Abstract
Proton MR spectroscopy (1H MRS) and dynamic contrast-enhanced (DCE) MR imaging provide functional information, including vascular volume, vascular permeability, and choline (Cho) metabolism. In this study, we applied these two imaging modalities to quantitatively characterize 36 malignant breast lesions in 32 patients and analyzed the correlation between them. The Cho concentration was quantified by single-voxel 1H MRS using water as an internal reference. The measured Cho levels ranged from 0.32 to 10.47 mmol/kg, consistent with previously reported values. In 25 mass type lesions, the Cho concentration was significantly correlated with tumor size (r = 0.69, p < 0.0002). In addition, the Cho level was found to be significantly higher in lesions presenting as mass type lesions compared to non-mass type diffuse enhancements (p = 0.035). The enhancement kinetics from tissues covered within each MRS voxel were measured and analyzed with a 2-compartmental model to obtain pharmacokinetic parameters Ktrans and kep. A significant correlation was found between the Cho level and pharmacokinetic parameter kep (r = 0.62, p < 0.0001), indicating that tissues with a high Cho level have higher wash-out rates in DCE MR imaging. The results suggest a correlation between choline metabolism and angiogenesis activity, which might be explained by the association of Cho with cell replication and angiogenesis required to support tumor growth.
Keywords: proton MR spectroscopy, dynamic contrast-enhanced MRI, quantification, breast cancer, choline
1. Introduction
High-resolution anatomic MRI and dynamic contrast-enhanced MR imaging (DCE) MR imaging have become an established clinical modality for detection and diagnosis of breast lesions (1). In March 2007, the American Cancer Society issued new guidelines recommending annual MRI screening for women who have a lifetime risk higher than 20% for developing breast cancer. DCE MR imaging is a well established technique for monitoring contrast enhancement kinetics, which may, in turn, reveal characteristics of tumor microvasculature. Malignant tumors often show a pattern of rapid wash-in followed by wash-out. This is known to be associated with a high vascular volume and/or high vascular permeability due to angiogenesis, which is needed to support tumor growth (2). Angiogenesis will provide a higher blood supply, and the newly formed vessels have wide endothelial junctions hence a higher vascular permeability. Conversely, benign tumors are more likely to show a slow wash-in followed by a persistent enhancement pattern due to a lower degree of angiogenesis (3).
In vivo proton MR spectroscopy (1H MRS) is a non-invasive technique that can provide tumor metabolic information, which has also been shown to have potential to aid in diagnosis and management of breast tumors (4–7). Combined application of DCE MR imaging and 1H MRS has been used in diagnosing breast lesions in several studies (8, 9). The addition of MRS yields a higher specificity relative to that using DCE MR imaging alone. Despite their wide application, however, the quantitative correlation between the vascular and metabolic information obtained with DCE MR imaging and MRS has not been thoroughly investigated (10).
In this study, we performed quantitative in vivo 1H MRS and DCE MR imaging to investigate the correlation between Cho concentration and DCE MR imaging model parameters in malignant breast lesions. For 1H MRS, we applied single-voxel MRS with an internal reference method using the water signal to quantify Cho concentration. This method can compensate for the partial volume of adipose tissue in the selected MRS voxel, allowing measurement of the molal concentration (mmol/kg) of water-soluble metabolites. Cho concentration was also correlated with tumor size. For analysis of DCE MR imaging, we used the Generalized Kinetic Model based on two compartments (11) to obtain the transfer constant (Ktrans) and the rate constant (kep). The rate constant is the ratio of the transfer constant to the extravascular extracellular space (EES) fractional volume (ve). The measured Cho concentration was correlated with contrast enhancement parameters, including percentage enhancement at 2 minutes after injection (SE%-2min), Ktrans, and kep. The lesions presented as mass type and non-mass type were analyzed in separate groups and results compared.
2. Materials and methods
2.1. Subjects
Thirty two patients (29–91 years old, mean 48) with biopsy-confirmed breast cancer were included in this study. Based on the morphological pattern of enhancement, all lesions were categorized into one of two groups: mass type lesion and non-mass type enhancements according to the ACR BIRADS lexicon (12). Twenty one patients had mass type lesions with well defined borders, and 11 had non-mass type lesions showing diffuse enhancements without clearly defined borders. In the mass group, 17 of 21 patients had a solitary mass and 4 had at least 2 differentiable masses (1.6 – 5.0 cm, median 3.0 cm). All together, 25 mass lesions and 11 non-mass lesions were studied. This study was approved by the University of California Irvine Institutional Review Board, and informed consent was obtained from each patient.
2.2. MRI/MRS protocol
All patients were scanned on a clinical 1.5T whole body system (Eclipse; Philips Medical System, Cleveland, Ohio) with the standard MRS acquisition software provided by the manufacturer. A body coil was used for transmission, and a dedicated four-channel phased-array breast coil (USA Instruments, Aurora, Ohio) was used for receiving. All patients were examined in prone position with their breasts cushioned in rubber foam to reduce motion. After the scout scan, sagittal view T1-weighted pre-contrast images were acquired from the breast of concern using a spin echo (SE) sequence with TR/TE = 1000/12ms, matrix size = 256×256, field of view (FOV) = 22 cm, and 34 slices with 3–4 mm thickness. For DCE MR imaging a 3-dimensional RF-FAST (Fourier Acquired Steady State) pulse sequence was prescribed. This sequence yields T1-weighting by spoiling the transverse steady state via RF phase cycling. Thirty-two axial slices with 4 mm thickness were used to cover both breasts. The imaging parameters were TR/TE = 10/3.6 ms, flip angle = 20°, acquisition matrix size = 256×128, and FOV varying between 32 and 38 cm. The scan time was 42 seconds per acquisition. The sequence was repeated 16 times (e.g., 16 frames) for dynamic acquisitions, 4 for pre-contrast and 12 for post-contrast sets. The contrast agent (Ominscan®, 1 cc/10 lbs body weight) was manually injected during continuing acquisition, at the beginning of the 5th acquisition, and was timed to finish in 12 seconds to make the bolus length consistent for all patients. Immediately following the contrast, 10 cc saline was injected to flush in all contrast media.
The pre-contrast images acquired at the 3rd frame were subtracted from the post-contrast images acquired at the 6th frame to generate subtraction images on the scanner console. The subtraction images were used for placing the volume of interest in the viable tumor region showing the strongest perfusion (i.e. early enhancement) for the subsequent MRS. Localized single-voxel 1H MR spectra were acquired from the enhanced lesion. In 4 patients with multiple separate masses, MRS was acquired from two largest lesions. The spectroscopic voxel was carefully positioned to maximize the coverage of the contrast-enhanced lesions while minimizing the inclusion of adipose tissue. The voxel size was either 1.8 × 1.8 × 1.8 cm3 or 2.0 × 2.0 × 2.0 cm3. Localization was obtained using the point-resolved spin-echo sequence (PRESS) (13, 14) followed by voxel shimming. The typical water peak linewidth (FWHM) ranged from 8 to 17 Hz. The spectra were acquired with water suppression and fat attenuation via three CHESS pulses (15, 16) with 60 Hz bandwidth and frequency-selective presaturation pulse (FATSAT), respectively. The following acquisition parameters were used: TR = 2000 ms, TE = 270 ms, 128 acquisitions, spectral width = 1953 Hz, and 2048 data points. A fully relaxed, unsuppressed spectrum (TR/TE = 2000/270 ms, 24 acquisitions) was also acquired to measure the amplitude of the water peak in the localized volume as the internal reference. The MRS scan time was approximately 6 minutes. After including the additional time for voxel placement and shimming, the total scan time for the entire sequence could be completed within 15 minutes.
2.3. Data analysis
2.3.1. Tumor size measurement
The total of 36 lesions consisting of 25 mass and 11 non-mass types were studied. A radiologist determined the size measurement based on the maximum intensity projection (MIP) of the subtraction images. The longest dimension and the longest perpendicular dimension of the MIP were measured. The equivalent one dimensional tumor size was calculated by taking square root of their product.
2.3.2. Analysis of MRS data: preprocessing, fitting, and quantification
The jMRUI software package (17) was used for time-domain analysis. For the unsuppressed spectra used to measure the water peak, each free induction decay signal was first zero-filled to 4096 points. After Fourier transformation, automatic (or manual) phasing was used to correct every signal with the zero-order phase of its water peak. The maximum peak of the water signal was assigned to 4.7 ppm, implicitly setting the polymethylene lipid peak at 1.32 ppm. For preprocessing and quantification of the water signal, we selected a frequency range of 4.2 – 5.2 ppm. In order to measure the Cho peak from the water and fat suppressed spectrum, we performed a preprocessing that consisted of zero-filling of 4096 points, Gaussian apodization of 5 Hz, Fourier transformation, and phase correction of the transformed spectrum. A narrow frequency range (e.g., 2.92 – 3.52 ppm) was selected for analyzing Cho peak to quantify its amplitude.
AMARES (Advanced Method for Accurate, Robust and Efficient Spectral fitting) (18), a widely used quantitation tool for MRS data, was employed to fit the spectra. In this study, a Gaussian lineshape model was chosen for quantifying the Cho peak. Soft constraints were imposed for a faster and more accurate quantitation during spectral fitting. Linewidths for the Cho peak were allowed to vary between 1 and 10 Hz. The frequency constraint range was restricted to ± 0.2 ppm (e.g., 3.12 – 3.32 ppm). After the zero and first order phases were switched off, the frequency-selective option (19) was applied, weighting the first 20 points of the time domain signal by the first quarter of a squared sine function. The Cramer-Rao lower bound (CRLB) was used as a measure of fitting accuracy (20). Uncertainty in the estimated Cho concentration was the standard deviation (SD) of the Cho signal amplitude as estimated using the CRLB. In the water unsuppressed spectra, water peak was fit at 4.7 ppm.
Absolute quantification of Cho concentration was acquired using the water peak as an internal reference. All acquisitions were recorded at maximum receiver gain which made corrections for different receiver setting unnecessary. Hence, the absolute Cho concentration was calculated based on Eq. [1],
| [1] |
where [Cho] is the concentration of the Cho metabolite in units of mmol/kg, SCho is the signal amplitude of the Cho, and SH2O is the signal amplitude of the unsuppressed water in the localized spectrum. The terms nCho and nH2O represent the number of 1H nuclei in each respective molecule. The ratio of SCho and SH2O amplitudes can be changed to molar concentration by correcting for the number of 1H nuclei per molecule and the molecular weight of water, MWH2O. and are the numbers of data acquisitions for water unsuppressed and suppressed spectra. The parameters fT1 and fT2 are the correction factors for T1 and T2 relaxation times: fT1 = 1 − exp(−TR/T1) and fT2 = exp(−TE/T2). T2 relaxation times were 269 ms for Cho and 97 ms for water; T1 relaxation times were 1513 ms for Cho and 746 ms for water (21).
2.3.3. Analysis of DCE MR Imaging data
For the analysis of DCE MR imaging, the averaged enhancement time course from tissues contained in each MRS voxel was measured. In the case of a 2.0 × 2.0 × 2.0 cm3 voxel, it encompassed 5 axial slices in DCE MR imaging (each 4 mm thick) along the superior-inferior direction with 13 × 13 (anterior-posterior × medial-lateral) pixels on each slice. The mean signal intensity time course from the resulting 845 pixels was measured and converted into a fractional enhancement time course with respect to the mean of baseline. The fractional enhancement time course was fit to the Generalized Kinetic Model (11) using a fixed biexponential decay for the arterial input function (AIF) (22). We assumed that the fractional enhancement is linearly proportional to the contrast concentration when fitting it to the model. The limitations and uncertainties of tracer kinetic modeling and the estimated model parameters, are well summarized in an article by Padhani et al. (23). The measurement of parameter Ktrans in units of [1/min] requires conversion from enhancement to absolute [Gd] concentration. The other model parameter, kep (the rate constant) is in the absolute unit of min−1, and is directly comparable to those reported in the literature.
Maximum enhancement was usually reached between 2–3 minutes after injection of the contrast agent. The percentage enhancement from dynamic frame #8 (the 4th post-contrast time frame), approximately at 2.5 minutes after injection (for simplicity, noted as SE%-2min hereafter) was obtained for correlation analysis. Together with Ktrans (sensitive to the initial wash-in phase) and kep (sensitive to wash-out phase), they represented three key features of each DCE kinetic curve.
2.4. Statistical Analysis
Statistical analysis was performed using the Microcal software package (Microcal Origin® Version 6.0 for Windows; Microcal Software Inc., MA). Pearson’s linear regression was employed to determine whether the Cho level was correlated with lesion size. The correlation between Cho concentration and the 3 DCE MR imaging parameters (SE%-2min, Ktrans, and kep) among all 36 lesions were also investigated. Correlation coefficient r and p values were reported. The significance level was set at p < 0.05.
3. Results
3.1. Group characteristics
Table 1 summarizes the group mean of Cho concentration and DCE MR imaging parameters. The mean lesion size was 2.8 cm for the mass type group (n = 25) and 6.3 cm for the non-mass group (n = 11). For these 36 lesions, the measured Cho levels ranged from 0.32 to 10.47 mmol/kg (mean, 3.20 mmol/kg), which are well within the previously published in vivo Cho concentrations (24). The mass type group had a significantly higher mean Cho concentration than the non-mass type (3.82 vs. 1.79 mmol/kg, p = 0.035). However, no significant group differences were observed in the DCE MR imaging parameters SE-2min%, Ktrans and kep (p = 0.88, 0.24, and 0.51, respectively).
Table 1.
Summary of findings (mean ± SD) for tumor size, Cho concentration, and DCE MR imaging parameters in 2 groups of lesions, presented as either mass or diffuse type.
| Tumor morphology | Tumor Size (cm)* | Cho conc. (mmol/kg)† | DCE-MRI parameters |
||
|---|---|---|---|---|---|
| Max% | Ktrans (1/min) | kep (1/min) | |||
| Mass (N=25) | 2.8 ± 0.6 | 3.82 ± 2.88 | 123 ± 38 | 0.26 ± 0.17 | 1.21 ± 0.71 |
| Diffuse (N=11) | 6.3 ± 0.9 | 1.79 ± 1.53 | 121 ± 44 | 0.19 ± 0.11 | 1.06 ± 0.64 |
Note.-Tumor size (cm2) was calculated as the product of long dimension × short dimension.
p < 0.0002
p = 0.035, where significance level was set at p < 0.05.
3.2. Correlation between Cho-level and MRI parameters
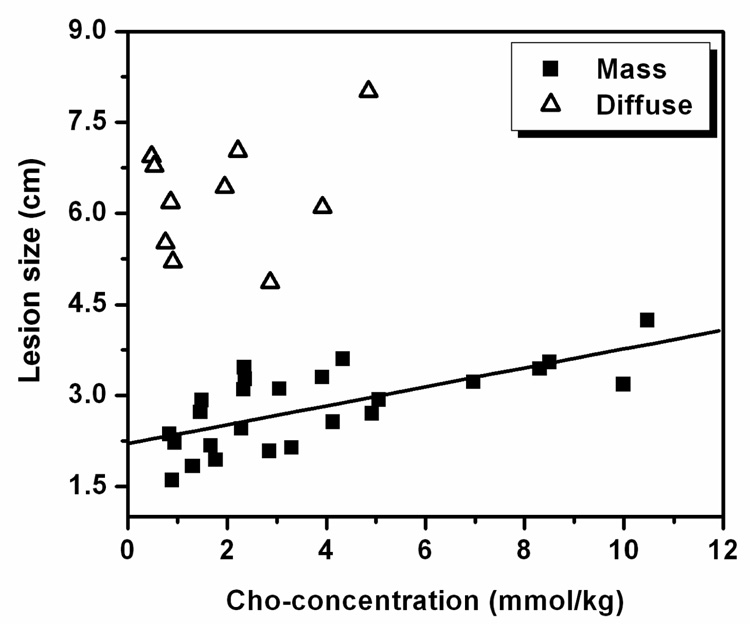
The comparison between Cho concentration and lesion size among mass and non-mass groups is shown in Figure 1. The linear regression analysis for the mass type group (n = 25) showed a significant correlation (r = 0.69 and p < 0.0002). The data from the non-mass group (n = 11) are also shown in Figure 1, but no significant correlation was observed (no regression line shown).
Figure 1.
Correlation between Cho concentration and equivalent 1-dimensional tumor size. A significant linear correlation was found for 25 cases presenting as mass type lesion (r = 0.69, p < 0.0002). The other 11 cases presenting as diffuse-type lesion are overlaid. Despite their relatively larger size, they have lower Cho concentrations.
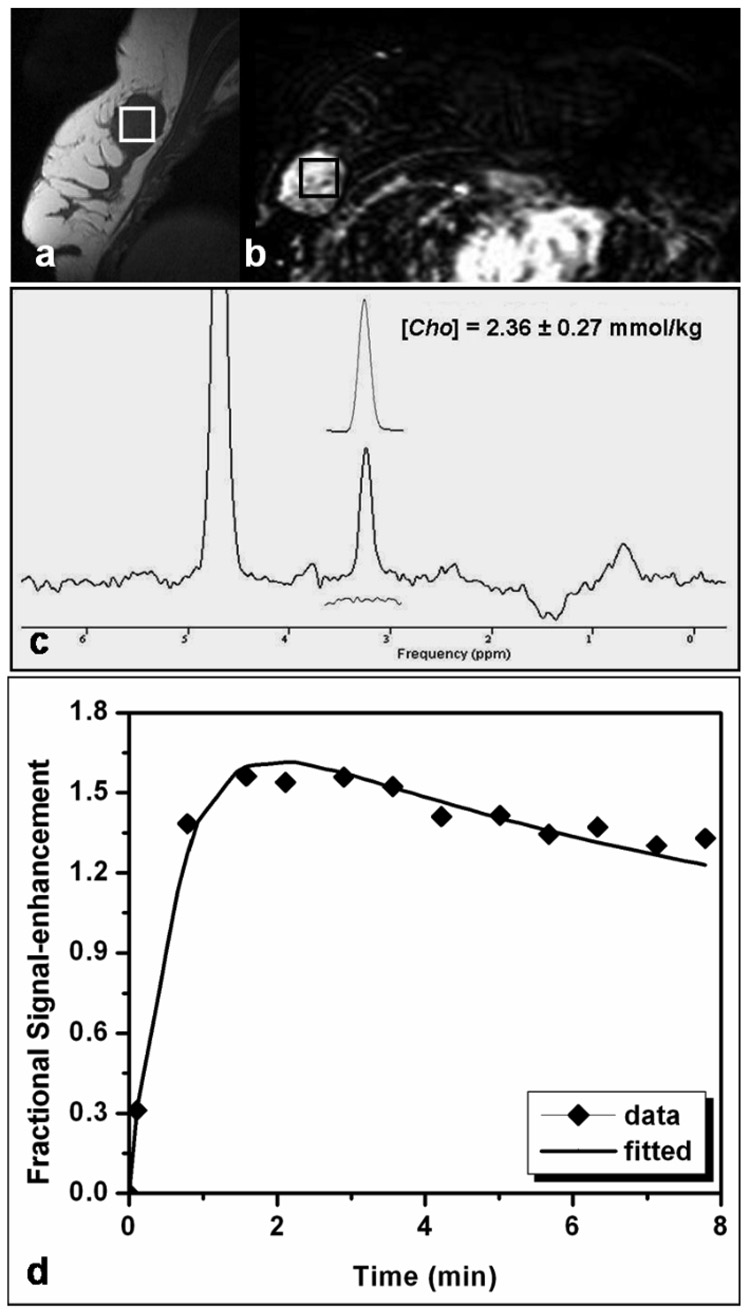
Figure 2 shows results of MRS and DCE MR imaging from one patient with a solitary mass. The lesion size was measured as 3.6 (S-I) × 2.5 (A-P) × 2.5 (L-R) cm3. The spectroscopic voxel (2 × 2 × 2 cm3) was carefully positioned to maximize the coverage of the hypointense lesion on the pre-contrast sagittal slice image bisecting the MRS voxel (Fig. 2a), and also on the contrast-enhanced lesion on the axial subtraction image (Fig. 2b). The elevated Cho peak at 3.22 ppm is clearly visible in the water-fat suppressed spectrum (Fig. 2c). The Gaussian model fitting of the Cho peak yields [Cho] = 2.36 ± 0.27 mmol/kg. The corresponding MRS voxel averaged enhancement time course fitted with the kinetic model is shown in Figure 2d.
Figure 2.
MRI and MRS measurements in a patient with a mass type lesion. The pre-contrast sagittal image of one lesion showed low signal intensity. The spectroscopic voxel (size, 2 × 2 × 2 cm3) is placed in the hypointense lesion on the pre-contrast sagittal image (a) and on the contrast-enhanced lesion in the subtraction image (b). A Cho peak at 3.22 ppm is clearly visible in the water-fat suppressed spectrum (c). The Gaussian model fitting of the Cho peak produces a measurement of [Cho] = 2.36 ± 0.27 mmol/kg. The estimated model fit is shown above the full spectrum and the residue is shown underneath. The corresponding DCE MR imaging kinetics from the MRS voxel labeled in Fig. 2(a) and 2(b) is shown in Fig. 2(d). The symbol is the experimentally measured enhancement percentage, and the line is the best-fitting result using the 2-compartmental pharmacokinetic model.
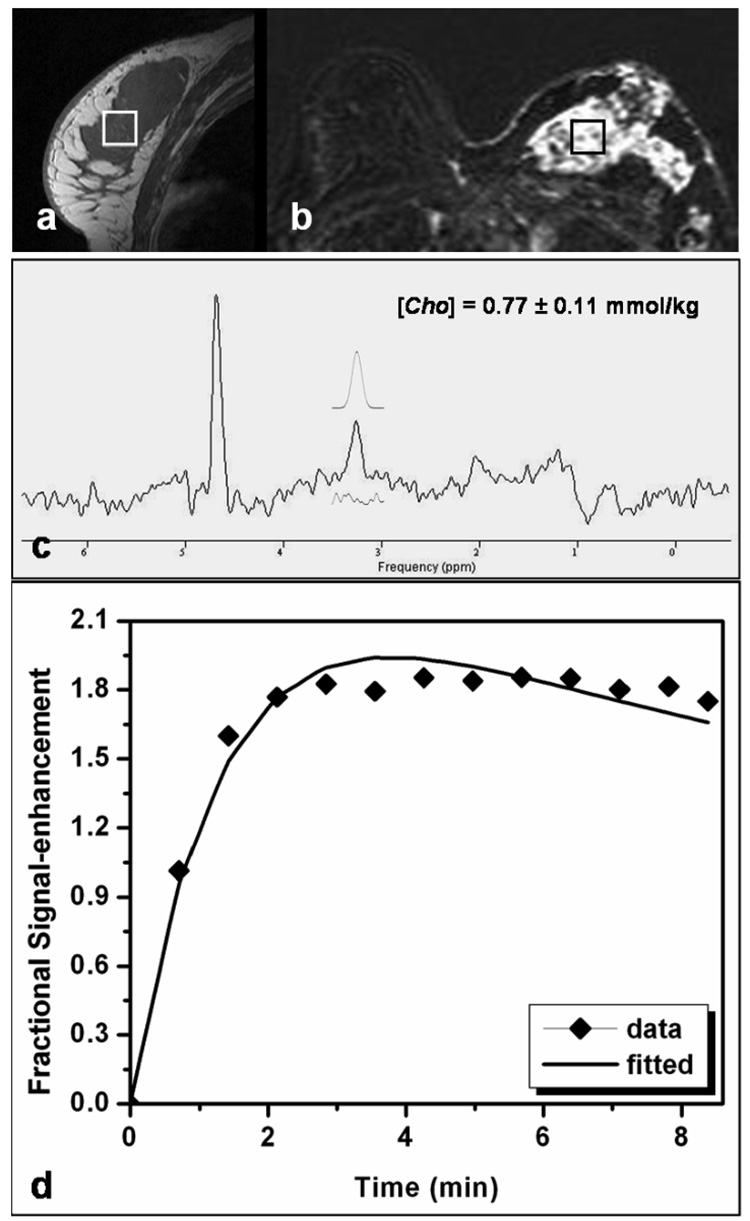
Figure 3 shows a case study from another patient with a non-mass lesion displaying a diffuse enhancement pattern. One large hypointense lesion was shown on the pre-contrast sagittal image (Fig. 3a) showing heterogeneous enhancements on the contrast-enhanced subtraction image (Fig. 3b). The spectroscopic voxel (2 × 2 × 2 cm3) was positioned over the enhanced lesion noted on the subtraction image. A Cho peak was clearly detected in the water-fat suppressed spectrum, with [Cho] = 0.77 + 0.11 mmol/kg (Fig. 3c). The corresponding MRS voxel enhancement time course fitted with the kinetic model is shown in Figure 3d.
Figure 3.
MRI and MRS measurements in a patient with a diffuse-type lesion. The spectroscopic voxel (size, 2 × 2 × 2 cm3) is positioned within the large hypointense lesion on the pre-contrast sagittal image (a) as well as in the enhanced lesion on the subtraction image (b). A Cho peak is visible on the water-fat suppressed spectrum (c), with [Cho] = 0.77 ± 0.11 mmol/kg. Among all subjects, this patient had the lowest Cho concentration. The corresponding DCE MR imaging kinetics is shown (d), symbols for measured enhancement percentage, and the line for the best fitting results.
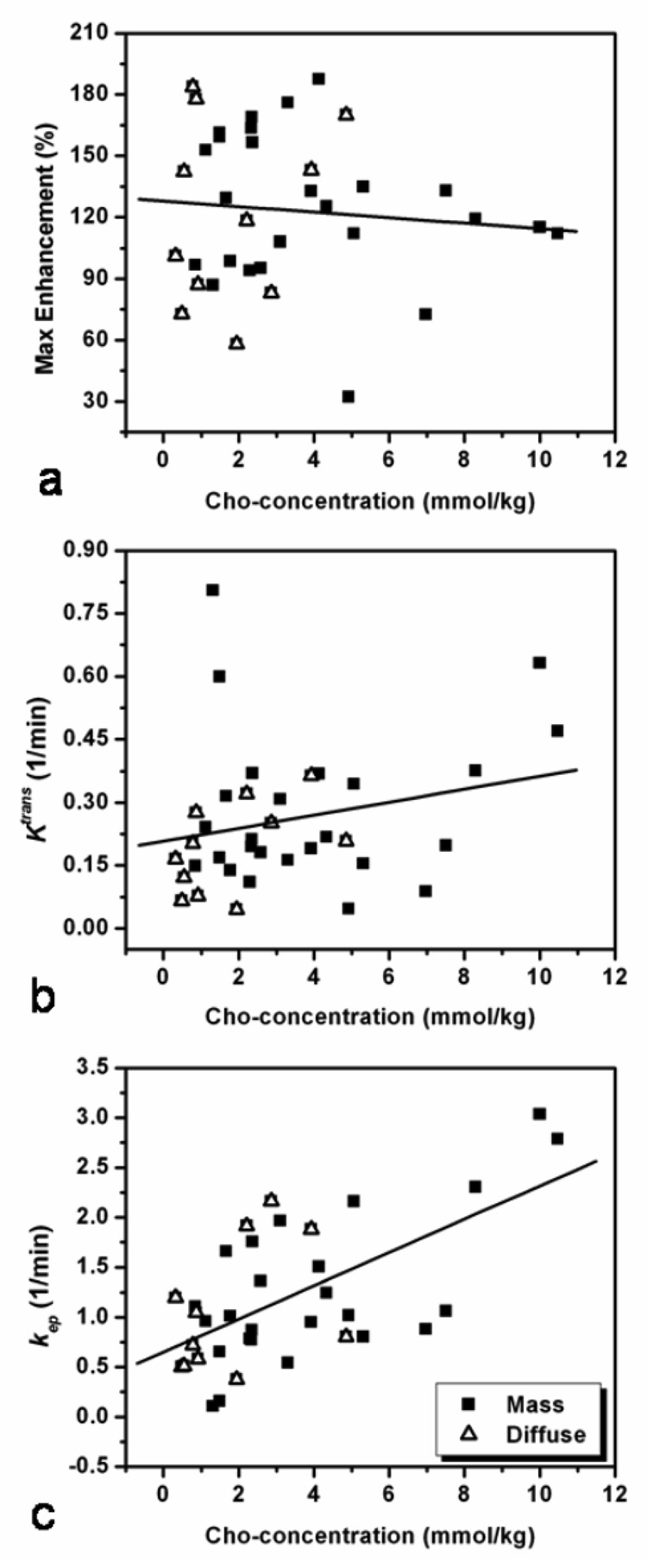
The DCE MR imaging parameters (SE-2min%, Ktrans, and kep) were analyzed for correlation with the Cho level by pooling all lesions together (n = 36). Figure 4 shows the scattered plots between Cho concentration and DCE MR imaging parameters. Lesions of different morphology (mass or diffuse) are labeled with different symbols. A statistically significant correlation between [Cho] and the rate constant kep (r = 0.62, p < 0.0001) was found, but not with SE%-2min (r = −0.008, p = 0.639) or Ktrans (r = 0.216, p = 0.211).
Figure 4.
Correlation between the Cho and DCE MR imaging parameters SE-2min%, Ktrans, and kep among 36 lesions in 32 patients. Lesions of different morphology, mass or diffuse, are labeled with different symbols, but analyzed together. The Cho level is not correlated with the SE-2min% (r = −0.008, p = 0.639) (a), or wash-in rate Ktrans (r = 0.216, p = 0.211) (b), but has a significant correlation with the wash-out rate kep (r = 0.62, p < 0.0001) (c).
4. Discussion
In vivo 1H MRS and DCE MR imaging can provide metabolic and vascular functional information which may both be acquired during the same MR imaging session. In this study we performed quantitative analyses of DCE MR imaging and single-voxel 1H MRS acquired from breast cancer and investigated the association between them. Our findings indicate a statistically significant correlation between Cho concentration and the DCE parameter, the rate constant kep (r = 0.62, p < 0.0001). The association between these two parameters may be explained on the basis of 2 factors: a) Cho is associated with active cell replication; and b) Tumor growth requires active angiogenesis, resulting in leaky immature vessels (i.e. with wide endothelial junctions). Correlation with the signal enhancement SE%-2min and Ktrans was not significant. kep is related to wash-out, and is known to be a better DCE parameter compared to wash-in or enhancement intensity differentiating between benign and malignant lesions (25).
Early in vivo 1H MRS studies demonstrated that elevated Cho peak at 3.2 ppm is observed in neoplastic tissues (4, 5, 7, 26–27). 1H NMR spectra acquired from biopsied tissues demonstrated that the Cho resonance peak is comprised of multiple signals, such as phosphocholine, glycerophosphocholine, and free choline (28, 29). However, these three signals cannot be resolved in vivo at 1.5T, at which only a single resonance peak, representing the aggregate of all choline containing compounds, is observed. Among these, the primary component is phosphocholine, a precursor of cell membrane synthesis. Thus, the elevated Cho level in breast cancer maybe associated with increased membrane synthesis due to ongoing tumor cell replication.
Several previously published studies have measured the absolute Cho concentration in vivo. For example, Roebuck et al. (4) found Cho levels ranging 0.4 – 5.8 mmol/L in seven patients with confirmed malignant breast tumors. Bakken et al. (30) reported 2.0 mM of choline-containing compounds found in a single breast cancer patient at 1.5T. Finally, Bolan et al. (24) reported Cho measurements of 0.4 – 10 mmol/kg in malignant breast tissue spectra at 4.0T. Our Cho levels, obtained from 36 spectra of 32 patients, ranged between 0.32 – 10.47 mmol/kg, consistent with the values reported in the aforementioned studies.
The large range in Cho concentrations may reflect the heterogeneous nature of breast lesions. Gribbestad et al. (31) reported that phosphatidylcholine, a precursor of choline-derived phospholipids, also showed a large variation even among the same tumor types. The increase of choline has often been reported in breast cancer and is regarded as a marker for elevated proliferation rates (32). Singer et al. (33) also reported that the metastatic breast cancer cell line 21MT-2 had a significantly higher concentration of choline than did the primary breast cancer cell lines 21PT and 21NT.
In addition to the intrinsic heterogeneous nature of breast tumors, the limitations of in vivo 1H MRS detection may also contribute to a complicated Cho distribution pattern. For example, Cho detection maybe difficult in diffusive enhancement type cancers because of the intermingling of tumor cells with adipose tissue. In this study, diffusive enhancement type cancer showed a much lower overall Cho level than mass type cancers (p = 0.035). This result is consistent with previous findings by Su et al. correlating the number of Cho-positive voxels measured using chemical-shift imaging with different tumor morphology (10). We also found a significant correlation between the Cho concentration and tumor size among 25 mass type lesions (r = 0.69, p < 0.0002). This could be explained by either by the association of cell replication with tumor size, or could be due to the detection limitation of MR spectroscopy. The latter may be because Cho detection in larger tumors was less prone to fat contamination. Given these issues, the correlation between Cho and tumor size warrants further investigation.
Despite the common use of MRS and DCE MR imaging in breast cancer, the association between them was rarely reported. A feasibility study by Jacobs et al. (34) suggested that proton spectroscopy may be a promising technique for classifying breast lesions when DCE MR imaging alone cannot make a differential diagnosis of the enhancing lesions. Further investigation is needed to determine whether and how combined parameters (e.g., Cho and kep) measured using 1H MRS and DCE MR imaging can be used to aid in characterizing of breast lesions. Huang et al. (8) reported that the combined protocol with DCE MR imaging, 1H MR spectroscopy, and perfusion MR imaging could reach the highest sensitivity and specificity in the diagnosis of breast cancer. Meisamy et al. (9) demonstrated that addition of MRS information can improve diagnostic accuracy of both experienced and inexperienced radiologists.
As MRS is a more established diagnostic tool for prostate cancer, several studies reporting an association between 1H MRS and DCE MR imaging in prostate cancer have been published, including Liney et al. (35), van Dorsten et al. (36), and Noworolski et al. (37). The characteristics of normal tissue and cancer in the periphery zone and central gland were different, and the results could not be compared to breast cancer.
The single-voxel 1H MRS technique has limitations in terms of lesion coverage, and the results may be affected by tumor heterogeneity. Any adipose tissue included in the MRS voxel makes B0 shimming more difficult due to the susceptibility problem. A non-homogeneous static field may affect the performance of chemically selective fat suppression and water suppression, in localized MRS. As was done in our study, a long echo time and fat suppression could be used to suppress the lipid sideband, nonetheless, improved water and lipid suppression techniques are greatly needed. Field homogeneity may be degraded because of metal clips left in the lesions after biopsy, which have yet to be studied in detail. Maril et al. (38) suggested that using both phase maps and multi-voxel MRS can provide an effective means to correct inhomogeneities in the breast. Bolan et al. (39) reported a TE-averaging method, which causes coherent cancellation of sideband artifacts by averaging spectra acquired at several different TEs values. These methods maybe helpful for improving Cho detection accuracy.
Another technical limitation is the pharmacokinetic model fitting with an acceptable arterial input function. The clinical protocol requires coverage of the whole breast with a relatively high spatial resolution and a typical temporal resolution of 1–2 minutes. In our study, we sacrificed spatial resolution to allow for a temporal resolution of 42 sec. While this was sufficient to measure a smooth enhancement kinetic curve (Fig. 2d and Fig. 3d), it was not sufficient to measure the AIF for each individual subject. We used the Generalized Kinetic Model (11) with a fixed biexponential decay for AIF determined by Tofts et al. based on a healthy population (22). This was the most common approach, as described in detail by Padhani et al. (23). However, Buckley et al. (40) suggested that this approach leads to a systematic overestimation of the transfer constant in tumors. Reliable methods for measuring AIF using a higher temporal resolution may allow an accurate estimation of these model-based kinetic parameters as reported recently by Parker et al. (41). It should be noted, however, that Parker et al. sacrificed the spatial resolution of DCE MR imaging in pursuit of a high temporal resolution (4.97 s) while employing other techniques such as elliptical k-space sampling to improve temporal resolution. The trade-off between spatial and temporal resolutions makes individual measurement of AIF clinically impractical. Therefore, the use of population AIF is a convenient and reasonable approach for DCE analysis.
Our approach of directly fitting the fractional enhancement time course to the model assumes a linear relationship between the signal enhancement and concentration of contrast agent, which is only true at low concentration levels and/or when exchange effects are not important. Workie et al. (42, 43) validated the linear relationship for 3D-GRE images under condition of low Gd-DTPA concentrations (11). Our fitted transfer constant is also equal to apparent Ktrans as defined in their work. As noted in Figure 2d and Figure 3d, although the fitted curve in general followed the experimental data points, there were minor undershoot and overshoot problems. As described in Yankeelov et al. (44) and Li et al. (45), the fitting quality may be improved using the “shutter-speed model”, which considered water exchange effects by introducing a parameter, the intracellular water molecule lifetime (τi).
5. Conclusion
Our study demonstrates the combined use of quantitative 1H MRS and DCE MR imaging in characterizing malignant breast lesions in patients. The measured Cho levels showed a wide variation (range, 0.32 – 10.47 mmol/kg), but consistent with previous in vivo results. The corresponding contrast enhancement kinetics was measured for each 1H MRS voxel. In the 36 lesions investigated, the mass type lesions showed a higher Cho concentration compared to that of non-mass type lesions. A significant correlation was found between the Cho concentration and DCE MR imaging parameter kep. The result may be explained by the association between the high rate of cell replication and angiogenesis required to support tumor growth.
Acknowledgement
The authors wish to thank Mr. Byron Feig for the linguistic editing of this manuscript, and Mrs. Becky Semon, RT for her aid in MRI data acquisition.
This paper was supported in part by NIH/NCI Grant No. CA90437 and the California Breast Cancer Research Program No. 9WB-0020 and No. 12FB-0031.
REFERENCES
- 1.Kuhl CK, Schild HH. Dynamic image interpretation of MRI of the breast. J Magn Reson Imaging. 2000;12:965–974. doi: 10.1002/1522-2586(200012)12:6<965::aid-jmri23>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 3.Kuhl CK, Mielcareck P, Klaschik S, et al. Dynamic breast MR imaging: are signal intensity time course data useful for differential diagnosis of enhancing lesions? Radiology. 1999;211:101–110. doi: 10.1148/radiology.211.1.r99ap38101. [DOI] [PubMed] [Google Scholar]
- 4.Roebuck JR, Cecil KM, Schnall MD, Lenkinski RE. Human breast lesions: characterization with proton MR spectroscopy. Radiology. 1998;209:269–275. doi: 10.1148/radiology.209.1.9769842. [DOI] [PubMed] [Google Scholar]
- 5.Kvistad KA, Bakken IJ, Gribbestad IS, Ehrnholm B, Lundgren S, Fjosne HE, haraldseth O. Characterization of neoplastic and normal human breast tissues with in vivo1H MR spectroscopy. J Magn Reson Imaging. 1999;10:159–164. doi: 10.1002/(sici)1522-2586(199908)10:2<159::aid-jmri8>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Jagannathan NR, Singh M, Govindaraju V, Raghunathan P, Coshic O, Julka PK, Rath GK. Volume localized in vivo proton MR spectroscopy of breast carcinoma: variation of water-fat ratio in patients receiving chemotherapy. NMR Biomed. 1998;11:414–422. doi: 10.1002/(sici)1099-1492(199812)11:8<414::aid-nbm537>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 7.Jagannathan NR, Kumar M, Seenu V, Coshic O, Dwivedi SN, Julka PK, Srivastava A, Rath GK. Evaluation of total choline from in-vivo volume localized proton MR spectroscopy and its response to neoadjuvant chemotherapy in locally advanced breast cancer. Br J Cancer. 2001;84:1016–1022. doi: 10.1054/bjoc.2000.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang W, Fisher PR, Dulaimy K, Tudorica LA, O’Hea B, Button TM. Detection of breast malignancy: diagnostic MR protocol for improved specificity. Radiology. 2004;232:585–591. doi: 10.1148/radiol.2322030547. [DOI] [PubMed] [Google Scholar]
- 9.Meisamy S, Bolan PJ, Baker EH, et al. Adding in vivo quantitative 1H MR spectroscopy to improve diagnostic accuracy of breast MR imaging: preliminary results of observer performance study at 4.0T. Radiology. 2005;236:465–475. doi: 10.1148/radiol.2362040836. [DOI] [PubMed] [Google Scholar]
- 10.Su MY, Baik HM, Yu Hon, Chen JH, Mehta R, Nalcioglu O. Comparison of choline and pharmacokinetic parameters in breast cancer measured by MR spectroscopic imaging and dynamic-contrast enhanced MRI. Technol Cancer Res Treat. 2006;5:401–410. [PubMed] [Google Scholar]
- 11.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging. 1997;7:91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 12.American College of Radiology. ACR Breast Imaging Reporting and Data System, Breast Imaging Atlas. Reston, Va: American College of Radiology; 2003. [Google Scholar]
- 13.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann. N.Y. Acad Sci. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 14.Ordidge RJ, Bendall MR, Gordon RE, Connelly A. Volume selection for in vivo spectroscopy. In: Govil G, Khetrapal CL, Sarans A, editors. Magnetic resonance in biology and medicine. New Delhi, India: Tata-McGraw-Hill; 1985. pp. 387–397. [Google Scholar]
- 15.Hasse A, Frahm J, Hanicke W, Mattaei 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol. 1985;30:431. doi: 10.1088/0031-9155/30/4/008. [DOI] [PubMed] [Google Scholar]
- 16.Doddrell DM, Galloway G, Brooks W, Filed J, Bulsing J, Irving M, Baddeley H. Water signal elimination in vivo, using suppression by mistimed echo and repetitive gradient episodes. J Magn Reson. 1986;70:176–180. [Google Scholar]
- 17.Naressim A, Couturier C, Devos JM, Janssen M, Mangeat C, de Beer R, Graveron-Demilly D. Java-based graphical user interface for the MRUI quantitation package. MAGMA. 2001;12:141–152. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]
- 18.Vanhamme L, van den Boogaart A, Huffel SV. Improved method for accurate and efficient quantification of MRS data with use of prior knowledge. J Magn Reson. 1997;129:35–43. doi: 10.1006/jmre.1997.1244. [DOI] [PubMed] [Google Scholar]
- 19.Knijn A, de Beer R, van Ormondt D. Frequency-selective quantification in the time domain. J Magn Reson. 1992;97:444–450. [Google Scholar]
- 20.van den Bos A. In: Handbook of Measurement Science. Sydenham PH, editor. Vol. 1. Wiley, Chichester; 1982. p. 33. [Google Scholar]
- 21.Baik HM, Su MY, Yu Hon, Mehta R, Nalcioglu O. Quantification of choline-containing compounds in malignant breast tumors by 1H MR spectroscopy using water as an internal reference at 1.5T. Magn. Reson. Mater Phy. 2006;19:96–104. doi: 10.1007/s10334-006-0032-4. [DOI] [PubMed] [Google Scholar]
- 22.Tofts PS, Kermode AG. Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magn Reson Med. 1991;17:357–367. doi: 10.1002/mrm.1910170208. [DOI] [PubMed] [Google Scholar]
- 23.Padhani AR, Hayes C, Assersohn L, Powles T, Makris A, Suckling J, Leach MO, Husband JE. Prediction of clinicopathologic response of breast cancer to primary chemotherapy at contrast-enhanced MR imaging: initial clinical results. Radiology. 2006;239:361–374. doi: 10.1148/radiol.2392021099. [DOI] [PubMed] [Google Scholar]
- 24.Bolan PJ, Meisamy S, Baker EH, Lin J, Emory T, Nelson M, Everson LI, Yee D, Garwood M. In vivo quantification of choline compounds in the breast with 1H MR Spectroscopy. Magn Reson Med. 2003;50:1134–1143. doi: 10.1002/mrm.10654. [DOI] [PubMed] [Google Scholar]
- 25.Su MY, Yu HJ, Carpenter PM, McLaren CE, Nalcioglu O. Pharmacokinetic parameters analyzed from MR contrast enhancement kinetics of multiple malignant and benign breast lesions detected in the same patients. Technol Cancer Res Treat. 2005;4:255–263. doi: 10.1177/153303460500400305. [DOI] [PubMed] [Google Scholar]
- 26.Yeung DK, Cheung HS, Tse GM. Human breast lesions: characterization with contrast-enhanced in vivo proton MR spectroscopy-initial results. Radiology. 2001;220:40–60. doi: 10.1148/radiology.220.1.r01jl0240. [DOI] [PubMed] [Google Scholar]
- 27.Tse GM, Cheung HS, Pang LM, et al. Characterization of lesions of the breast with proton MR spectroscopy: comparison of carcinomas, benign lesions, and phyllodes tumors. AJR Am J Roentgenol. 2003;181:1267–1272. doi: 10.2214/ajr.181.5.1811267. [DOI] [PubMed] [Google Scholar]
- 28.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipids metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 29.Sitter B, Sonnewald U, Spraul M, Fjosne HE, Gribbestad IS. High-resolution magic angle spinning MRS of breast cancer tissue. NMR Biomed. 2002;15:327–337. doi: 10.1002/nbm.775. [DOI] [PubMed] [Google Scholar]
- 30.Bakken IJ, Gribbestad IS, Singstad TE, Kvistad KA. External standard method for the in vivo quantification of choline-containing compounds in breast tumors by proton MR spectroscopy at 1.5 Tesla. Magn Reson Med. 2001;46:189–192. doi: 10.1002/mrm.1175. [DOI] [PubMed] [Google Scholar]
- 31.Gribbestad IS, Fjosne HE, Haugen OA, Nilsen G, Krane J, Petersen SB, Kvinnsland S. In vitro proton NMR spectroscopy of extracts from breast carcinoma and non-involved breast tissue. Anticancer Res. 1993;13:1973–1980. [PubMed] [Google Scholar]
- 32.Ting Y-LT, Sherr D, Degani H. Variations in energy and phopholipid metabolism in normal and cancer human mammary epithelial cells. Anticancer Res. 1996;16:1381–1388. [PubMed] [Google Scholar]
- 33.Singer S, Souza K, Thilly WG. Pyruvate utilization, phosphocholine and adenosine triphosphate (ATP) are makers of human breast tumor progression: A 31P and 13C Nuclear Magnetic Resonance (NMR) spectroscopy study. Cancer Res. 1995;55:5140–5145. [PubMed] [Google Scholar]
- 34.Jacobs MA, Barker DPhil PB, Atgani P, Ouwerkerk R, Bhujwalla ZM, Bluemke DA. Combined dynamic contrast enhanced breast MR and proton spectroscopic imaging: a feasibility study. J Magn Reson Imaging. 2005;21:23–28. doi: 10.1002/jmri.20239. [DOI] [PubMed] [Google Scholar]
- 35.Liney GP, Turnbull LW, Knowles AJ. In vivo magnetic resonance spectroscopy and dynamic contrast enhancing imaging of the prostate gland. NMR Biomed. 1999;12:39–44. doi: 10.1002/(sici)1099-1492(199902)12:1<39::aid-nbm543>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 36.van Dorsten FA, van der Graaf M, Engelbrecht MR, van Leenders GJ, Verhofstad A, Rijpkema M, de la Rosette JJ, Barentsz JO, Heerschap A. Combined quantitative dynamic contrast-enhanced MR imaging and 1H MR spectroscopy imaging of human prostate cancer. J Magn Reson Imaging. 2004;20:279–287. doi: 10.1002/jmri.20113. [DOI] [PubMed] [Google Scholar]
- 37.Noworolski SM, Henry RG, Vigneron DB, Kurhanewicz J. Dynamic contrast-enhanced MRI in normal and abnormal prostate tissues as defined by biopsy, MRI, and 3D MRSI. Magn Reson Med. 2005;53:249–255. doi: 10.1002/mrm.20374. [DOI] [PubMed] [Google Scholar]
- 38.Maril N, Collins CM, Greenman RL, Lenkinski RE. Strategies for shimming the breast. Magn Reson Med. 2005;54:1139–1145. doi: 10.1002/mrm.20679. [DOI] [PubMed] [Google Scholar]
- 39.Bolan PJ, DelaBarre L, Baker EH, Merkle H, Everson LI, Yee D, Garwood M. Eliminating spurious lipid sidebands in 1H MRS of breast lesions. Magn Reson Med. 2002;48:215–222. doi: 10.1002/mrm.10224. [DOI] [PubMed] [Google Scholar]
- 40.Buckley DL. Uncertainty in the analysis of tracer kinetics using dynamic contrast-enhanced T1-weighted MRI. Magn Reson Med. 2002;47:601–606. doi: 10.1002/mrm.10080. [DOI] [PubMed] [Google Scholar]
- 41.Parker GJM, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, Jackson A, Watson Y, Davies K, Jayson GC. Experimentally-derived functional form for population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med. 2006;56:993–1000. doi: 10.1002/mrm.21066. [DOI] [PubMed] [Google Scholar]
- 42.Workie DW, Dardzinski BJ, Graham TB, Laor T, Bommer WA, O’Brien KJ. Quantification of dynamic contrast-enhanced MR imaging of the knee in children with juvenile rheumatoid arthritis based on pharmacokinetic modeling. Magn Reson Imaging. 2004;22:1201–1210. doi: 10.1016/j.mri.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Workie DW, Dardzinski BJ. Quantifying dynamic contrast-enhanced MRI of the knee in children with juvenile rheumatoid arthritis using an arterial input function (AIF) extracted from popliteal artery enhancement, and the effect of the choice of the AIF on the kinetic parameters. Magn Reson Med. 2005;54:560–568. doi: 10.1002/mrm.20597. [DOI] [PubMed] [Google Scholar]
- 44.Yankeelov TE, Rooney WD, Huang W, Dyke JP, Li X, Tudorica A, Lee JH, Koutcher JA, Springer CS., Jr Evidence for shutter-speed variation in CR bolus-tracking studies of human pathology. NMR Biomed. 2005;18:173–185. doi: 10.1002/nbm.938. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Huang W, Yankeelov TE, Tudorica A, Rooney WD, Springer CS. Shutter-speed analysis of contrast reagent bolus-tracking data: preliminary observations in benign and malignant breast disease. Magn Reson Med. 2005;53:724–729. doi: 10.1002/mrm.20405. [DOI] [PubMed] [Google Scholar]