Abstract
Costello syndrome is a rare congenital anomaly syndrome associated with mental retardation and predisposition to benign and malignant tumors, caused by heterozygous missense mutations in the HRAS oncogene. Previously, all molecularly analyzed mutations appeared de novo, and most arose in the paternal germline. A single patient with somatic mosaicism for a Costello syndrome causing HRAS mutation has been reported. Here we describe the first documented transmission of an HRAS mutation from a parent with somatic mosaicism to a child with typical Costello syndrome. Prior to the identification of the underlying gene mutation in Costello syndrome, this family had been identified clinically. The proband was subsequently found to carry a G12S HRAS germline mutation. Testing of the parents for parental origin identified his father as mosaic for the same HRAS mutation. The mother was found not to carry an HRAS mutation. The causative familial mutation is identified as a c. 34G>A, which is the most common mutation in the HRAS gene in patients with Costello syndrome. The father carries the mutation in 7–8% of his alleles. This is the second case of mosaicism observed in Costello syndrome and the first direct molecular evidence of father to son transmission of the disease-causing mutation. Our observation underlines the importance of parental evaluation, and may have implications for genetic counseling and clinical practice.
Keywords: Ras oncogenic mutation, allelic specific amplification, MAPK dysregulation
INTRODUCTION
Costello syndrome [OMIM 218040] typically encompasses severe failure-to-thrive, cardiac abnormalities including tachyarrhythmia and hypertrophic cardiomyopathy, a predisposition to papillomata and malignant tumors, and neurologic abnormalities including nystagmus, hypotonia, developmental delay and mental retardation [Gripp and Lin, 2006]. A clinical diagnosis can now be confirmed through the identification of an HRAS mutation [Aoki et al., 2005] and the diagnosis should be reconsidered if no HRAS mutation is seen [Gripp et al., 2007; Kerr et al., 2008] More than 80% of patients with Costello syndrome share the same underlying HRAS mutation, resulting in a G12S amino acid change [Aoki et al., 2005; Estep et al., 2006; Gripp et al., 2006a; Kerr et al., 2006; Sol-Church et al., 2006 , van Steensel et al., 2006; Zampino et al., 2007]. The mutations were de novo in all analyzed cases, and in the vast majority occurred in the paternal germline [Sol-Church et al., 2006; Zampino et al., 2007]. Only one case of somatic mosaicism for an HRAS mutation in Costello syndrome has been confirmed [Gripp et al., 2006b]. We report here on a second individual with a mosaic HRAS mutation, resulting in transmission to a child with Costello syndrome.
SUBJECTS AND METHODS
Informed consent was obtained based on the protocol approved by the institutional review board at the A. I. duPont Hospital for Children (#2005-051).
Patient 1
The patient is a 21-year-old male in whom a clinical diagnosis of Costello syndrome was made in early childhood based on the typical medical history of severe feeding difficulties and failure to thrive requiring the use of a feeding tube until age 4 ½ years. He had developmental delay. On his most recent exam his height was 150 cm (50th centile for 12.5 years). He had deep palmar creases, joint laxity and nasal papillomata. His facial findings and his hyperkeratotic skin were typical for Costello syndrome.
Cardiac hypertrophy was detected at age 17 years. He has had hypertrophic cardiomyopathy for nearly 9 years, which has steadily increased and remains stable at moderate severity. He has no syncope or palpitations.
The most recent echocardiogram performed at age 20 years showed mild concentric left ventricular hypertrophy with additional focal thickening in the mid-septal region. The peak Doppler gradient averaged 100 mm hg (range 90–120 mm Hg, mean 48 mm). Treatment has included atenolol, and he is currently treated with a calcium channel blocker, verapamil (180 mg orally daily, 4.3 mg/kg/day). Echocardiograms have not shown congenital heart defects or valve dysplasia, nor did he have serious arrhythmia in infancy. Electrocardiograms have been obtained on a regular basis (at least annually) and have shown increased ventricular voltages, without significant repolarization abnormalities, and increased atrial complexes. Holter monitoring performed at ages 18 and 19 years showed rare ventricular couplets and triplets. The most recent one performed at age 21 years showed similar rare ventricular and atrial ectopy with mild nonsustained ventricular tachycardia. He had a treadmill exercise stress test at age 20 years which was successful up to stage 3, and limited due to his lack of cooperation.
Patient 2
The father of Patient 1 came to medical attention years ago when physical findings suggestive of mosaic Costello syndrome were noted by the physician (L.A.D) treating Patient 1 [Bodkin et al., 1999]. Patient 2 had a history of feeding problems and severe failure to thrive in early childhood. He has hyperkeratosis on both hands and his left foot, but not on the right foot. Papillomata were present on the right, but not the left perianal region. Nasal papillomata had been removed. Hyperpigmentation, thick ear lobes, and patches of curly hair in his otherwise straight hair were noted. His medical history includes kidney stones in his 20’s; and bilateral inguinal hernia repair in his 50’s. He also had coronary artery disease, which was treated with stenting, ballooning and pharmacotherapy. A cardiac echo performed in his 50s did not show structural anomalies, in particular no hypertrophy.
The patient had developmental delay, and was academically challenged in school, succeeding with family support. Prior to confirmation by DNA analysis, he expressed awareness that this son and he had similar physical and developmental Costello syndrome traits.
To respect the family's preference for privacy , no patient's pictures are included in this report.
Molecular Techniques
DNA from the patients, Patient 1’s mother and an unrelated normal control individual was extracted from buccal cells using the PureGene DNA Isolation Kit (Gentra Systems, Minneapolis, MN, USA). A second sampling of the buccal swab was collected on Patient 2 to validate the original result. Parent-child biologic relationships were confirmed using the AmpFlSTR® Profiler Plus™ kit from Applied Biosystems (Foster City, CA), as previously described [Sol-Church et al., 2006].
All primers are listed in Table 1. For the G12S mutation screen, a discrete 575bp region of HRAS containing exon 2 was amplified using primers F1 and R1. Two additional overlapping PCR products were obtained for Patient 2 using primers F2 and R2 (490bp), and F3 and R1 (649bp). The latter product was subjected to a second round of PCR amplification using primers F3 and R1, in order to rule out preferential allelic amplification. Additional regions of the HRAS gene were screened using primers that amplified exons 3, exons 4–5, and exon 6 (supplemental Table 1). All PCR products were purified and sequenced in both directions using the ABI BigDye Terminator Cycle Sequencing Ready Reaction kit v 3.1 and analyzed on an ABI3130XL Genetic Analyzer. All sequences were analyzed by direct observation of the electropherogram and using Sequencher 4.8 (Gene Codes Corporation, Ann Harbor MI).
Table 1.
HRAS Primers
| Forward | Sequence | Reverse | Sequence | Size (bp) | Ta (°C) |
|---|---|---|---|---|---|
| F1 | ATTTGGGTGCGTGGTTGA | R1 | CCTCTAGAGGAAGCAGGAGACA | 575 | 58 |
| F2 | GCAGGAGACCCTGTAGGAGGAC | R2 | CGGTATCCAGGATGTCCAACAG | 490 | 60 |
| F3 | ACCTGTTCTGGAGGACGGTAA | R1 | CCTCTAGAGGAAGCAGGAGACA | 649 | 59 |
| F3 | ACCTGTTCTGGAGGACGGTAA | RW | TATATACTCTTGCCCACACCGCC | 462 | 60 |
| F3 | ACCTGTTCTGGAGGACGGTAA | RM | ACTCTTGCCCACACCGCT | 457 | 60 |
| F4 | TCTAATTTGGGTGCGTGGTTGA | RW | TATATACTCTTGCCCACACCGCC | 392 | 60 |
| F4 | TCTAATTTGGGTGCGTGGTTGA | RM | ACTCTTGCCCACACCGCT | 387 | 60 |
| F5 | Biotin-AGGAGCGATGACGGAATATAAGC | R5 | TCTATAGTGGGGTCGTATTCGTCC | 117 | 61 |
| R6 | TCTTGCCCACACCGC | ||||
| 3F | GAGAGGCTGGCTGTGTGAACTC | 3R | TCTGCTCCCTGAGAGGTGGAA | 410 | 59 |
| 4F | GCTTCCTGTGTGTGTTTGCCA | 4R | CCAGTAGCCCCACTAAGACTCAGA | 768 | 59 |
| 6F | GGCACCTGTTGGTTCTGAGTCTT | 6R | TTCCTTCCTCCTCCTTCCGTCT | 639 | 59 |
Allele specific primers RW and RM were designed to selectively amplify either the wild type G12 or the mutated S12 alleles. Patient 1’s alleles were amplified using F4/RW and F4/RM and sequenced using F4. Longer allele specific PCR products were generated for Patient 2 using F3/RW and F3/RM, and the sequencing performed using primer F3. Standard PCR conditions were used and supplemental Table 1 lists the products size and annealing temperature of each primer set.
SgrA1 digestion of the 575 bp and 649 bp products derived from Patient 1 and Patient 2 was performed as follows. Unpurified products were incubated with the restriction endonuclease for 3 hours at 37°C, separated by gel electrophoresis on a 2% agarose gel and photographed under UV light. For the sample derived from Patient 2, the SgrA1-resistant 649 bp band containing the mutated allele was gel purified and subjected to a second round of PCR amplification, followed by purification and sequencing.
Allelic quantification was performed using pyrosequencing technology. Briefly, a 177 bp biotinylated PCR product was generated using primers F5 and R5. Following immobilization of the biotin tagged single stranded PCR product onto Streptavidine Sepharose HP (Amersham Pharmacia Biotech, Uppsala, Sweden) sequencing primer R6 was annealed, and pyrosequencing was initiated using the PSQ96 Gold Reagent Kit following the manufacturer’s recommendation (Pyrosequencing AB, Uppsala, Sweden). Data analysis was performed using the PSQ96 allelic quantitation (AQ) option.
RESULTS
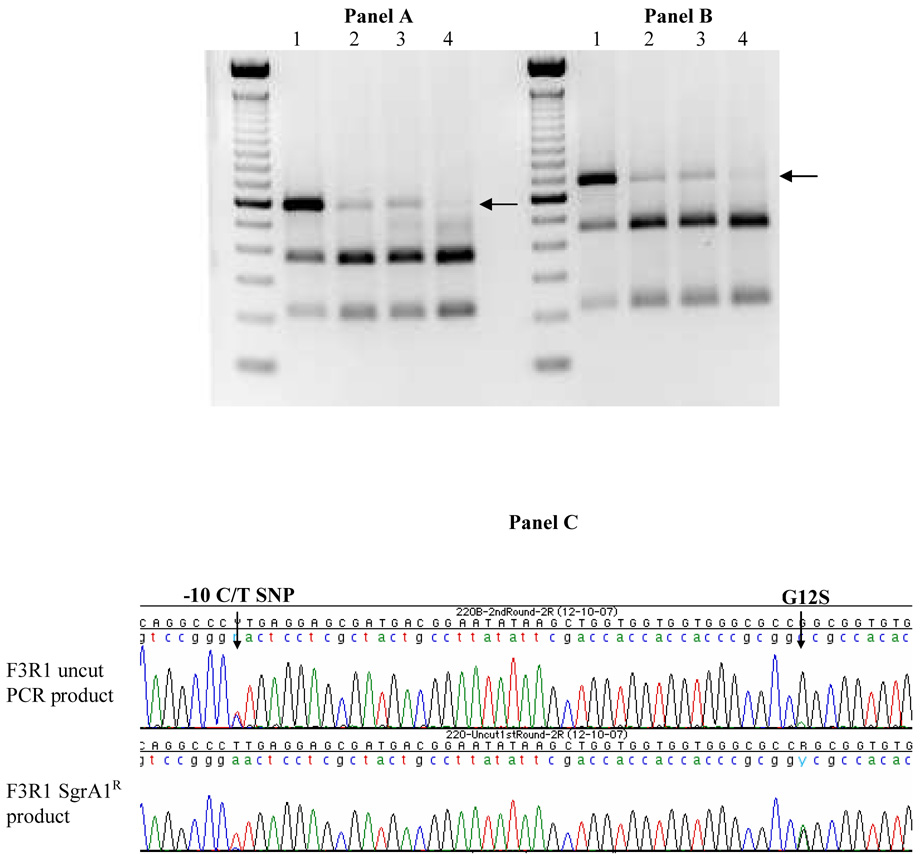
Patient 1 and 2’s DNAs were sequenced across a region of the HRAS gene containing the first translated exon (exon 2) and flanking intronic regions. Patient 1 carries a known SNP (−10C>T) and a germline c.34G>A mutation, resulting in a heterozygous G12S change in the ras protein (Figure 1A). Both DNA samples isolated from his father, Patient 2, showed the heterozygous SNP as well as evidence of the G12S mutation, albeit at a lower level suggesting somatic mosaicism (Figure 1B and 1C). Results for Patient 2 were confirmed using independently amplified overlapping PCR products (data not shown). The mother of Patient 1 is homozygous C at the −10 SNP and carries no mutation in exon 2 (Figure 1D). The SNP is an informative marker for this family as Patient 1’s −10C allele can be traced back to his mother, while the −10T allele is inherited from the father (Patient 2). Preferential amplification of either allele in Patient 2 was ruled out by performing a second round of amplification using the 649 bp PCR product as a template and primers F3 and R1. No other mutation was detected in exons 3, 4, 5 and 6 in either patient. As previously reported in Sol-Church et al. [2006] the G12S mutation leads to loss of a cognate site for endonuclease SgrA1. Restriction endonuclease analysis of PCR products using SgrA1 was performed on the patients and a control sample (Figure 2). Compared to the normal control, Patient 1 and Patient 2 carry alleles that are resistant to hydrolysis, thus confirming the presence of the mutation in both individuals (Figure 2, panels A and B). Consistent with Patient 2’s low level mosaicism observed by direct sequencing, the father’s undigested product is less compared to Patient 1. Patient 2’s 649bp SgrA1-resistant product seen in Figure 2B (lane 2) was gel purified and sequenced. The sequencing profiles depicted in Figure 2C indicate that this restriction endonuclease resistant product is enriched in mutant allele and shows no preferential amplification when subjected to second round amplification. Interestingly, the −10T allele is also enriched suggesting that this allele may be present in cis with the CS-causing mutation.
Figure 1. Genotyping status of HRAS in family members as detected by direct sequencing.
Buccal cell genomic DNA was collected from Patient 1 (A), Patient 2 (B and C, two independently collected samples) and Patient 1’s mother (D). Patient 1 shows a heterozygous c.34G>A change, causing a G12S mutation, while Patient 2 carries low levels of the mutated allele. Father (B) and child (A) are heterozygous C/T at the −10 SNP, while the mother (D) is homozygous C and carries a wild type c.34G base.
Figure 2. Confirmation of the presence of the G12S allele in Patient 2 by restriction endonuclease analysis.
Panel A and B: Restriction digest analysis of genomic DNAs isolated from Patient 1 (lane 1), Patient 2 (lanes 2 and 3), and a wild type control (lane 4). Genomic DNAs were amplified with primers F1R1 (panel A) or F3R1 (panel B) and subjected to SgrA1 digestion. The arrows indicate the location of the SgrA1 resistant DNA fragments (SgrA1R) that were gel purified and sequenced. Panel C: Sequencing results of Patient 2’s F3R1 PCR before (uncut) and after digestion with SgrAI.
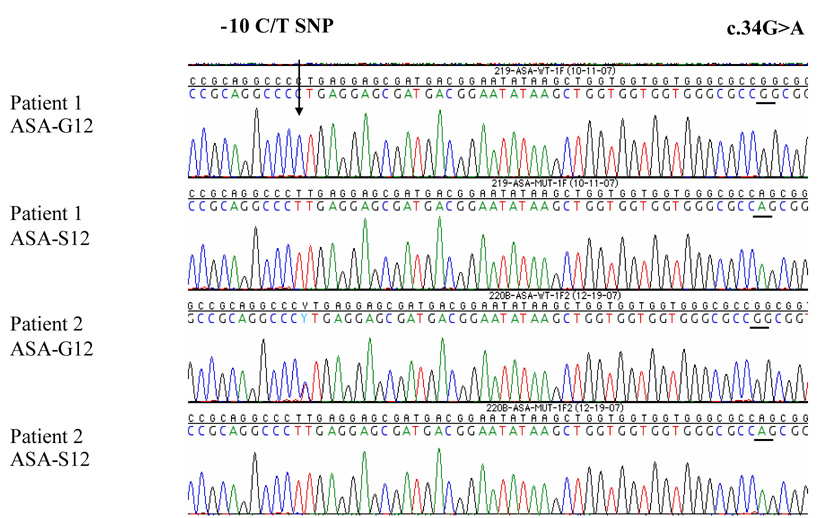
Pyrosequencing was used to quantify the fraction of mutated alleles in the DNA from both affected individuals (supplemental Fig. 4). Patient 1’s observed allelic ratio (48.2%) is close to that expected of a germline mutation (50%), while only 7.5% of Patient 2’s alleles are mutated, indicating somatic mosaicism. Patient 2’s buccal sample consists of a mixed population of cells: most cells are wild type and carry a normal HRAS gene and a small fraction of mutant “CS” cells carry the heterozygous G12S mutation. Allele specific amplification (ASA) was performed to verify the paternal origin of Patient 1’s germline mutation, and also to clarify whether or not the father’s mutation had occurred randomly on his chromosome homologs. During ASA specific primers were used to selectively amplify either the wild type (ASA-G12) or the mutant (ASA-S12) alleles (Figure 3). Sequencing results show that Patient 1’s wild type amplicons carry in cis the −10C allele, which he has inherited from his mother. Conversely, his mutated allele carries the −10T paternal allele thus demonstrating that the germline mutation was inherited from the father.
Figure 3. Parental origin of the germiline G12S mutation in Patient 1 and evidence of mosaicism in Patient 2.
Allelic specific amplification (ASA) was used to amplify specific alleles in Patients 1 and 2. The location of the informative SNP at bp-10 is indicated. Each chromatogram depicts the sequence of either the wild type (G12) or mutated S12 alleles for both patients. The ASA base permitting allelic discrimination is underlined.
As expected from a mosaic population of cells, Patient 2’s wild type allele revealed the presence of the heterozygous C>T SNP, contributed from both normal and mutant cells. In contrast, Patient 2’s mutant allele generated via ASA-S12, clearly identifies in cis the presence of the homozygous −10T SNP allele. These results unequivocally demonstrate that the clonal nature of the mutated cells in the father arose from a single hit to the SNP T-allele.
DISCUSSION
Before the gene defect for Costello syndrome was identified, Bodkin et al [1999] postulated somatic mosaicism in the father of a male with typical Costello syndrome. The father had a history of feeding problems, a patchy distribution of skin and hair abnormalities, and nasal papillomata. The authors suggested that this apparent male-to-male transmission was consistent with autosomal dominant inheritance, in contrast to the previously considered recessive inheritance pattern. Since then, a discrete number of heterozygous germline mutations in the oncogene HRAS have been reported to cause Costello syndrome. Currently, 13 different DNA variants of the HRAS gene, affecting amino acid 12, 13, 22, 58, 63, 117 or 146, have been reported in Costello syndrome patients [reviewed in Sol-Church and Gripp, 2008 in press]. Substitutions of glycine 12 or 13 in the ras protein account for 95% of these mutations. The most common changes (G12S, G12A) result in the typical phenotype, while milder cases can be due to mutations resulting in presumably less functional impairment of the abnormal protein product [Gripp et al., 2008]. Another mechanism resulting in an apparently mild Costello syndrome phenotype despite a typical G12S HRAS mutation is somatic mosaicism [Gripp et al., 2006b]. The first molecular evidence of mosaicism in Costello syndrome was seen in a female with developmental delay, short stature, sparse hair, coarse facial features, and nasal papillomata. She had irregular skin hyper- and hypopigmentation, a finding not typical for Costello syndrome, but often associated with mosaicism for chromosome abnormalities. Standard assays performed on this patient’s white blood cell derived DNA did not show an HRAS mutation. However, buccal cell derived DNA revealed a sequence change qualitatively consistent with the G12S mutation. Allelic quantitation demonstrated the presence of this mutation in ~25–30% of the cells’ alleles. It was noted that this patient with somatic mosaicism had regular periods, whereas most females with Costello syndrome have delayed or incomplete puberty or amenorrhea due to central hypogonadism [White et al 2005]. Thus, this patient was considered at risk for offspring with Costello syndrome [Gripp et al., 2006b].
Through molecular analysis of the family originally reported by Bodkin et al 1999, we have now shown for the first time that parental somatic mosaicism can result in an offspring with typical Costello syndrome. Similar to the case reported here, parental somatic mosaicism could explain the findings in the family with mild mental retardation, short stature, macrostomia, thick lips, hyperlax and hyperpigmentated skin and facial papillomata in the mother of siblings with presumed typical Costello syndrome [Ioans and Fryns, 2002]. However, while this mother’s findings could be consistent with somatic mosaicism for a typical Costello syndrome mutation, they may also be consistent with the “attenuated phenotype” apparently associated with rare Costello syndrome causing mutations [Gripp et al., 2008]. In contrast to the families reported here and by Ioans and Fryns, no physical findings suggestive of parental somatic mosaicism were noted in the few other reported siblings with Costello syndrome [Zampino et al., 1993; Johnson et al., 1998] Thus, parental germline mosaicism appears to be a more likely explanation than parental somatic mosaicism for the recurrence in these families. Detailed molecular studies will need to be performed in these families in order to confirm the underlying mechanism.
Costello syndrome in a mosaic individual may be difficult to recognize on clinical grounds, but it should always be considered when evaluating parents of affected individuals. The incidence of somatic mosaicism for Costello syndrome is unknown, but the identification of two mosaic individuals has important consequences for clinical diagnosis, molecular testing and genetic counseling. In addition to the obvious concern for the risk to offspring, somatic mosaicism may have important consequences regarding medical problems related to the HRAS mutation. For example, patients with Costello syndrome are at increased risk for transitional cell carcinoma of the bladder, presumably due to the HRAS mutation representing the first “hit” in carcinogenesis. An individual with somatic mosaicism may or may not carry the HRAS mutation in the transitional cell epithelium of the bladder, and the presence of cells with two wild type alleles may mitigate the tumor risk. We do not know if the risk for bladder cancer in our Patient 2 is increased compared to the general population. In order to better understand the consequences of a somatic HRAS mutation, long term follow up of these rare patients will be required, and it is important to keep in mind the variability inherent in mosaic states.
Acknowledgments
The authors wish to thank the family for allowing us to share this information. We also thank our colleagues, Drs. Lars Erickson and Steven Colan, who have contributed to the care of Patient 1. KSC is supported by a grant from the NIH National Center for Research Resources, Center of Biomedical Research Excellence (COBRE) program (1 P20 RR020173) and by the Nemours Foundation.
Footnotes
Web Resources
The URL for data presented herein is as follows: Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, Matsubara Y. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- Bodkin NM, Mortimer ES, Demmer La. Male-to-male transmission of Costello syndrome consistent with autosomal dominant inheritance. Am J Hum Genet. 1999;65 Suppl:A143. [Google Scholar]
- Estep AL, Tidyman WE, Teitell MA, Cotter PD, Rauen KA. HRAS mutations in Costello syndrome: Detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am J Med Genet, Part A. 2006;140(1):8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- Gripp K, Lin A. [1997–2006];Seattle: Copyright, University of Washington; Costello Syndrome in: GeneReviews at GeneTests: Medical Genetics Information Resource [database online] 2006 Available at www.genetests.org.
- Gripp KW, Lin AE, Stabley DL, Nicholson L, Charles I, Scott CI, Jr, Doyle D, Aoki Y, Matsubara Y, Zackai EH, Lapunzina P, Gonzalez-Meneses A, Holbrook J, Agresta CA, Gonzalez IL, Sol-Church K. HRAS Mutation Analysis in Costello Syndrome: Genotype and Phenotype Correlation. Am J Med Genet Part. 2006a;A140:1–7. doi: 10.1002/ajmg.a.31047. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Stabley DL, Nicholson L, Hoffman JD, Sol-Church K. Somatic Mosaicism for an HRAS mutation causes Costello Syndrome. Am J Med Genet. 2006b;140A:2163–2169. doi: 10.1002/ajmg.a.31456. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Lin AE, Nicholson L, Allen W, Cramer A, Jones KL, Kutz W, Peck D, Rebolledo MA, Wheeler PG, Wilson W, Al-Rahawan MM, Stabley DL, Sol-Church K. Further Delineation of the Phenotype Resulting from BRAF or MEK1 Germline Mutations Helps Differentiate Cardio-facio-cutaneous Syndrome from Costello Syndrome. Am J Med Genet. 2007;143A:1472–1480. doi: 10.1002/ajmg.a.31815. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Innes MA, Axelrad ME, Gillan TL, Parboosingh JS, Davies C, Leonard NJ, Lapointe M, Doyle D, Catalano S, Nicholson L, Stabley DL, Sol-Church K. Costello syndrome associated with novel germline HRAS mutations: An attenuated phenotype? Am J Med Genet. 2008;146A:683–690. doi: 10.1002/ajmg.a.32227. [DOI] [PubMed] [Google Scholar]
- Ioan DM, Fryns JP. Costello syndrome in two siblings and minor manifestations in their mother. Further evidence for autosomal dominant inheritance? Genet Couns. 2002;13(3):353–356. [PubMed] [Google Scholar]
- Johnson JP, Golabi M, Norton ME, Rosenblatt RM, Feldman GM, Yang SP, Hall BD, Fries MH, Carey JC. Costello syndrome: phenotype, natural history, differential diagnosis, and possible cause. J Pediatr. 1998;133:441–448. doi: 10.1016/s0022-3476(98)70284-7. [DOI] [PubMed] [Google Scholar]
- Kerr B, Allanson J, Delrue MA, Gripp KW, Lacomb D, Lin AE, Rauen KA. The diagnosis of Costello syndrome: Nomenclature in Ras/MAPK pathway Disorders. Am J Med Genet. 2008;146A:1218–1220. doi: 10.1002/ajmg.a.32273. [DOI] [PubMed] [Google Scholar]
- Kerr B, Delrue MA, Sigaudy S, Perveen R, Marche M, Burgelin I, Stef M, Tang B, Eden T, O'Sullivan J, De Sandre-Giovannoli A, Reardon W, Brewer C, Bennett C, Quarrell O, McCann E, Donnai D, Stewart F, Hennekam R, Cave H, Verloes A, Philip N, Lacombe D, Levy N, Arveiler B, Black G. Genotype-phenotype correlation in Costello syndrome; HRAS mutation analysis in 43 cases. J Med Genet. 2006;43:401–405. doi: 10.1136/jmg.2005.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sol-Church K, Gripp KW. Molecular basis of Costello syndrome. In: Zenker M, editor. Noonan Syndrome and Related Disorders. Monogr Hum Genet. Vol 17. Basel: Karger; 2008. pp. 1–10. In press. [Google Scholar]
- Sol-Church K, Stabley DL, Nicholson L, Gonzalez IL, Gripp KW. Paternal Bias in Parental Origin of HRAS Mutations in Costello Syndrome. Hum Mutat. 2006;27(8):736–741. doi: 10.1002/humu.20381. [DOI] [PubMed] [Google Scholar]
- van Steensel MA, Vreeburg M, Peels C, van Ravenswaaij-Arts CM, Bijlsma E, Schrander-Stumpel CT, van Geel M. Recurring HRAS mutation G12S in Dutch patients with Costello syndrome. Exp Dermatol. 2006;15(9):731–734. doi: 10.1111/j.1600-0625.2006.00474.x. [DOI] [PubMed] [Google Scholar]
- White SM, Graham JM, Jr, Kerr B, Gripp K, Weksberg R, Cytrynbaum C, Reeder JL, Stewart FJ, Edwards M, Wilson M, Bankier A. The adult phenotype in Costello syndrome. Am J Med Genet. 2005;136A:128–135. doi: 10.1002/ajmg.a.30747. [DOI] [PubMed] [Google Scholar]
- Zampino G, Mastroiacovo P, Ricci R, Zollino M, Segni G, Martini-Neri ME, Neri G. Costello syndrome: further clinical delineation, natural history, genetic definition, and nosology. Am J Med Genet. 1993;47(2):176–183. doi: 10.1002/ajmg.1320470210. [DOI] [PubMed] [Google Scholar]
- Zampino G, Pantaleoni F, Carta C, Cobellis G, Vasta I, Neri C, Pogna EA, DeFeo E, Delogu A, Sarlozy A, Atzeri F, Selicorni A, Rauen KA, Cytrynbaum CS, Weksberg R, Dallapiccola B, Ballabio A, Gelb BD, Neri G, Tartaglia M. Diversity, parental germline origin, and phenotypic spectrum of de novo HRAS missense changes in Costello syndrome. Hum Mutat. 2007;28:265–272. doi: 10.1002/humu.20431. [DOI] [PubMed] [Google Scholar]