpuscle (RC) (glomerulus), early proximal tubule (PT), anlage of and immature loop of Henle (H) and distal tubule. The ureteric bud gives rise to the collecting duct, which can be divided into the medullary collecting duct (MCD), cortical collecting duct (CCD), and the ureteric tip region. In addition the interstitium (stromal cells) can be divided into the medullary interstitium (MI) and the cortical and nephrogenic interstitium (CI). Diagram modified from (Little et al., 2007).
We employed a combination of LCM, and FACS using transgenic mice with component specific GFP expression, to isolate precise cell populations of the developing kidney. Microarrays were then used to provide quantitative and universal readouts of gene expression levels. Most kidney development components were isolated from E15.5 kidneys by LCM, using unique structural characteristics and/or specific lectin staining patterns for identification (Fig. 2). We also used Six2-GFP and Meis1-GFP transgenic mice, which show specific GFP expression in the cap mesenchyme and cortical interstitium, respectively, allowing the isolation of these components from E15.5 kidneys by FACS (Supplementary Fig. S1, Fig S2). In addition, we have previously described the gene expression patterns of the E11.5 ureteric bud and E11.5 metanephric mesenchyme (Schwab et al., 2006a), as well as the E12.5 renal vesicles (Potter et al., 2007) with the results re-analyzed here in the context of the entire kidney development dataset. Each component was examined at least in triplicate, using independent biological samples for each Affymetrix MOE430 version 2 microarray. Target amplification of the RNA from laser captured or FACS purified cells was carried out using the Epicentre two round in vitro transcription system, which we have previously shown gives robust results, with very high correlation coefficients for replicates (Potter et al., 2007; Schwab et al., 2006a). The resulting gene expression patterns, from 54 microarrays, were analyzed with GeneSpring software. This dataset provides a global view of kidney organogenesis, describing the expression levels of almost all genes in the different developmental stages of nephron formation, including comprehensive transcription factor, growth factor and receptor gene expression patterns.
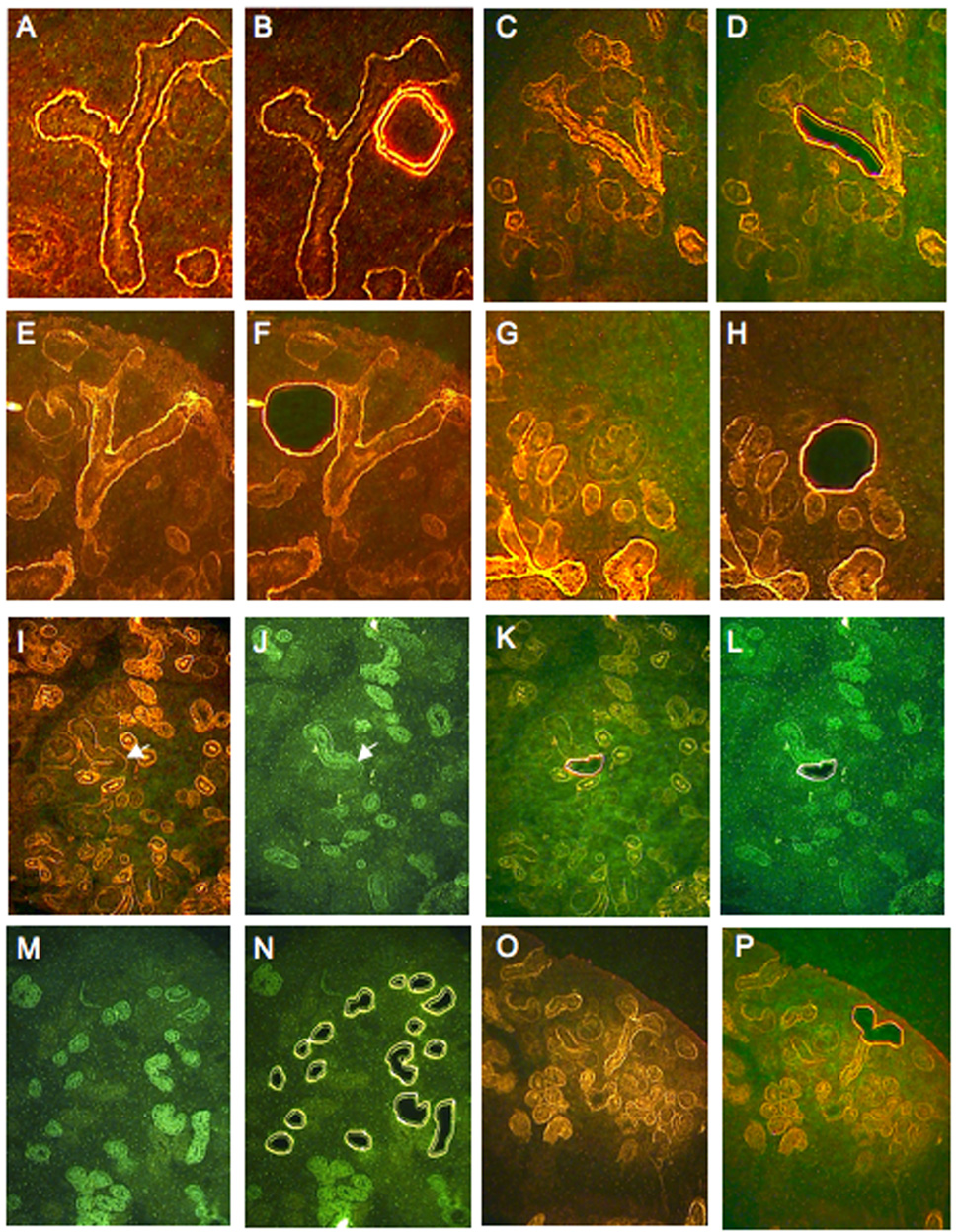
Fig. 2. Laser capture of kidney development components.
(A) Y-shaped branching ureteric bud with underlying renal vesicle on the right side. (B) Laser cut surrounds renal vesicle. Branching ureteric bud (C) with cortical collecting duct region removed by laser capture (D). (E) Branching ureteric tip with underlying S-shaped body on left side. (F) Sshaped body removed by laser capture. (G,H) Renal corpuscle (glomerulus) removed by laser capture. (I) PNA stains epithelial structures of the cortex. (J) LTA specifically stains proximal tubules. (K,L) Isolation of anlage of loop of Henle. PNA only stained tubule, distal to region of tubule showing LTA staining, is removed by laser capture. (M,N) LTA stains proximal tubules, many of which are removed by laser capture. (O,P) Ureteric tree terminal branch, removed by laser capture. Panels A and B are E12.5 kidney, while all other panels are E15.5. Panels J, L, M and N are LTA stained while all others are PNA stained. For illustrations of laser capture of other components refer to GUDMAP.ORG.
Expression analysis
To detect informative gene expression patterns and features present in the renal development atlas, we used a variety of comparative and analytical approaches. One goal was to identify individual genes and groups of genes that are likely to be important for renal development based on their component specific expression. To do this, microarray data was subjected to GC-RMA normalization and probesets were filtered for raw signal strength to identify those likely to be well-expressed in at least two replicates of one individual sample type, to yield a combined pool of 21,799 probesets selected for further pattern analysis. Given the possibility that there could be some variation in the composition of some of the individual laser captured tissue elements (see Discussion), we then used a liberal approach to identify the most compartment-specific probesets for each compartment, based on the relative rank of their normalized expression versus the median of their expression across the entire set of 54 samples. Each compartment was averaged and we identified the 1000 top-ranked probesets for each compartment and pooled these lists. Because of overlaps, the 15 lists of 1000 probesets each combined to make 7629 probesets (termed the 7K list).
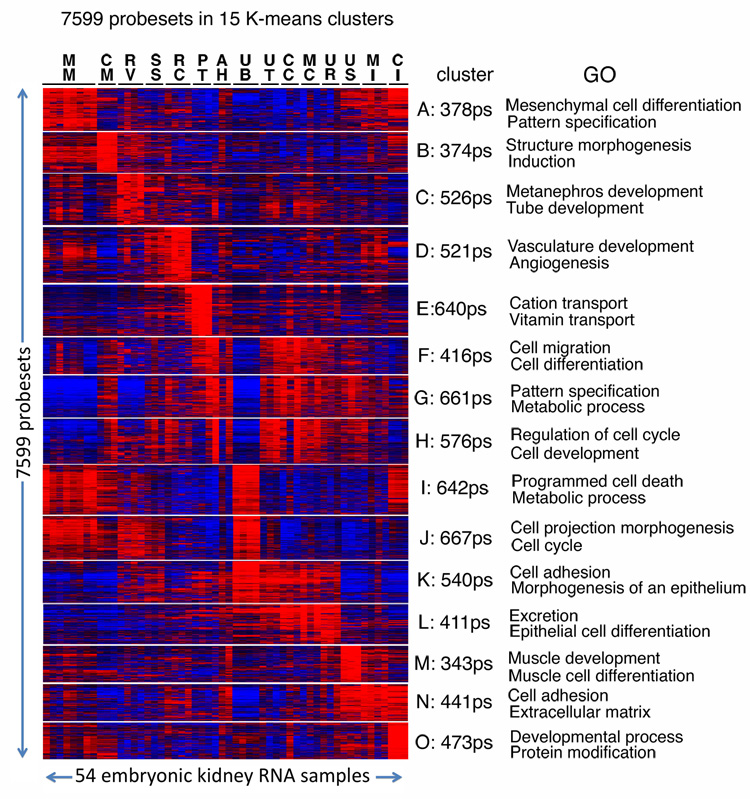
The 7K list of differentially expressed genes was subjected to K-means clustering, producing the 15 clusters shown in the heat map of Fig. 3. In addition, GO ontology terms were analyzed to identify enriched biological processes within each cluster. The samples of Fig. 3 are ordered to reflect in part the developmental sequence of nephron formation. Examination of this heat map shows, as would be predicted, that structures of the kidney that are closely related share a high degree of correlative gene expression. For example cluster B shows genes with the strongest differential expression in the cap mesenchyme, and many of these genes also show elevated expression in the preceding developmental structure, the metanephric mesenchyme, and the subsequent structure, the renal vesicle. Cluster N illustrates the close gene expression relationships of the medullary interstitium, nephrogenic interstitum, and ureteral smooth muscle compartments. Indeed, in general relatively few compartments exhibit highly restricted gene expression patterns. The primary exceptions are the proximal tubule (cluster E) and ureteral smooth muscle (cluster M), which show extremely specific expression of transporter and muscle development gene sets, respectively, as they undergo early differentiation. The related clusters F, G and H illustrate a set of genes largely off early (E11.5 metanephric mesenchyme, E11.5 ureteric bud, E12.5 renal vesicle) and active late (all other structures are E15.5) in development. And cluster J genes show a mirror image expression pattern, on in early and off in late development. These genes with early/late differential expression are often involved in cell cycle, programmed cell death, cell migration, and cell differentiation. Another mirror image pattern is apparent in clusters K and L, which include genes with elevated expression in the forming collecting ducts. The genes of cluster K are most active in the early E11.5 ureteric bud and the terminal regions of the E15.5 collecting ducts, which are still undergoing branching morphogenesis. Appropriately many of these genes are involved in cell adhesion and epithelial morphogenesis. The genes of cluster L, however, are most highly expressed in the more differentiated urothelium and medullary collecting duct. Many of these genes function in excretion and epithelial cell differentiation. For an expanded view of each cluster heat map, complete lists of all genes and individual gene ontologies, see Supplementary Table S1. The results of a detailed enrichment analysis for the sets of genes in each cluster, including ontologies, phenotypes, domains, and evolutionarily conserved promoter transcription factor binding sites are presented in Supplementary Table S2.
Fig. 3. Gene expression relationships of kidney development components.
This heat map shows probesets with the most component specific expression. Each horizontal line represents a probeset, with red indicating high expression and blue indicating low expression in the various components. Compartments are: MM, E11.5 metanephric mesenchyme. CM, E15.5 cap mesenchyme. RV, E12.5 renal vesicle. SS, E15.5 S-shaped body. RC, E15.5 renal corpuscle. PT, E15.5 proximal tubules. AH, E15.5 Anlage of and immature loop of Henle. UB, E11.5 ureteric bud. UT, E15.5 ureteric tip region. CCD, E15.5 cortical collecting duct. MC, E15.5 medullary collecting duct. UR, E15.5 urothelium. US, ureteral smooth muscle layer. MI, medullary interstitium. CI, cortical and nephrogenic interstitium. Genes are divided into 15 K-means clusters. Two GO biological processes are shown for each cluster (P < 0.001). For complete lists of genes for each cluster and additional GO terms see Supplementary Materials.
Validation
To assess the quality of the microarray data we first performed an in silico, or historical validation. The data was checked against gene expression patterns that had been previously defined by in situ hybridizations. There was excellent agreement, with ret and Wnt11 transcripts, for example, in the E11.5 ureteric bud and E15.5 ureteric tip regions, respectively, MafB transcripts in the E15.5 S-shaped body and renal corpuscle, Six2 transcripts in the E11.5 metanephric mesenchyme and E15.5 cap mesenchyme, the Lhx1, Hes5 and Dll1 genes showing strong expression in the E12.5 renal vesicle, Cited1 in the E15.5 cap mesenchyme, Gata3 in the E11.5 ureteric bud and its derivatives, WT1 showing highest expression in the cap mesenchyme and renal corpuscle, and Foxd1 highly restricted to the cortical (and nephrogenic) interstitium (cortical stroma).
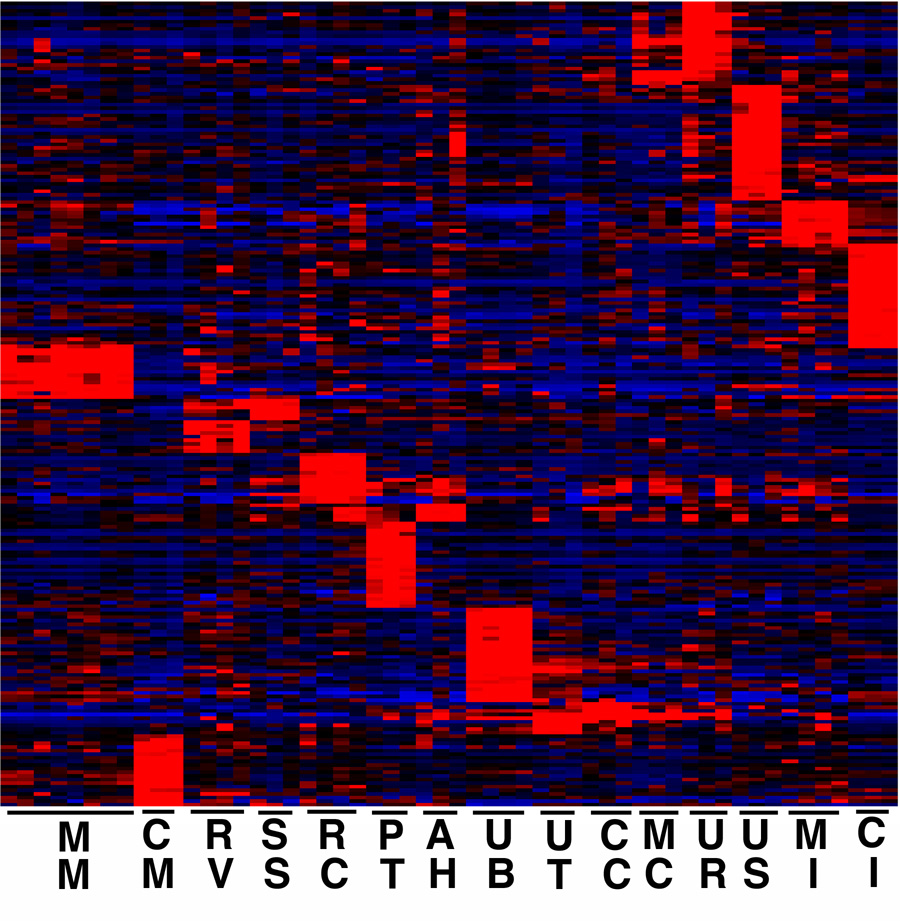
To further validate the microarray data we carried out in situ hybridizations, using a stringent screen of the microarray data to identify the 223 genes predicted to have the most compartment specific expression (Fig. 4). The names of these genes, as well as quantitative measures of their expression levels in all compartments are provided in Supplementary Table S3. As already noted, some components of the developing kidney, such as the differentiating proximal tubules, showed a large number of genes with very specific expression, but for most developmental compartments the number of genes with truly restricted expression was quite small, in part because of the overlapping gene expression patterns of structures in a developmental sequence. This has also been observed in Drosophila, where it has been referred to as “anlage in statu nascendi” (Tomancak et al., 2002), with genes important in the development of a discrete structure often showing earlier and in some cases wider expression in anlage.
Fig. 4. Heat map of genes with most component specific expression.
The top 223 genes with the most restricted expression patterns. Compartments are: MM, E11.5 metanephric mesenchyme. CM, E15.5 cap mesenchyme. RV, E12.5 renal vesicle. SS, E15.5 S-shaped body. RC, E15.5 renal corpuscle. PT, E15.5 proximal tubules. AH, E15.5 Anlage of and immature loop of Henle. UB, E11.5 ureteric bud. UT, E15.5 ureteric tip region. CCD, E15.5 cortical collecting duct. MC, E15.5 medullary collecting duct. UR, E15.5 urothelium. US, ureteral smooth muscle layer. MI, medullary interstitium. CI, cortical and nephrogenic interstitium. For gene lists with associated expression patterns see Supplementary Materials.
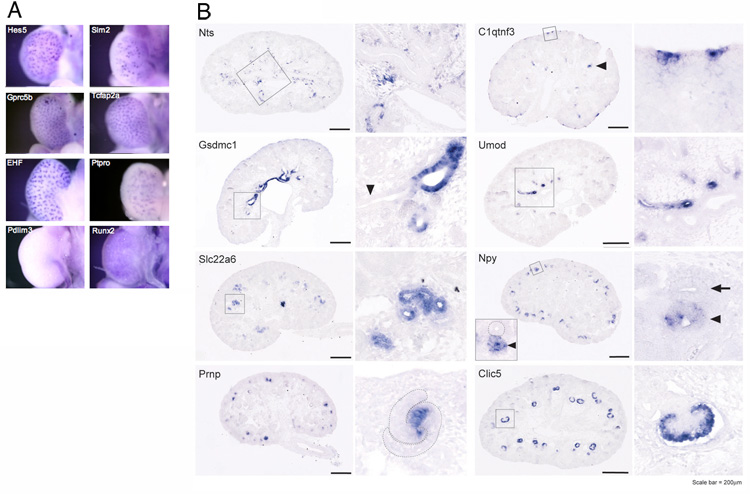
The results of both whole mount and section in situ hybridizations provided confirmation of the microarray data. The whole mount in situ hybridization patterns for the Hes5, Sim2, Gprc5b, Tcfap2a, Ehf, Ptpro, Pdlim3 and Runx2 genes were consistent with the microarray results (Fig. 5A). In addition, section in situ hybridizations (SISH) were performed for 38 genes on E15.5 (TS23) metanephroi sections to further confirm the microarray results. Section in situ hybridizations provide higher resolution results than whole mount in situ hybridizations, providing a more rigorous validation. The tested genes were selected from the top 10 most enriched genes per compartment from the microarray analysis and included at least two representative genes for each of 14 compartments. Fig. 5B shows examples of gene expression for eight genes in eight different compartments. Of genes tested 13/38 showed expression enriched in the predicted compartment, but with transcripts also detected in other compartments. Another 13/38 of genes showed expression restricted to the appropriate compartment. For example Gsdmc1, which was enriched in medullary collecting duct by microarray, was indeed restricted to the medullary collecting duct by SISH but absent in the cortical collecting duct and all other compartments. In addition, Nts transcripts, which microarrays indicated were enriched in the medullary interstitium, showed strongest expression by SISH in the medullary interstitium, and additional expression in the cortical interstitium, and was not detected in any other compartment (Fig. 5B). For 12/38 genes there was no detectable signal, or low level ubiquitous expression or non-specific background signal. Because microarrays are more sensitive they can detect some low transcript level expression differences that would not be seen by in situ hybridizations. Indeed, for eight of the twelve genes giving no detectable or background signal the microarray expression levels were very low, ranging from 6 to 7.4 raw log scale expression, indicating that the in situ hybridizations were simply not sensitive enough for validation. And of the remaining four genes that did not validate by section in situ hybridization, two of these did indeed validate by whole mount in situ hybridization, indicating a likely technical problem for these two section in situ hybridizations. Therefore only 2/38, or 5%, of tested genes gave sufficient expression levels for in situ detection and failed to validate the microarray predicted patterns. These results are highly supportive of the microarray data. For a complete listing of section in situ hybridization results see Supplementary Table S4.
Fig. 5. In situ hybridization validation of microarray results.
E15.5 kidneys.A. Whole mount in situ hybridizations. Observed expression patterns are consistent with microarray results predicting strongest expression for Hes5 in S-shaped body and renal vesicle, Sim2 in S-shaped body, Gpcr5b in ureteric bud, Tcfap2a and EHF in ureteric bud, Ptpro in the renal corpuscle, Pdlim3 in the ureteral smooth muscle, and Runx2 in the cortical stroma. B. Section in situ hybridizations provide higher resolution. Each gene is represented by two images, a global view of the metanephric kidney and an enlarged high magnification image (the area magnified is outlined). The subcompartment the gene was identified as being enriched in, from the microarray analysis, is indicated in parentheses following the gene symbol. Nts (medullary interstitium) was expressed in the medullary interstitium in a specific region surrounding the collecting ducts, as well as in a subset of the cortical interstitium adjacent to the medulla, also surrounding the collecting ducts. C1qtnf3 (cortical and nephrogenic interstitium) was expressed in a small subset of the nephrogenic interstitium, the renal interstitium located within the nephrogenic zone of the metanephros, as well as in a small subset of cortical renal tubules (arrowhead). Gsdmc1, (also known as Mlze) (medullary collecting duct) was specifically expressed in the medullary collecting duct and expression was absent from the cortical collecting duct (arrowhead). Umod (anlage of and immature loop of Henle) was specifically expressed in the immature loop of Henle. Slc22a6 (early proximal tubule) was specifically expressed in the early proximal tubule. Npy (renal vesicle) was expressed in renal vesicles (arrowheads) and in the lower limb of comma-shaped bodies (the ureteric tip is indicated by an arrow or is outlined). Prnp (S-shaped body) was expressed in the medial segment of S-shaped bodies (outlined in the enlarged image) and in the upper limb of comma-shaped bodies. Clic5 (stage III-IV renal corpuscle) was specifically expressed in the visceral epithelium (podocyte layer) of stage III and IV renal corpuscles. Scale bar=200µm.
Although microarray results are global, sensitive and quantitative they do not provide information concerning possible subcompartment restricted expression: they do not define the distribution of expression within a compartment. In several instructive cases we found that the validating in situ hybridizations revealed expression confined to a subregion of the compartment. For example, Prnp is expressed in the S-shaped body as predicted, but only in a very restricted segment of the S-shaped body (Fig. 5B). Likewise C1qtnf3 expression was limited to discrete subregions of the interstitium (Fig. 5B). These results illustrate how the combination of microarray analysis, to screen for genes with more restricted expression, and in situ hybridization, to define precise expression domains, can identify novel subcompartment gene expression patterns.
Genetic program of nephrogenesis
The dataset provides a global description of the genetic program of nephrogenesis. As mentioned previously, the progression involves metanephric mesenchyme giving rise to cap mesenchyme, which produces renal vesicles, which become S-Shaped bodies, which form the final structures of the nephron, the renal corpuscle or glomerulus, the proximal tubule, loop of Henle, and distal tubule. The microarray results give the comprehensive gene expression states of key intermediates of nephrogenesis. In addition, by considering the differences in gene expression between sequential components, the data provide a view of the changes in gene expression that drive nephron formation.
The cap mesenchyme is the condensed mesenchyme that abuts the branching ureteric tips. The cells of the cap mesenchyme give rise to the bulk of the cellular constituents of the nephron, excepting the endothelial and mesangial cells of the renal corpuscle (glomerulus) (Boyle et al., 2008; Kobayashi et al., 2005). By examining the progression or changes in gene expression between the cap mesenchyme and renal vesicle we gain insight into the initial events of nephron formation. Using stringent filtering parameters [T-Test, with Benjamini and Hochberg multiple testing correction FDR <0.02 (Benjamini and Hochberg, 1995), raw rank expression >30 percentile, and fold change >3] we identified 1,043 genes with differential expression. Expected differences are observed, including increased Lhx1 and Fgf8, and decreased Six2 and Cited2 expression in the renal vesicle. In addition the data provides a rich analysis of the genetic program of this early stage of nephrogenesis. For example, a gene ontology analysis finds 91 genes with transcription regulation function, including Six4, Hes5, Hoxc4, Hoxc8, Emx2, Tcf3, Tcf4, Sox4, Nfat5, Sim1, Irx3, and 80 more, with significantly altered expression. Similarly 161 genes are identified with established function in developmental processes, including increased expression of Notch1, Dll1, DKK1 and Mdk1 in the renal vesicle. Other functional lists include genes previously implicated in pattern specification (34 genes), cell differentiation (63 genes), and anatomical structure morphogenesis (59 genes). In summary, the result is a comprehensive view of the changing pattern of gene utilization as the cap mesenchyme becomes renal vesicle.
In a similar manner it is possible to examine each developmental stage of nephron formation. For example, analysis of the RV to S-shaped body transition (T-Test, raw rank expression >30% in 4/8 samples, fold change >3, and P <0.05) identifies 1,072 genes with differential expression. This gene list reveals changes in expression of genes including growth factors, receptors, transcription factors, and other families of genes important in driving progression of the renal vesicle to the S-shaped body. Expected changes in gene expression are observed, including the early expression of MafB, a podocyte marker, in the S-shaped body. The list of differently expressed genes yields a global view of the complex changes in gene expression that propel the transformation of a renal vesicle into an S-shaped body. Discrete changes in signaling pathways and transcription factor programs can be dissected out. For example, in the BMP signaling pathway we observe the upregulation of Smad5, Bmp7, twisted gastrulation, and RGMB, and the downregulation of Smad6, Bmp2, follistatin, Bmpr1b, and Bmper, providing a view of the complex changes in BMP signaling that take place. By comparing the gene expression patterns of the sequential elements of nephrogenesis it is possible to derive a global view of the genetic program that drives this process. See Supplementary Table S5 and Supplementary Table S6 for gene lists.
The genetic circuitry of kidney development
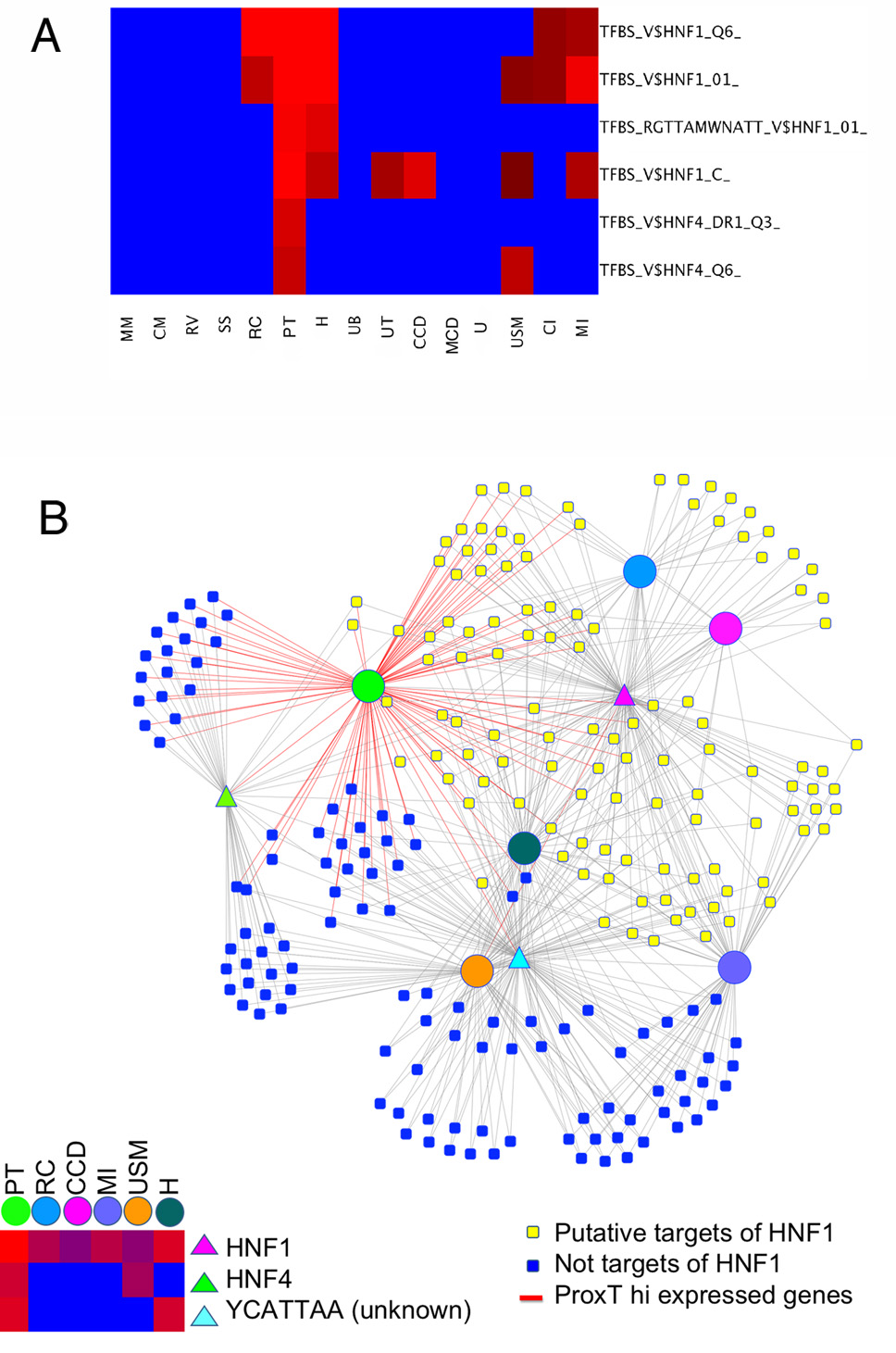
The microarray expression data can be used to help define the genetic regulatory network of kidney development. Coordinately expressed genes have a higher probability of being regulated by shared transcription factors. By determining the distribution of evolutionarily conserved (human, mouse, rat and dog) transcription factor binding sites in the promoters of genes co-expressed in a given compartment it is possible to define a regulatory signature. For example, we observe a very statistically strong over abundance of HNF1β binding sites in the promoters of genes showing compartment specific expression in the proximal tubule. HNF1β is a homeodomain transcription factor that binds DNA as a homodimer or as a heterodimer with the closely related HNF1α (Cereghini, 1996). Both HNF1α and HNF1β bind to very similar or identical target sequences (Baumhueter et al., 1988). Although named hepatocyte nuclear factors they are also expressed during kidney development (Lazzaro et al., 1992). The heat map of Fig. 6A shows the enrichment of HNF1 binding sites in the promoters of proximal tubule highly expressed genes, using several related defined HNF1 consensus binding sites, giving highly significant P-values (10−10), with weaker P-values suggesting possible involvement in other compartments as well, including renal corpuscles and medullary interstitium.
Fig. 6. Occurrence of transcription factor binding sites in promoters of highly expressed genes.
A. This heat map shows the very high frequency of HNF1 and HNF4α binding sites in the highly expressed genes of certain compartments. Red indicates high and blue shows low statistical significance. Four related defined binding sites were used for HNF1 and two for HNF4α. For example, P-values for V$HNF1_Q6_ were 10−4.9 for RC, 10−10 for PT, 10−6 for H, 10−2.3 for CI and 10−2.7 for MI, while not significant for any other compartment. This strongly suggests that HNF1 drives the expression sets of downstream target genes carrying promoter binding sites, in certain compartments. B. Diagram illustrating relationships of compartments (circles), transcription factors (triangles) and candidate target genes (squares). Lower left shows heat map of significance of binding sites in highly expressed genes in compartments, summing the data shown in panel A. Yellow squares are targets of HNF1, while blue are not. The overlapping set of targets expressed in multiple compartments is apparent. MM, E11.5 metanephric mesenchyme, CM, E15.5 cap mesenchyme, RV, E12.5 renal vesicle, S, E15.5 S-shaped body, RC E15.5 renal corpuscle (glomerulus), PT, E15.5 proximal tubules, H, E15.5 Loop of Henle and distal tubule, UB, E11.5 ureteric bud, UT, E15.5 tipregion of collecting ducts, CCD, E15.5 cortical collecting ducts, MCD, E15.5 medullary collecting duct, U, E15.5 urothelium, USM, E15.5 ureteral smooth muscle layer, CI, E15.5 cortical and nephrogenic interstitium (cortical stroma), MI, E15.5 medullary interstitium, (medullary stroma). YCATTAA is a conserved binding site for which the corresponding transcription factor is not yet known.
This analysis yields a list of 44 candidate target genes with highly enriched expression in the proximal tubules and strong consensus binding sites for HNF1 in the promoter. Examination of this gene list (see Supplementary Data) provides validation of the approach and deeper insight into the genetic circuitry of proximal tubule development. For example one of the genes identified is PKHD1, mutation of which causes cystic kidney disease in rodent and man (Ward et al., 2002). Further, both chromatin immunoprecipitation studies (Gresh et al., 2004) and detailed promoter analysis (Hiesberger et al., 2004) showed that this gene is indeed directly regulated by HNF1. It has also been shown that kidney specific mutation of HNF1β in mice results in polycystic kidney disease and reduced expression of PKHD1 (Gresh et al., 2004; Hiesberger et al., 2004). Another gene on the list of candidate HNF1 targets is the organic anion transporter 3 (hOAT3/SLC22A8), known to play a major role in the excretion of a variety of organic ions. Previous studies of the promoter of this gene have also shown it is directly regulated by HNF1 alpha and beta (Kikuchi et al., 2006). Another candidate target revealed is TMEM27 (collectrin, a homologue of ACE2). The promoter of this gene shows the presence of three evolutionarily conserved HNF1 binding sites (Supplementary Fig. S3). Of interest the TMEM27 mutant mouse has defects in proximal tubule amino acid transport (Akpinar et al., 2005). In addition, this gene has been shown to be a direct downstream target of both HINF1α (Fukui et al., 2005) and HINF1α (Zhang et al., 2007).
Finally, another noteworthy downstream candidate target of HNF1 is HNF4α. This suggests an interesting genetic hierarchy, with HNF1 regulating the downstream transcription factor HNF4α, which in turn also shows a very strong P value for regulation of a further downstream set of 19 target genes in the proximal tubule (Supplementary Data) (Fig. 6A). This is illustrated as a graphical network in Fig. 6B, with candidate HNF1 targets in yellow, shown with compartment-specific expression, and the two largely distinct subsets of candidate HNF4α targets expressed in the proximal tubule and ureteral smooth muscle compartments.
This same approach can be used to dissect genetic regulatory networks of additional developmental components. For example, analysis of the promoters of genes that exhibit elevated expression in the cap mesenchyme reveals a significant enrichment for Tcf/Lef binding sites, suggesting possible downstream targets of Wnt signaling. The Tcf/Lef binding site is well-defined, unusually long, and remarkably conserved (van Noort and Clevers, 2002), lending considerable statistical power to the analysis. A total of 122 genes with elevated expression in the cap mesenchyme carry the Tcf/Lef binding sequence in their proximal promoter, including multiple Hox genes (Hoxc4, Hoxc5, Hoxc8, Hoxc10, Hoxa5, Hoxa10, and Hoxd10), and other transcription factors including Meis2, Sox11, Six3, and Eya1. These 122 genes represent candidate downstream effectors of Wnt signaling in the cap mesenchyme.
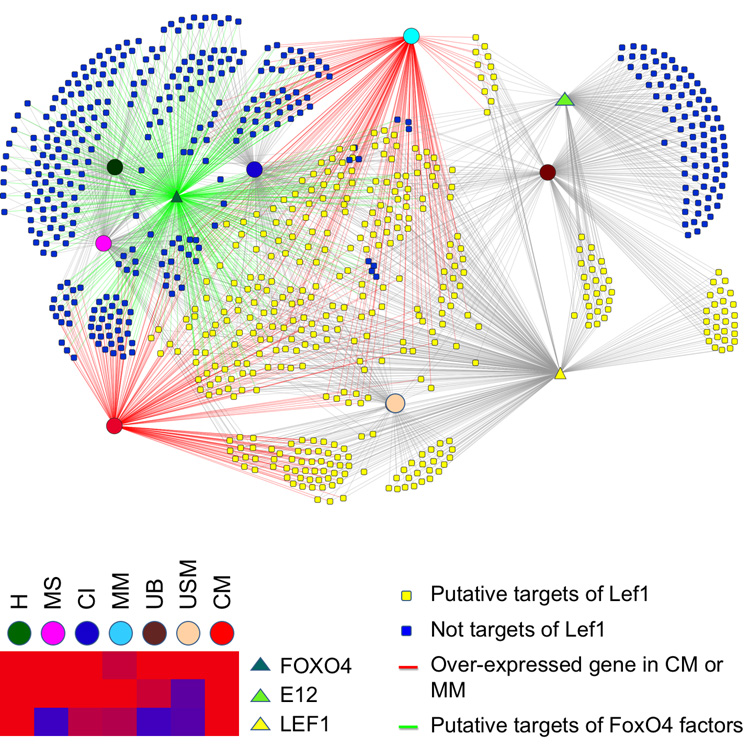
Wnt signaling has been shown to be important in several aspects of kidney development. Analysis of the expression data, in conjunction with identification of evolutionarily conserved Tcf/Lef binding sites in promoters, suggests overlapping sets of targets in different developmental compartments (Fig. 7). In part the different targets are likely the result of co-expression of different sets of interacting transcription factors. For example, β-catenin can interact with both Tcf/Lef and the Foxo transcription factors, in a competitive manner, with each capable of inhibiting the binding of β-catenin to the other (Almeida et al., 2007; Essers et al., 2005; Hoogeboom et al., 2008). As shown by these comparative expression/promoter enrichment analyses, β-catenin-Tcf/Lef and β-catenin-Foxo targets appear to be largely non-overlapping sets of genes (Fig. 7).
Fig. 7. Compartment relationships of candidate Lef1 and Foxo4 target genes.
Candidate target genes are associated with compartments with high expression (circles) and transcription factor (triangle) binding sites in promoters by lines. Genes in yellow are candidate Lef1 targets. Overlapping sets of compartment expression, and promoter combinatorial codes of TFBS contributing to some compartment specificity, are shown.
Discussion
In this report we present an atlas of gene expression in the developing kidney. This quantitative, sensitive and global definition of the gene expression profiles of the major components of the developing kidney provides a foundation for further analysis of the genetic mechanisms of nephrogenesis. For example, this atlas of normal gene expression provides a baseline that can be used to better understand abnormalities in kidney development present in mutant mice. This could be through improved in situ hybridization analysis, now equipped with new sets of molecular markers, or through a LCM-microarray based analysis of the mutant phenotype, allowing a more universal characterization of altered gene expression patterns (Potter et al., 2007; Schwab et al., 2006a). The atlas also promotes the generation of additional useful genetic tools for the analysis of kidney development, with for example component specific expression of Cre and/or GFP. Global expression analysis of developmental processes can also reveal potential functional redundancies and guide genetic analyses. Mutant screens and targeted mutation studies often fail to realize expected developmental defects. The microarray atlas data presented here can help identify co-expressed genes with overlapping function.
This work considerably extends previous microarrays studies examining kidney development. Microarrays were first used to examine changing gene expression patterns of entire rat (Stuart et al., 2001) and then mouse (Challen et al., 2005; Schwab et al., 2003) kidneys as a function of developmental time. Some spatial definition of microarray expression profiles has been added by physical dissection of E11.5 metanephric mesenchyme and ureteric bud (Schwab et al., 2006b; Stuart et al., 2003), and in two cases by FACS sorting, of entire E15.5 UB (Challen et al., 2005) using Hoxb7-GFP, and of mesenchyme using Sal1-GFP positive cells (Takasato et al., 2004). Nevertheless, the study presented here represents the first comprehensive microarray analysis of kidney development, using laser capture or FACS to allow the analysis of all the major elements of nephrogenesis.
It is important to note some of the limitations of the microarray-based dataset. First, over 60% of genes can be alternatively processed, and the microarrays used in this study do not reveal patterns of exon usage. Second, several of the components examined in this study consist of mixtures of different cell types. One extreme example is the S-shaped body, which includes cells that are differentiating into podocytes (visceral epithelium), proximal tubules and other diverse structures. The individual laser-captured sections will include different proportions of these variant cell types, and the final microarray results will represent an average of the gene expression patterns of the different cells collected. It is to be expected therefore, that there will be some heterogeneity in the microarray readouts of the biological triplicates performed for each component. Ideally one would like to extend the microarray analysis to the level of single cell types. This is becoming possible as the combination of microarray data and in situ hybridization results identify genes with increasingly restricted domains of expression. This facilitates the generation of transgenic mice with more localized GFP expression, which can then be used in combination with laser capture microdissection and/or FACS to produce gene expression profiles of more restricted cell types.
The microarray-based atlas that we have developed also provides deeper insight into the mechanisms of kidney development, and organogenesis in general. One emerging theme is that while some genes do display extremely component specific expression, their number is surprisingly small. Different developmental compartments generally exhibit extensive overlap in gene expression patterns, with the differences more quantitative than qualitative in nature. The results suggest an analog model of organogenesis, with differences in gene expression levels often more important than differences in gene expression on/off states. The combinatorial codes of gene expression that drive compartment specific development appear to have an important quantitative element. Microarrays, with their ability to define quantitative gene expression levels, are particularly powerful tools for the characterization of such analog gene expression codes.
It is also possible to use the expression data to begin to provide a global view of the genetic circuitry of kidney development. The microarray results provide a comprehensive analysis of the expressed transcription factors, as well as all other genes expressed. It is possible to begin to connect individual transcription factors with their targets by looking for the presence of evolutionarily conserved transcription factor binding sites within promoters of expressed genes. For example we show a very strong statistical association between the expression of Hnf1 in the developing proximal tubules and the presence of a well-conserved Hnf1 binding sites in the promoters of many genes with proximal tubule enriched expression. However, it is important to note that this microarray expression based dissection of genetic circuitry is not all inclusive, as some target genes will carry transcription factor binding sites outside of the scanned 4 Kb proximal promoter region, some genes will be regulated by non-canonical binding sites, and only a few hundred transcription factor binding sites have been well-defined. In addition, the presence of a highly conserved binding site alone does not guarantee biological function for a transcription factor. Ideally one would like to combine microarray expression analysis with global chromatin immunoprecipitation studies to better define transcription factor positioning in the genome, but with the laser capture approach primarily used in this report the limiting amounts of material available make this impractical. Nevertheless, this one example, examining HNF1β targets in the proximal tubules, demonstrates the power of a bioinformatics based approach. By combining observed coordinate expression with analysis of evolutionarily conserved transcription factor binding sites within promoters we find a number of genes previously confirmed to be regulated by HNF1β, and identify many additional candidate targets.
The microarray atlas presented here establishes a precedent for the global analysis of organogenesis, similar in scope to the Allen Brain Atlas (Lein et al., 2007), only focused on a developing organ, and made using a different set of tools. The LCM/FACS-microarray strategy offers several advantages over an approach using only in situ hybridizations. First it is very sensitive, as microarrays can detect transcripts present in only a few copies per cell. Second, it is more quantitative, as single channel microarrays give a numerical measure of gene expression level, while in situ hybridizations give only a relative staining intensity. In addition, using microarrays is far more efficient and cost effective than performing tens of thousands of in situ hybridizations. Nevertheless, there is a significant price to be paid, as the microarray analysis of laser captured components does not provide the potentially single cell spatial resolution achievable with in situ hybridization.
In summary, we describe an atlas of gene expression patterns of the developing kidney generated by using either LCM or FACS to purify components of the developing kidney, followed by hybridization to microarrays to provide comprehensive and quantitative gene expression profiles. This resource provides a global definition of the gene expression program of nephrogenesis, setting the stage for further genetic dissection of this remarkable process.
Experimental Procedures
Sample collection, laser capture microdissection, RNA purification and target amplification
Outbred CD1 E12.5 and E15.5 kidneys (metanephroi) were rapidly dissected from embryos and stored briefly in ice cold PBS, rinsed quickly in OCT, placed in a mold with OCT and snap frozen in liquid nitrogen cooled isopentan, and stored in liquid nitrogen until sectioning. A Microm HM500 cryostat was used to cut 9 micron sections, which were collected on Arcturus PEN membrane glass slides, which were pretreated with poly lysine (Sigma). Laser capture microdissection was performed with an Arcturus Veritas instrument, using both UV cutting (setting 3) and infrared capture lasers, with CapSure HS LCM caps, which were carefully monitored for the presence of non-specific material, which was ablated if possible, or the cap discarded. RNA purification and target amplification was performed as described (GUDMAP.org). In brief, RNA was purified with Qiagen RNeasy Micro kits with 100 µl of RLT having 30 ng of added polyinosine carrier. We typically used a minimum total of approximately one thousand cell sections per RNA preparation, which usually required pooling of about 30–50 component sections. For target amplification we used the TargetAmp 2-round aminoallyl amplification kit (Epicentre). We added an additional 3/4 µl of semi-random primer (SBI) at step one, before speedvac concentration to 3 µl. We also added 1 µl of this primer for part F, step one, in addition to the Epicentre primer. We included a 1 min room temp incubation to allow the primer to anneal. Complete protocols are available at GUDMAP.org (http://www.gudmap.org/Research/Protocols/Potter.html). All animals were housed in an IACUC approved facility, and handled with institutional animal care committee approved protocols.
Identification of specific kidney components
Early proximal tubules were laser captured based on their specific LTA (Lotus tetragonolobus agglutinin) lectin staining. The medullary collecting duct, cortical collecting duct and collecting duct distal to the last branch point were isolated by combining molecular markers (positive DBA, Dolichos biflorus agglutinin and PNA, peanut agglutinin lectin staining), location in kidney and structure. Other components of the developing E15.5 kidney, including S-shaped body, urothelium of the ureter, medullary interstitium, and forming muscle layer surrounding the urothelium (ureteral mesenchyme) were identified based on unique position or structure, combined with the pan-epithelial lectin stain PNA. The loop of Henle (including cortical anlage of the loop of Henle and medullary immature loop of Henle) was identified as the tubule distal to the proximal tubule, PNA positive and LTA negative, in sections that included a continuous proximal tubule and loop of Henle.
In situ hybridizations
Whole mount and section in situ hybridizations were performed as previously described (Little et al., 2007) and as described on the GUDMAP gene expression database (http://www.gudmap.org, see http://www.gudmap.org/Research/Protocols/Little.html and http://www.gudmap.org/Research/Protocols/McMahon.html). For section in situ hybridization outbred CD1 embryos of unknown sex were harvested at 15.5dpc (TS23). Embryonic kidneys were collected, fixed in fresh 4% paraformaldehyde in PBS at 4°C overnight, processed and embedded in paraffin and sectioned at 7mm. Digoxigenin (DIG)-labelled antisense riboprobes were used for RNA section in situ hybridization. Briefly, following dewaxing and rehydration, sections were fixed with 4% paraformaldehyde in PBS, washed with PBS, assembled into slide chambers and inserted into the Tecan Freedom Evo150 robot. Sections were then permeabilised at 25°C with Proteinase K (10µg/ml) for 10 minutes, fixed with 4% paraformaldehyde and acetylated (0.1M triethanolamine, 0.65%HCl and 0.25% (v/v) acetic anhydride). Sections were immersed in pre-hybridisation solution whilst the chamber racks were heated from 25°C to 65°C. Hybridisation occurred at 65°C for 10 h with 0.5 µg/ml of probe in hybridisation buffer (50% formamide, 10% dextran sulphate, 1× Denhardt’s, 0.2mg/ml yeast tRNA, 0.5 mg/ml salmon sperm). After washing with 50% Formamide, 1×SSC sections were treated with 2 µg/mL RNase A for 15minutes then washed with a series of SSC stringency washes. Sections were blocked for 60minutes (20% sheep serum, 2% Blocking Reagent in 1×MBST(100mM Maleic acid, 150mM NaCl, 0.1% Tween-20, pH7.5)) and incubated with 1:2000 of anti-DIG-alkaline phosphatase Fab fragments overnight at 4°C. Sections were washed with 1×MBST followed by NTMT (0.1M NaCl, 0.1M Tris.HCl pH9.6, 50mM MgCl2, 0.1% Tween20). Chromogenic substrate BM Purple was used to detect the in situ alkaline phosphatase activity. Once the signal had reached optimal intensity, the slides were fixed in 4% paraformaldehyde/PBS at 25°C for 20 min followed by PBS washes in order to preserve the in situ hybridization signal. Images were captured with an Olympus BX51, CC12 camera then analyzed using the .slide OlyVia software. All ISH experiments included two control probes for both strong (Wnt4) and weak (Shh) expression to indicate the level of sensitivity of detectable gene expression within each experiment.
Data analysis
In order to assess the relative enrichment of different groups of genes that derive from the GUDMAP dataset with respect to various gene associations, we undertook a comparative approach using a modified form of Gene Set Enrichment Analysis. Each list of genes from the 7K clusters or 12K clusters were separately examined for its relative enrichment for genes that are associated with Gene Ontology, Pathways, and Transcription Factor Binding sites (TFBS) as derived from analyses performed using the MSigDB (http://www.broad.mit.edu/gsea/msigdb/) reference database (Subramanian et al., 2005). The enrichment p-value was converted to an intensity score by the S=(−log(pvalue)). Specifically for TFBS, we mined the catalog of human, mouse, rat and dog conserved regulatory motifs in promoters (4 Kb) (Xie et al., 2005), using ToppGene server (Chen et al., 2007), for each of the kidney compartment-enriched gene lists. For obtaining a network view of putative shared conserved TFBSs and the corresponding gene sets we used Cytoscape (Shannon et al., 2003), a JAVA-based bioinformatics software package for visualizing, modeling and analyzing molecular and genetic interaction networks. Cytoscape provides a unique in silico approach to examine and display transcriptional networks based on putative TFBS and target gene interactions characterized from either previous studies or computational predictions (Ideker et al., 2002; Shannon et al., 2003; Spirin and Mirny, 2003). We loaded the more than 2,800 interactions representing the seven kidney compartments with enriched genes along with their enriched conserved TFBSs derived using the ToppGene server (Chen et al., 2007). As the number of known gene-TFBS pairs expands, such a Cytoscape-assisted network approach for analyzing large genomic datasets will dramatically increase our understanding of the underlying transcriptional regulatory networks and could assist in the discovery of many potential targets for biological follow-up studies or experimental validations. Additionally, the network based approach facilitates identification of the modular nature of transcriptional regulation through cooperativity between different transcription factors targeting a group of genes enriched in one or more than tissue compartments.
The results from each gene selection method were concatenated and reentered as a matrix of data. The data in this new interpretation was clustered to find similar patterns in the different methods. By examining the original data with this new approach, interesting and informative patterns can be seen that are based on gene-gene relationships within a cluster or highly ranked gene list rather than based on the aggregation of individual gene behaviors. All microarray data is available at GUDMAP.org and GEO (GSM144585-93, GSM152245-9, GSM207260, GSM203834-6, GSM207261-3, GSM144596-621, GSM152250-52).
Tissue processing for confocal microscopy
Kidneys were dissected in phosphate buffered saline (PBS). The kidneys or the organ explants were rocked for 1–2 h in 2% paraformaldehyde in PBS, washed twice with PBS, and then rocked for 1–2 h in 100% methanol. The tissues were washed twice with cold PBS containing 0.05% Tween-20 (PBT). Kidneys were bisected. Primary antibodies, diluted to 1:250 to 1:400, were added to the tissues in 400 µL of PBT containing 2% goat serum and incubated overnight with rocking. Tissues were washed with 5 exchanges of PBT over 8 h with rocking. The secondary antibodies, diluted to 1:400 in PBT containing 2% goat serum, were added and incubated overnight. The tissues were again washed with 5 exchanges of PBT over 8 h. Fluorescein-conjugated Dolichos biflorus agglutinin (DBA, Vector), used to mark ureteric bud branches, was diluted 1:60 in PBT and added to the tissues and incubated over night. The tissue was washed for 5–10 min and mounted in a depression slide in PBT before they were examined by confocal microscopy. The entire procedure was performed at 4 °C with pre- cooled reagents.
We used the following primary antibodies: anti-WT1 (c-19, Santa Cruz), anti-Uvomorulin (E-cadherin, Sigma). The secondary antibodies were Alexa 555-conjugated anti-rabbit and Alexa 633-conjugated anti-rat secondary antibodies (Molecular Probes).
Supplementary Material
Acknowledgments
We acknowledge Han Sheng Chiu, Emmanuelle Lesieur and Darrin Taylor for technical assistance. This work was supported by NIH NIDDK grants to ML (DK070136, DK070136), AM (DK070181) and SP (DK070251).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akpinar P, Kuwajima S, Krutzfeldt J, Stoffel M. Tmem27: a cleaved and shed plasma membrane protein that stimulates pancreatic beta cell proliferation. [see comment] Cell Metabolism. 2005;2:385–397. doi: 10.1016/j.cmet.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Almeida M, Han L, Martin-Millan M, O'Brien CA, Manolagas SC. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting beta-catenin from T cell factor- to forkhead box O-mediated transcription. J Biol Chem. 2007;282:27298–27305. doi: 10.1074/jbc.M702811200. [DOI] [PubMed] [Google Scholar]
- Baumhueter S, Courtois G, Crabtree GR. A variant nuclear protein in dedifferentiated hepatoma cells binds to the same functional sequences in the beta fibrinogen gene promoter as HNF-1. EMBO Journal. 1988;7:2485–2493. doi: 10.1002/j.1460-2075.1988.tb03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Developmental Biology. 2008;313:234–245. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB Journal. 1996;10:267–282. [PubMed] [Google Scholar]
- Challen G, Gardiner B, Caruana G, Kostoulias X, Martinez G, Crowe M, Taylor DF, Bertram J, Little M, Grimmond SM. Temporal and spatial transcriptional programs in murine kidney development. Physiol Genomics. 2005;23:159–171. doi: 10.1152/physiolgenomics.00043.2005. [DOI] [PubMed] [Google Scholar]
- Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- Fukui K, Yang Q, Cao Y, Takahashi N, Hatakeyama H, Wang H, Wada J, Zhang Y, Marselli L, Nammo T, et al. The HNF-1 target collectrin controls insulin exocytosis by SNARE complex formation.[see comment] Cell Metabolism. 2005;2:373–384. doi: 10.1016/j.cmet.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Gresh L, Fischer E, Reimann A, Tanguy M, Garbay S, Shao X, Hiesberger T, Fiette L, Igarashi P, Yaniv M, et al. A transcriptional network in polycystic kidney disease. EMBO Journal. 2004;23:1657–1668. doi: 10.1038/sj.emboj.7600160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesberger T, Bai Y, Shao X, McNally BT, Sinclair AM, Tian X, Somlo S, Igarashi P. Mutation of hepatocyte nuclear factor-1beta inhibits Pkhd1 gene expression and produces renal cysts in mice. Journal of Clinical Investigation. 2004;113:814–825. doi: 10.1172/JCI20083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogeboom D, Essers MA, Polderman PE, Voets E, Smits LM, Burgering BM. Interaction of FOXO with beta -catenin inhibits beta -catenin/TCF activity. J Biol Chem. 2008 doi: 10.1074/jbc.M706638200. [DOI] [PubMed] [Google Scholar]
- Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;1 18 Suppl:S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kusuhara H, Hattori N, Shiota K, Kim I, Gonzalez FJ, Sugiyama Y. Regulation of the expression of human organic anion transporter 3 by hepatocyte nuclear factor 1alpha/beta and DNA methylation. Molecular Pharmacology. 2006;70:887–896. doi: 10.1124/mol.106.025494. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kwan KM, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development. 2005;132:2809–2823. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- Lazzaro D, De Simone V, De Magistris L, Lehtonen E, Cortese R. LFB1 and LFB3 homeoproteins are sequentially expressed during kidney development. Development. 1992;114:469–479. doi: 10.1242/dev.114.2.469. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, et al. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr Patterns. 2007;7:680–699. doi: 10.1016/j.modgep.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SS, Hartman HA, Kwan KM, Behringer RR, Patterson LT. Laser capture-microarray analysis of Lim1 mutant kidney development. Genesis: the Journal of Genetics & Development. 2007;45:432–439. doi: 10.1002/dvg.20309. [DOI] [PubMed] [Google Scholar]
- Schwab K, Hartman HA, Liang HC, Aronow BJ, Patterson LT, Potter SS. Comprehensive microarray analysis of Hoxa11/Hoxd11 mutant kidney development. Developmental Biology. 2006a;293:540–554. doi: 10.1016/j.ydbio.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Schwab K, Hartman HA, Liang HC, Aronow BJ, Patterson LT, Potter SS. Comprehensive microarray analysis of Hoxa11/Hoxd11 mutant kidney development. Dev Biol. 2006b;293:540–554. doi: 10.1016/j.ydbio.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Schwab K, Patterson LT, Aronow BJ, Luckas R, Liang HC, Potter SS. A catalogue of gene expression in the developing kidney.[see comment] Kidney International. 2003;64:1588–1604. doi: 10.1046/j.1523-1755.2003.00276.x. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart RO, Bush KT, Nigam SK. Changes in global gene expression patterns during development and maturation of the rat kidney. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5649–5654. doi: 10.1073/pnas.091110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart RO, Bush KT, Nigam SK. Changes in gene expression patterns in the ureteric bud and metanephric mesenchyme in models of kidney development. Kidney Int. 2003;64:1997–2008. doi: 10.1046/j.1523-1755.2003.00383.x. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasato M, Osafune K, Matsumoto Y, Kataoka Y, Yoshida N, Meguro H, Aburatani H, Asashima M, Nishinakamura R. Identification of kidney mesenchymal genes by a combination of microarray analysis and Sall1-GFP knockin mice. Mech Dev. 2004;121:547–557. doi: 10.1016/j.mod.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biology. 2002;3 doi: 10.1186/gb-2002-3-12-research0088. RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort M, Clevers H. TCF transcription factors, mediators of Wnt-signaling in development and cancer. Dev Biol. 2002;244:1–8. doi: 10.1006/dbio.2001.0566. [DOI] [PubMed] [Google Scholar]
- Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, Kubly V, Cunningham JM, Bacallao R, Ishibashi M, et al. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein.[see comment] Nature Genetics. 2002;30:259–269. doi: 10.1038/ng833. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wada J, Yasuhara A, Iseda I, Eguchi J, Fukui K, Yang Q, Yamagata K, Hiesberger T, Igarashi P, et al. The role for HNF-1beta-targeted collectrin in maintenance of primary cilia and cell polarity in collecting duct cells. PLoS ONE. 2007;2:e414. doi: 10.1371/journal.pone.0000414. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.