Abstract
The thyroid stimulating hormone receptor (TSHR) belongs to the glycoprotein hormone receptor subfamily of seven-transmembrane spanning receptors. TSHR is expressed in thyroid follicular cells and is activated by TSH, which regulates growth and function of these cells. Recombinant TSH is used in diagnostic screens for thyroid cancer, especially in patients after thyroid cancer surgery. Currently, no selective small molecule agonist of the TSHR is available. To screen for novel TSHR agonists, we miniaturized a cell-based cAMP assay into 1536-well plate format. This assay uses a HEK293 cell line stably expressing the TSHR and a cyclic nucleotide gated ion channel (CNG), which functions as a biosensor. From a quantitative high-throughput screen of 73,180 compounds in parallel with a parental cell line (without the TSHR), 276 primary active compounds were identified. The activities of the selected active compounds were further confirmed in an orthogonal HTRF cAMP-based assay. 49 compounds in several structural classes have been confirmed as small molecule TSHR agonists that will serve as starting compounds for chemical optimization and studies of thyroid physiology in health and disease.
Keywords: Thyroid-stimulating hormone TSH, TSHR, TSHR agonist, quantitative high throughput screening, qHTS, HTS, probe identification, CNG, PubChem
INTRODUCTION
TSH is an α/β heterodimeric glycoprotein hormone secreted from the anterior pituitary gland, which belongs to the glycoprotein hormone family including Chorionic Gonadotropin (CG), Luteinizing Hormone (LH), and Follicle Stimulating Hormone (FSH)1. The actions of TSH are mediated by a seven-transmembrane receptor, which upon TSH binding, couples preferentially to the G-alpha (s) protein (Gs) resulting in activation of adenylate cyclase and increase in cyclic adenosine 3’, 5’ monophosphate (cAMP) 1. TSHR is mainly expressed in thyroid follicular cells 2 and regulates their growth and function 3. Activation of the TSHR is used clinically in patients with thyroid cancer receiving thyroid hormone suppression therapy to screen for residual tumor after surgery 4. Since no selective small molecule agonist of the TSHR currently exists, recombinant TSH is used for this purpose, though recombinant TSH is expensive and must be administered intramuscularly. A small molecule TSHR agonist would potentially be more cost effective, especially if it is orally active, and would serve as an invaluable research tool for studying TSHR pharmacology and physiology.
Several assays have been used to measure the interaction between TSH and TSHR, but none have been ideal for identifying TSHR agonists via HTS. The traditional radiolabeled TSH receptor binding assay 5, 6 is sensitive but does not measure direct activation of the receptor. Measurement of the amount of intracellular cAMP generated upon the activation of the TSHR more directly reflects activity at the receptor; however, cAMP radioimmunoassays 7 and ELISAs 8 require multiple assay steps and washes, making them nontransferable to HTS. Indirect TSHR activation readouts, such as by the use of cAMP response element (CRE) reporter-gene assays or downstream gene expression measurements 9 are more amenable to HTS, but since they report activity at any point along the extended TSHR signaling pathway including gene transcription and protein synthesis, they may produce large numbers of actives which require time-consuming deconvolution to identify TSHR agonists.
Recently, several more HTS-friendly cAMP assays have been developed and used to screen Gs coupled GPCRs, including fluorescence polarization (FP) 10, homogeneous time-resolved fluorescence (HTRF) 11, enzyme fragmentation complementation (EFC) 12 and cyclic nucleotide gated ion channel (CNG)-coupled 13 approaches. The signal of cAMP change is amplified in the CNG coupled cAMP assay which can be monitored by membrane potential dye in live cells. Here, we describe the optimization, miniaturization, and screening of a CNG-coupled cAMP assay for the TSH receptor in 1536-well plate format. A quantitative high throughput screen (qHTS) of the optimized assay across a 73,180 member compound library identified a large number of active compounds, which were rapidly triaged using a series of experimental and computational counterscreens, taking advantage of the rich information content of qHTS-derived data. Several structural classes of small molecule TSHR agonists were identified.
MATERIALS AND METHODS
Materials
TSH, RO 20–1724, and forskolin were purchased from Sigma-Aldrich (St. Louis, MO). The membrane potential dye kit was purchased from BD Biosciences (Rockville, MD). The TSHR transfected HEK-293 cells coexpressed with a modified CNG channels and its parental cells (without the receptor) were obtained from BD Biosciences (named as ACT:One cells). The cell culture medium (DMEM) was purchased from Invitrogen (Carlsbad, CA) and fetal calf serum was from HyClone (Logan, UT).
Cell Culture and frozen cell preparation
Both TSHR transfected and parental HEK 293 cells (expressing a modified CNG channel) were maintained in DMEM medium containing 10% FBS, 100 units/ml Penicillin, 100 µg/ml Streptomycin, and 250 µg/ml Geneticin (Invitrogen) at 37°C in 5% CO2. For the TSHR expression cell line, additional 1 µg/ml Puromycin (Invitrogen) was added during the cell culture. The cells were seeded at a density of 3 to 4 million cells in a T175 flask (Corning, Corning, NY) containing 35 ml of media and were allowed to grow for 3 to 4 days to reach 80 – 90% confluence. A flask of HEK 293 cells at this density generally yielded 30 million cells total.
CNG channel coupled cAMP assay
Cells from four confluent T175 flasks were seeded into 25 to 30 T175 flasks at a density of 3 to 4 million cells per flask. After 3 days of growth, the flasks were harvested and the fresh cells were used for dispensing into 1536 well plates. This process was repeated on a daily basis for screening purposes. The cells were resuspended in 2% FBS DMEM medium at 500,000 cells/ml, and 4 µl of resuspended cells was dispensed into each well of black, clear bottom 1536-well tissue culture treated plates (Greiner Bio-One, Kremsmuenster, Austria) using a Multidrop Combi dispenser (Thermo Fisher Scientific, Waltham, MA). After overnight culture at 37°C with 5% CO2, the cells were generally 60 – 70% confluent in the 1536-well plates.
After overnight incubation, 4 µl of membrane potential dye containing 50 µM of the phosphodiesterase inhibitor RO 20–1724 was added to each well and the plates were incubated for 60 minutes at room temperature. A baseline measurement was then taken using the Envision Plate reader (PerkinElmer, Boston, MA) with excitation at 535/20 nm and emission at 590/20 nm with a gain of 150 and 5 flashes per well (PerkinElmer, Boston, MA). 20 nl of compound or positive control (30 nM TSH or 10 uM forskolin for TSHR or parental respectively) in DMSO was added to each well using a pintool (Kalypsys, San Diego, CA), and the plates were further incubated for 30 minutes at the room temperature. Plates were then measured again in the Envision plate reader with the same filters and settings as above.
Homogeneous Time Resolved Fluorescence (HTRF) cAMP Assay
All follow up compounds were assayed using a HTRF cAMP detection kit (Cisbio, Bedford, MA) on both TSHR cell line and parental cell line. Briefly, 750 cells were plated in 2.5 µl/well of complete media (DMEM containing 10 % FCS) in 1536 well solid bottom white plates and 20 nl/well compound in DMSO solution or controls was added. Following 30 minute incubation at room temperature, 2.5 µl/well of labeled d2 cAMP and 2.5 µl/well of anti-cAMP antibody (both diluted 1:20 in lysis buffer) were added to each well using a flying reagent dispenser (Aurora Discovery, San Diego). Plates were measured using the Envision plate reader (PerkinElmer, Boston, MA) with excitation at 330 nm and emissions of 615 nm and 660 nm.
Compound library preparation and quantitative high throughput screening
A library of 73,180 structurally diverse compounds was serially diluted 1:5 or 1: 2.236 in DMSO in 384-well plates to yield seven or fifteen concentrations (minimally 10 mM, 2 mM, 0.4 mM, 80 µM, 16 µM, 3.2 µM and 0.64 µM) and formatted into 1,536-well plates at 7 µl/well. A qHTS was performed as described previously 14 using a fully automated robotic screening system (Kalypsys, San Diego, CA). Final compound concentrations during cell incubation ranged from 1.60 nM to 25.0 µM.
Data analysis
Primary screen data were analyzed with customized software developed internally. The maximal response (100% activity) was determined by the response of 30 nM TSH and the basal response (0% activity) was measured by the DMSO control in the TSH screen. In the counter-screen with the parental cell line, 10 µM forskolin was added for the maximal response. The concentration-responses of all the compounds were analyzed using methods described in and structural clustering of active compounds was 14 performed using Leadscope Hosted Client (Leadscope Inc., Columbus, OH). The EC50 values of compounds in the confirmation and follow-up experiments were calculated from the concentration-response curves by nonlinear regression analysis using Prism software (GraphPad Software, San Diego, CA).
RESULTS AND DISCUSSION
Assay optimization and miniaturization
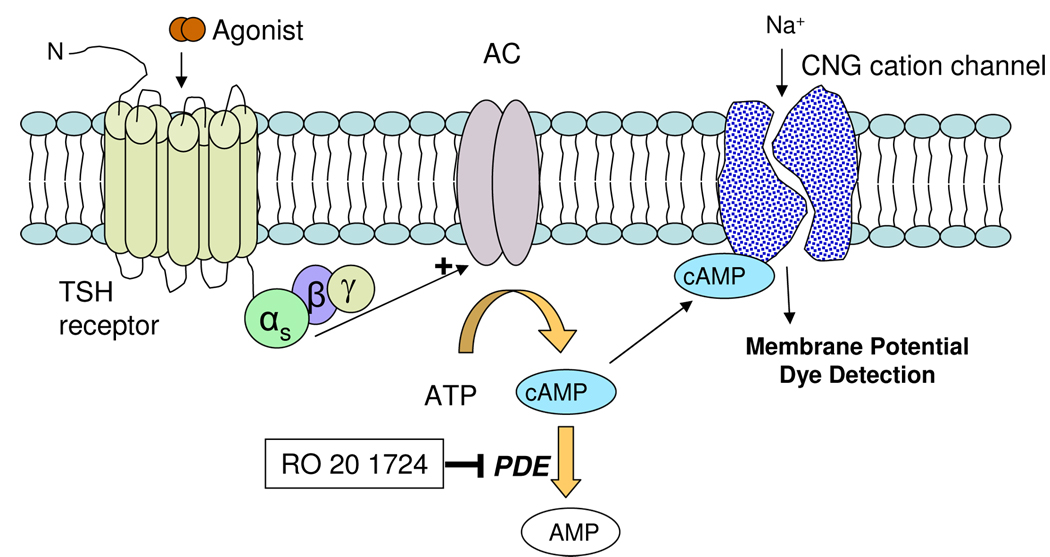
The assay used here allowed us to measure elevation in intracellular cAMP produced by TSH receptor activation in live cells. This HEK293 cell line stably expresses both the TSH receptor and a cyclic nucleotide-gated (CNG) cation channel 13, 15 (Fig. 1). Activation of the TSH receptor by an agonist results in Gs-adenylate cyclase coupling and an increase in intracellular cAMP, which binds to and activates the CNG channel. CNG channel activation produces an influx of Ca2+ and Na+ cations and subsequent membrane depolarization, which can be measured using a calcium-sensitive dye in a kinetic fluorescence plate reader, or even more conveniently, using a membrane potential dye 16 in a standard fluorescence plate reader.
Fig. 1. Schematic diagram of the detection mechanism of ACT:One cAMP assay (BD Biosciences, Rockville, MD).
The TSH receptor was cotransfected in the HEK 293 cells with a modified CNG channel, which serves as a biosensor to detect changes in the intracellular cAMP level. TSH stimulates the TSH receptor, which increases the intracellular cAMP level resulting in the activation of CNG cation channels. The membrane depolarization occurs after the influx of cations such as sodium and calcium, which can be detected by the membrane potential dye in the live cells.
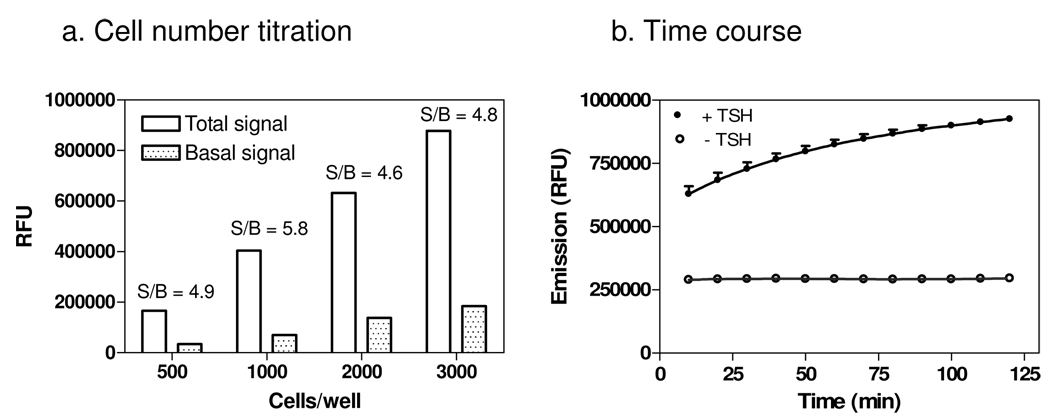
The original assay was developed in 96-well plate format. In order to adapt it to 1536-well plate format, the number of cells per well was first examined in the presence of TSH stimulation. The signal-to-basal ratio was similar with cell densities from 500 to 3000 cells/well; 1000 cells/well was selected since it had the highest signal-to-basal ratio (Fig. 2a). Time-course experiments showed that the signal-to-basal ratio was 2.67- and 2.92-fold at 30 and 60 min after addition of the agonist, respectively (Fig. 2b). Thus, an incubation time of 30 minutes after agonist addition was chosen for the assay in considerations of convenience for HTS and sensitivity of the compound screen. DMSO was also tested and it was found to have no effect on the assay at concentrations under (below?) 1%; the final DMSO concentration in the assay wells in the screen was 0.25%.
Fig. 2. The effects of cell density and agonist incubation time on the cAMP assay.
(a) Cell number titration in the presence and absence of 30 nM TSH. Cells were plated at 500, 1000, 2000 and 3000 cells/well in a 1536 well plate 24 hours before stimulation with 30 nM TSH for 60 minutes. (B) Time course of the agonist incubation time. Cells were seeded at 1000 cells/well in 1536 well plates and stimulated 24 hours later with 30 nM TSH and compared to cells without TSH stimulation. The fluorescence signals of the membrane potential dye were measured every 10 minutes from 10 to 120 minutes after the agonist addition.
With the assay conditions defined as 1000 cells/well and 30 minutes incubation after agonist addition, concentration-response to the native agonist TSH was tested in 1536-well plate format. The EC50 value of TSH was 1.40 nM in 1536-well format compared to 3.89 nM EC50 measured in 96-well format (Fig. 3), showing that the potency of TSH was similar in both assay formats.
Fig. 3. Concentration response of TSH determined from the cAMP assay.
In the 96-well plate assay, 60,000 cells/well were plated and incubated for 24 hours before the measurement. In the 1536-well plate assay, 1000 cells/well were plated in a 1536-well plate and incubated for 24 hours before the cAMP assay. The EC50 value of TSH were 3.89 and 1.90 nM determined from the 96-well and 1536-well plates respectively.
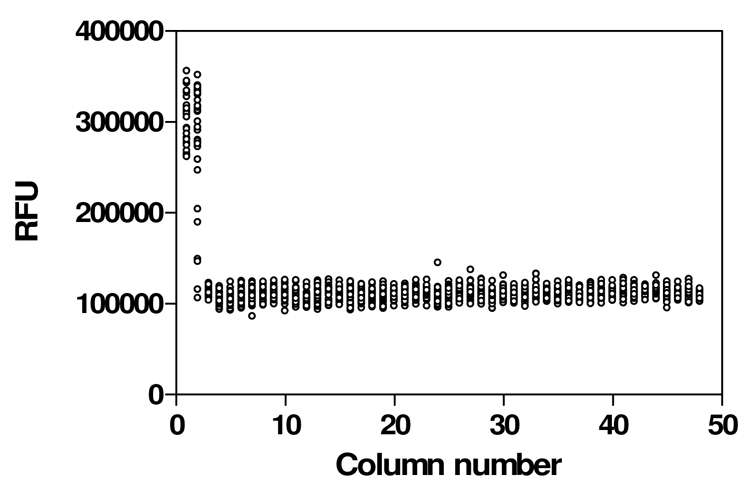
Assay parameters were then determined in the miniaturized 1536-well assay format using a DMSO plate. The signal-to-basal (S/B) ratio was 2.5 fold and the CV (coefficient of variation) and Z’ factor were 6.3% and 0.83, respectively (Fig. 4), measured in a fluorescence plate reader. These results indicated that the assay’s performance was adequate for qHTS, and that a kinetic reader was not needed for this cAMP assay.
Fig. 4. Scatter plot of the results from a 1536-well DMSO plate to establish assay performance parameters.
Cells were seeded at a density of 1000 cells/well. Columns 1–4 were controls: 30 nM TSH was added to the wells in column 1 to establish maximum signal; wells in column 2 received a TSH titration ranging from 0.1 nM to 1 µM in duplicate; columns 3 and 4 received DMSO as the negative control; DMSO was added to columns 5–48. Resulting signal-to-basal (S/B) ratio was 2.5, CV was 6.3 % and Z’ factor was 0.83.
Quantitative high throughput screen
73,180 compounds were screened using a fully automated screening system in the qHTS format 14. All the compounds were assayed at 7 or 15 concentrations, ranging from 0.32 nM to 25.0 µM. Response to the control agonists (TSH or forskolin) was measured in each plate and was stable across the entire assay (data not shown).
Freshly prepared cells were dispensed into 1536-well assay plates at 4 µl/well at a rate of 100 plates per hour and incubated at 37°C with 5% CO2 for 20 to 36 hrs before the robotic screening. The screen was initiated by addition of 4 µl/well membrane potential dye, followed by 60 min incubation at room temperature to allow for dye equilibration across the cell membrane. 20 nl/well compound or control was then added to the assay plates, which were incubated for additional 30 min before reading a fluorescence plate reader in bottom-read mode for fluorescence intensity (emission at 590 nm). Both TSHR and parental (lacking TSHR) lines were screened across 463 compound plates (total 926 plates). The average signal-to-basal ratio for the entire screen was 2.01, CV was 12.1%, and Z’ was 0.4 (Fig. 5). The relatively low Z’ was likely due to the use of HEK293 cells, which typically do not adhere well after treatment with membrane potential dyes 17; however, the Z’ was acceptable for HTS 18, particularly when performed in titration-response qHTS mode, which can produce reliable data in the presence of relatively low Z’values 14.
Fig. 5. HTS performance of 200 plates.
(a). Signal-to-basal (S/B) ratio. The total signals were obtained from the wells in the columns 1 and 2 (with 30 nM TSH) and the basal signals were collected from the wells in the column 4 which received DMSO. (b) Coefficient of variation (% CV), calculated as the standard deviation (s.d.) from the wells in the column 4 divided by the s.d. from the wells in the columns 1 and 2. (c) Z’ factor, calculated by Z’ = 1- (3*s.d. of total signal + 3*s.d. of basal signal)/(total signal – basal signal) 18.
qHTS data analysis
Since the screen was run in qHTS mode, concentration-response curves, potencies, efficacies, and structure-activity relationship data on all compounds were available immediately after the primary screen. Active compound series and singletons were initially denoted on the basis of high-confidence curve class (1.1, 1.2, and 2.1; details discussed in 14) and compound potency (< 10 µM). A total of 1012 compounds (1.4% of screened compounds) were designated as “provisional actives” from the primary screen.
Elimination of assay false positives
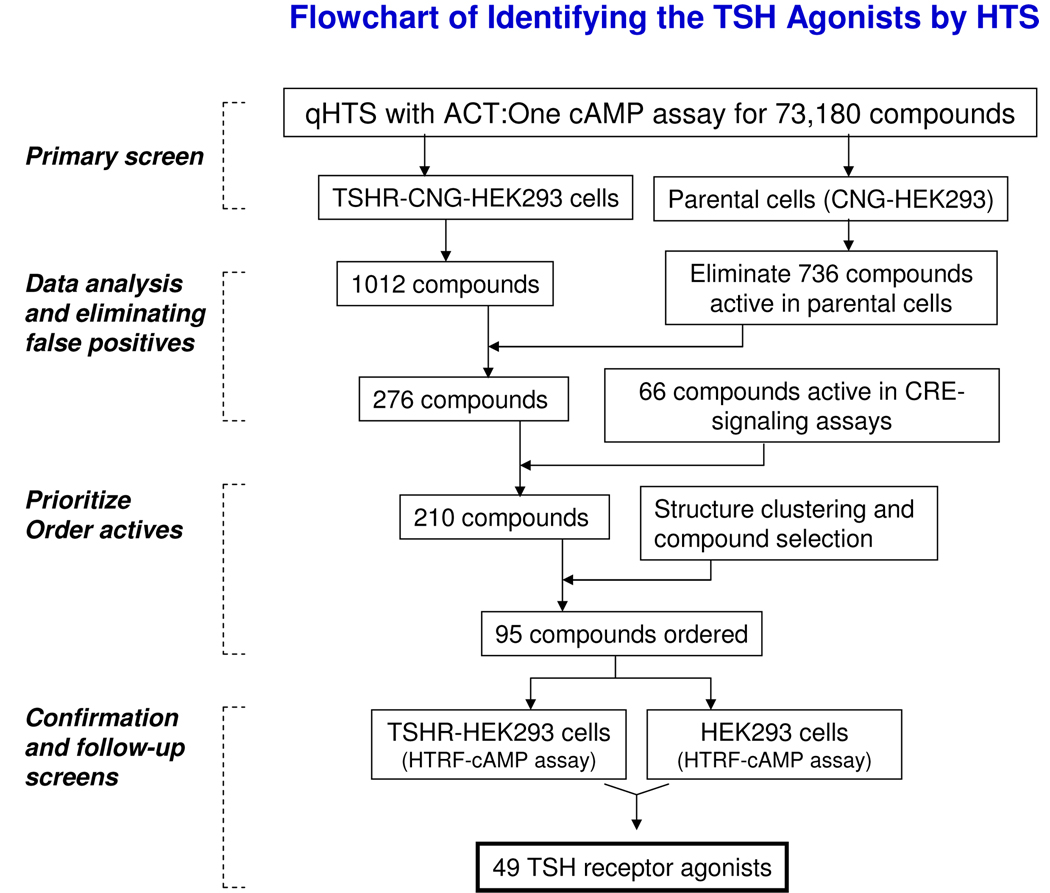
Large numbers of false positives are commonly found in cell-based phenotypic and cell signaling assays, and considerable time and effort are frequently required to identify the compounds acting via the desired mechanism (in this case, TSHR agonism). Though qHTS data is relatively refractory to many sources of false positives, we wished to eliminate other assay-related false positives before moving to the next stages of follow-up. These would include compounds that directly activated the CNG channel or cAMP cascade (e.g., adenylate cyclase, G-proteins, or other endogenous GPCRs expressed by HEK293 cells), fluorescent compounds, or noncompound assay fluorescence due dust or lint. We were able to rapidly eliminate compounds with undesired mechanisms using a progressive series of experimental and computational tests (Fig. 7).
Fig. 7. Flowchart for rapid triaging of primary hits to identify the TSH agonists.
The process of TSHR agonist discovery included the qHTS, false positive elimination, active compound selection and confirmation. For details, see text.
We first examined the activity of the 1012 compounds identified in the TSHR qHTS in the parental cell line which expresses the CNG channel without TSH receptor; 736 compounds were found to have activity that was not dependent on TSHR (i.e., similar activity was seen in both cell lines), and were eliminated. A further 66 further compounds showed activity in a Cyclic AMP Response Element (CRE)-signaling pathway assay previously screened in our laboratory (Xia et al., manuscript in preparation, and PubChem Assay ID 662), indicating direct activity on the cAMP cascade, and were also eliminated. This left 210 active compounds (0.29% of the screened compounds) which comprised 19 structural classes and 144 singletons. Of these, 95 compounds representing each of the scaffolds and the most potent, efficacious singletons were selected for confirmation and further studies. The primary screening results have been deposited into PubChem (AID pending, http://pubchem.ncbi.nlm.nih.gov/).
TSH agonist confirmation
The 95 selected compounds were tested for TSHR agonism in an alternate-readout HTRF cAMP assay, which measures cAMP concentrations in cell lysates using a cAMP antibody and the TR-FRET detection technology. The readout in the HTRF cAMP assay is distinct from that in the membrane potential dye measurement assay used in the qHTS, allowing us to further eliminate assay-related false positive compounds. Of the 95 compounds, 49 showed TSHR agonist activity in the HTRF cAMP assay. These 49 compounds were then tested in the parental cell line (without TSHR) in the same HTRF cAMP assay, and showed no activity (data not shown). Taken together, these results indicate that these 49 compounds are true small molecule TSHR agonists. Fig. 6 shows four representative compounds of these confirmed small molecule TSH agonists. Studies further characterizing these compounds in physiological and disease-relevant systems are currently in progress.
Fig. 6. Dose response curves and structures of four representative TSH receptor agonists.
Activities of compounds were normalized to the maximal response of TSH (30 nM) and the DMSO control in the TSH receptor cell line (closed circles) and the parental cell line (open triangles). All four compounds were inactive in the parental cell line (without the TSH receptor). (a) NCGC00057417 with an EC50 of 2.15 µM and 69.8% maximal response at 10 µM concentration. (b) NCGC00038940 with an EC50 of 2.57 µM and 46.6 % maximal response at 10 µM concentration. (c) NCGC00054245 with an EC50 of 10.1 µM and 50.6 % maximal response at 10 µM concentration. (d) NCGC00026086 with an EC50 of 1.70 µM and 54.5 % maximal response at 10 µM concentration.
In conclusion, we have optimized a cAMP assay using a CNG cation channel as a biosensor in HEK293 cells expressing the TSHR and miniaturized it to a 1536-well plate format. This assay was used to screen a library of 73,180 compounds in qHTS format. A total of 210 compounds in 19 structural classes, representing an active rate of 0.29%, were identified as true actives after exclusion a large number of primary screen actives by experimental and computational counter-screening. TSHR agonist activity was confirmed in 49 of 95 tested compounds comprising several medicinally attractive structural classes in an orthogonal TSHR cAMP assay utilizing a different detection technology. These compounds, the first small molecule TSHR agonists identified by HTS, will serve as chemical probes for further studies of TSH receptor pharmacology, and as an important starting point in medicinal chemistry optimization to produce TSHR agonists for in vivo use.
Table 1.
A protocol for the TSH receptor cAMP assay
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Cells | 4 µL | 1000 TSHR or parental cells/well |
| 2 | Incubation | 18 – 30 hours | 37°C; 5% CO2 |
| 3 | Reagent | 4 µL | Membrane potential dye |
| 4 | Incubation | 30 min | at room temperature |
| 5 | Compound | 20 nL | Compound Libraries or control |
| 6 | Incubation | 30 min | at room temperature |
| 7 | Detector | Ex = 535 and | Envision plate reader |
| Em = 590 nm | |||
Acknowledgements
This research was supported by the Molecular Libraries Initiative of the NIH Roadmap for Medical Research and the Intramural Research Programs of the National Human Genome Research Institute and the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
References
- 1.Vassart G, Pardo L, Costagliola S. A molecular dissection of the glycoprotein hormone receptors. Trends in Biochemical Sciences. 2004;29(3):119–126. doi: 10.1016/j.tibs.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Latif R, Ando T, Daniel S, Davies TF. Localization and regulation of thyrotropin receptors within lipid rafts. Endocrinology. 2003;144(11):4725–4728. doi: 10.1210/en.2003-0932. [DOI] [PubMed] [Google Scholar]
- 3.Davies TF, Ando T, Lin RY, Tomer Y, Latif R. Thyrotropin receptor-associated diseases: from adenomata to Graves disease. J Clin Invest. 2005;115(8):1972–1983. doi: 10.1172/JCI26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandes JK, Day TA, Richardson MS, Sharma AK. Overview of the management of differentiated thyroid cancer. Current Treatment Options in Oncology. 2005;6(1):47–57. doi: 10.1007/s11864-005-0012-3. [DOI] [PubMed] [Google Scholar]
- 5.Chazenbalk GD, Nagayama Y, Kaufman KD, Rapoport B. The functional expression of recombinant human thyrotropin receptors in nonthyroidal eukaryotic cells provides evidence that homologous desensitization to thyrotropin stimulation requires a cell-specific factor. Endocrinology. 1990;127(3):1240–1244. doi: 10.1210/endo-127-3-1240. [DOI] [PubMed] [Google Scholar]
- 6.Tate RL, Schwartz HI, Holmes JM, Kohn LD. Thyrotropin receptors in thyroid plasma membranes. Characteristics of thyrotropin binding and solubilization of thyrotropin receptor activity by tryptic digestion. J Biol Chem. 1975;250(16):6509–6515. [PubMed] [Google Scholar]
- 7.Ludgate M, Gire V, Crisp M, et al. Contrasting effects of activating mutations of GalphaS and the thyrotropin receptor on proliferation and differentiation of thyroid follicular cells. Oncogene. 1999;18(34):4798–4807. doi: 10.1038/sj.onc.1202864. [DOI] [PubMed] [Google Scholar]
- 8.Demeure MJ, Doffek KM, Wilson SD. Defective thyrotropin receptor G-protein cyclic adenosine monophosphate signaling mechanism in the FTC human follicular thyroid cancer cell line. Surgery. 1997;122(6):1195–1201. doi: 10.1016/s0039-6060(97)90227-0. discussion 1201-1192. [DOI] [PubMed] [Google Scholar]
- 9.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35(4):605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 10.Prystay L, Gagne A, Kasila P, Yeh LA, Banks P. Homogeneous cell-based fluorescence polarization assay for the direct detection of cAMP. J Biomol Screen. 2001;6(2):75–82. doi: 10.1177/108705710100600203. [DOI] [PubMed] [Google Scholar]
- 11.Gabriel D, Vernier M, Pfeifer MJ, Dasen B, Tenaillon L, Bouhelal R. High throughput screening technologies for direct cyclic AMP measurement. Assay Drug Dev Technol. 2003;1(2):291–303. doi: 10.1089/15406580360545107. [DOI] [PubMed] [Google Scholar]
- 12.Weber M, Ferrer M, Zheng W, Inglese J, Strulovici B, Kunapuli P. A 1536-well cAMP assay for Gs- and Gi-coupled receptors using enzyme fragmentation complementation. Assay Drug Dev Technol. 2004;2(1):39–49. doi: 10.1089/154065804322966306. [DOI] [PubMed] [Google Scholar]
- 13.Reinscheid RK, Kim J, Zeng J, Civelli O. High-throughput real-time monitoring of Gs-coupled receptor activation in intact cells using cyclic nucleotide-gated channels. Eur J Pharmacol. 2003;478(1):27–34. doi: 10.1016/j.ejphar.2003.08.036. [DOI] [PubMed] [Google Scholar]
- 14.Inglese J, Auld DS, Jadhav A, et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103(31):11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao Y. LJ, Llorente I, DeBernardi M, Cao L. ACT:One technology: a live cAMP assay.. Poster presentation in the 10th Anniversary Conference of Society of Biomolecular Sciences; 2004. [Google Scholar]
- 16.Zheng W, Spencer RH, Kiss L. High throughput assay technologies for ion channel drug discovery. Assay Drug Dev Technol. 2004;2(5):543–552. doi: 10.1089/adt.2004.2.543. [DOI] [PubMed] [Google Scholar]
- 17.Thomas P, Smart TG. HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods. 2005;51(3):187–200. doi: 10.1016/j.vascn.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]