Abstract
Mammalian autonomic nervous system (ANS) development requires the combinatorial action of a number of transcription factors, which include Mash1, Phox2b and GATA3. Here we show that the bHLH transcription factor, Hand2 (dHAND), is expressed concurrently with Mash1 during sympathetic nervous system (SNS) development and that the expression of Hand2 is not dependent on Mash1. This suggests that these two bHLH factors work in parallel during SNS development. We also show that ectopic expression of Hand2 activates the neuronal program and promotes the acquisition of a phenotype corresponding to peripheral neurons including neurons of the SNS lineage in P19 embryonic carcinoma cells. We propose that Hand2 works in parallel with other members of the transcriptional network to regulate ANS developmental but can ectopically activate the program by a cross-regulatory mechanism that includes the activation of Mash1. We show that this function is dependent on its interaction with the histone acetyltransferase p300/CBP, indicating that Hand2 functions to promote ANS development as part of larger transcriptional complex.
Keywords: autonomic, sympathetic, Hand2, development, p300
INTRODUCTION
The development of the autonomic nervous system (ANS), and in particular the sympathetic division of the ANS (SNS), has long provided a model to investigate the cellular, molecular and genetic events leading to the differentiation of neurons (Reviewed in (Bertrand et al., 2002; Goridis and Rohrer, 2002). From these studies numerous signaling pathways and transcription factors have been identified as critical components of SNS development, yet many gaps remain in our knowledge. During SNS development, neural crest (NC) cells migrate from the dorsal region of the neural folds, traversing the ventral-medial sclerotome of the somites until they reach the dorsal aorta where they are induced to differentiate in response to BMP (Reissmann et al., 1996; Varley and Maxwell, 1996; Varley et al., 1998). BMP signaling activates a number of genes encoding transcription factors essential for SNS development including the paired homeodomain genes Phox2a and Phox2b (Lo et al., 1998; Schneider et al., 1999) and the basic helix-loop-helix factors (bHLH) Mash1 (Shah et al., 1996; Lo et al., 1998) and Hand2 (Howard et al., 2000).
The Phox2 genes are closely related factors expressed in all catecholaminergic neurons and are required for the specification of catecholaminergic neurons and regulation of catecholaminergic biosynthesis in differentiated neurons (Tiveron et al., 1996; Pattyn et al., 1997). Loss of either Phox2 gene leads to loss of the ANS by mid-gestation (Morin et al., 1997; Pattyn et al., 1999). When ectopically expressed, both Phox2a and Phox2b activate noradrenergic neuronal development in NC stem cells in vitro (Lo et al., 1999) and NC cells located along the peripheral nerves (Stanke et al., 1999). Phox2 target genes involved in regulating noradrenergic specification during early development remain unknown, but in differentiated neurons, the Phox2 genes bind directly to and regulate expression of the noradrenalin biosynthetic enzymes dopamine-ß-hydroxylase (DBH) and tyrosine hydroxylase (TH) genes, suggesting that the Phox2 genes are required for maintenance of the differentiated phenotype (Kim et al., 1998; Yang et al., 1998). Although the Phox2 genes are required for acquisition of the catecholaminergic phenotype, their expression in non-catecholaminergic lineages during development suggests they do not specify specific lineages of the ANS.
The bHLH transcription factors are key genes in the specification and differentiation of neurons and are essential during central (CNS), peripheral nervous system (PNS) and ANS development (Reviewed in (Jan and Jan, 1994; Anderson and Jan, 1998). The bHLH factor Mash1 is expressed in a number of neuronal lineages during development including the CNS and ANS (Guillemot and Joyner, 1993). In the ANS, loss of Mash1 does not prevent development of neuroblasts but a majority of the neuroblasts fail to differentiate and are subsequently lost. This suggests that Mash1 plays a role in the differentiation of ANS neurons (Guillemot et al., 1993; Sommer et al., 1995). However, Mash1 is expressed in a number of neuronal lineages outside the ANS including those of the eye, olfactory epithelium and CNS where it does not induce ANS differentiation (Guillemot and Joyner, 1993). The broad expression of Mash1 argues against a role for Mash1 in the specification of ANS neurons. Further support of this, comes from ectopic expression studies, which demonstrate that Mash1 promotes generic neuronal development and not the ANS program (Farah et al., 2000; Tomita et al., 2000).
Unlike other transcription factors found in the ANS, the bHLH transcription factor Hand2 is restricted to ANS during neural development (Srivastava et al., 1995; Dai et al., 2004). Hand2 is a member of the twist family of bHLH factors and can activate transcription through binding to a subset of E-boxes (Dai and Cserjesi, 2002). However, it can also function to regulate limb development without a DNA binding domain (McFadden et al., 2002). This suggests that Hand2 can activate genes by direct binding to their regulatory regions and also as part of a transcriptional complex independent of DNA binding.
The role of Hand2 in neuronal development has been investigated in vitro and in vivo but its function in neuronal development remains unclear. Ectopic expression of Hand2 in quail NC cultures enhances catecholaminergic differentiation, while down regulation of Hand2 leads to suppression of development (Howard et al., 1999). In vivo, ectopic expression of Hand2 in NC cells along peripheral nerves also induces the catecholaminergic differentiation program (Howard et al., 2000). Since NC derived catecholaminergic neurons are predominantly sympathetic, these results suggest that Hand2 specifies the sympathetic lineage. However, Hand2 is expressed in all three lineages of the ANS suggesting that Hand2 plays a role in development of the ANS, not just the sympathetic lineage.
Here, we address where in the transcriptional hierarchy regulating ANS development Hand2 fits. its developmental role, and the molecular mechanism through which it regulates neuronal development. We show that Hand2 and Mash1 are co-expressed during SNS development and that expression of the Hand2 gene is not dependent on Mash1, suggesting that these two bHLH factors function in parallel during ANS development. We also show that Hand2 can activate the ANS neuronal development in P19 embryonal carcinoma cells suggesting that it possesses the ability to activate the neuronal program and promote lineage-specific selection. Hand2 also activates Mash1 through recruitment of the histone acetyltransferase p300/CBP, suggesting that Hand2 functions as part of a transcriptional complex during neuronal development. Based on these results, we propose that Hand2 activates the ANS developmental program through the cross-activation of a network of transcription factors that act in parallel during development.
MATERIALS AND METHODS
Plasmids
pRC-RSV-HAND2 was generated by subcloning the EcoRI-BamHI fragment containing the Hand2 coding region from pRSET-HAND2 (Dai and Cserjesi, 2002) into pcDNA3-HisB (Invitrogen). The HindIII-XbaI fragment containing the epitope tag and Hand2 cDNA was subcloned into pRC-RSV (Invitrogen). To make pEGFPJH-HAND2, pEGFP2JH was constructed by modifying pHOOK2 (Invitrogen) with a replacement of the HOOK gene sequence with EGFP by inserting the 0.74kb SpeI (blunted)-SalI fragment from pBI-EGFP (Clontech) into the EcoRV-XhoI site of pHOOK2. To generate pEGFPJH-HAND2, the BglII-XbaI Hand2 fragment from pcDNA3-HisB-H2 was inserted into the BglII-XbaI sites of pEFGP2JH. The Mash1 expression vector, CS2+MT MASH1, was provided by D. Turner (University of Michigan, Ann Arbor) (Farah et al., 2000). CMVβ p300 wt was provided by D. Livingston, (University of Southern California, Los Angeles, CA) (Eckner et al., 1996) and CMV-12SE1A expressing E1A wild type protein by S. Chellappan (University of South Florida, Tampa, FL).
Cell Culture and generation of stable Hand2 expressing P19 cell lines
P19 embryonal carcinoma (P19-EC) cells were cultured in D-MEM with 7.5% calf serum and 2.5% fetal bovine serum (Gibco). P19-EC cell lines stably expressing Hand2 (P19-H2) were generated by transfection with pRC-RSV-HAND2 and selection of neomycin resistant clones using 100 µg/ml Geneticin (Gibco). Expression of Hand2 protein in cell lines was examined by Western blot analysis monitored by immunocytochemistry with Hand2 specific antibody (Dai and Cserjesi, 2002).
Growth and differentiation of P19-EC and P19-H2 cells were described previously (Bain et al., 1994) with the following modifications. After 4 days of aggregation, aggregates were collected, dispersed, and cultured for 2 days. The media was then replaced with media supplemented with 5 µg/ml cytosine arabinoside (Sigma) to select for non-dividing neurons.
Neuro-2a cells were cultured and transfected as described previously (Dai et al., 2004).
Immunocytochemistry
For immunostaining, cells were fixed in 3% paraformaldehyde for 10 min and washed with PBS. The cells were incubated with blocking solution (1% goat serum in PBS) for 10 min then incubated with primary antibodies in blocking solution for 1 hr. After washing, cells were incubated with fluoroscein conjugated anti-mouse and rhodamine-conjugated anti-rabbit secondary antibodies (Chemicon). Antibodies were obtained from: peripherin, R. Liem (Columbia University, New York); TuJ1 (Sigma); TH (Chemicon); Mash1, J. Johnson (University of Texas, Southwestern, Dallas); SSEA1, Developmental Studies Hybridoma Bank; Hand2 as described previously (Dai and Cserjesi, 2002).
RNA analysis
Total RNA was isolated from P19 cells and adult sympathetic ganglia using Trizole (Gibco) following manufacturers instructions. For Northern analysis, 15 µg of total RNA was separated in a formaldehyde-agarose gel and transferred to a Duralon membrane (Stratagene). Blots were hybridized with 32P-dCTP-labeled probes generated by random primer labeling (Ambion) in Quick-Hybrid Buffer (Stratagene). To detect endogenous Hand2 transcripts, the 3’ untranslated region of the Hand2 cDNA was used as a probe.
RT-PCR analysis was performed on 0.5 µg of total RNA as described previously (Morikawa and Cserjesi, 2004). Primer sequences are available upon request.
In situ hybridization
In situ hybridization of whole mount mouse embryos and sections were performed with digoxigenin-labeled probes as described previously (Dai et al., 2004). Plasmids used to generate in situ probes were pBSK-HAND2 (Dai et al., 2004), Mash1 (Guillemot and Joyner, 1993), cRet (cRet9-pBKSII), provided by F. Costantini (Columbia University, New York), and Phox2b (pcDNA3-Phox2b) provided by J. -F. Brunet (CNRS, Ecole Normale Supérieure, Paris).
RESULTS
Expression of Hand2 and Mash1 during ANS development
As a first step in understanding the potential role of Hand2 during mouse ANS development, we examined Hand2 expression in the developing ANS by in situ hybridization (Fig. 1). Hand2 is expressed in all lineages of the ANS, the sympathoadrenal, parasympathetic, and enteric (Fig. 1A). Hand2 expression was not detected in the CNS or in the sensory lineage of the PNS. In the SNS, Hand2 expression was first detected in the rostral region of the sympathetic trunk at 10 dpc in the NC cells coalescing by the dorsal aorta. Hand2 expression was not detected in migrating NC, the precursors of the ANS. The expression of Hand2 in post-migratory NC cells suggests it is first expressed after NC cells are specified to form the SNS, overlapping the expression of Mash1.
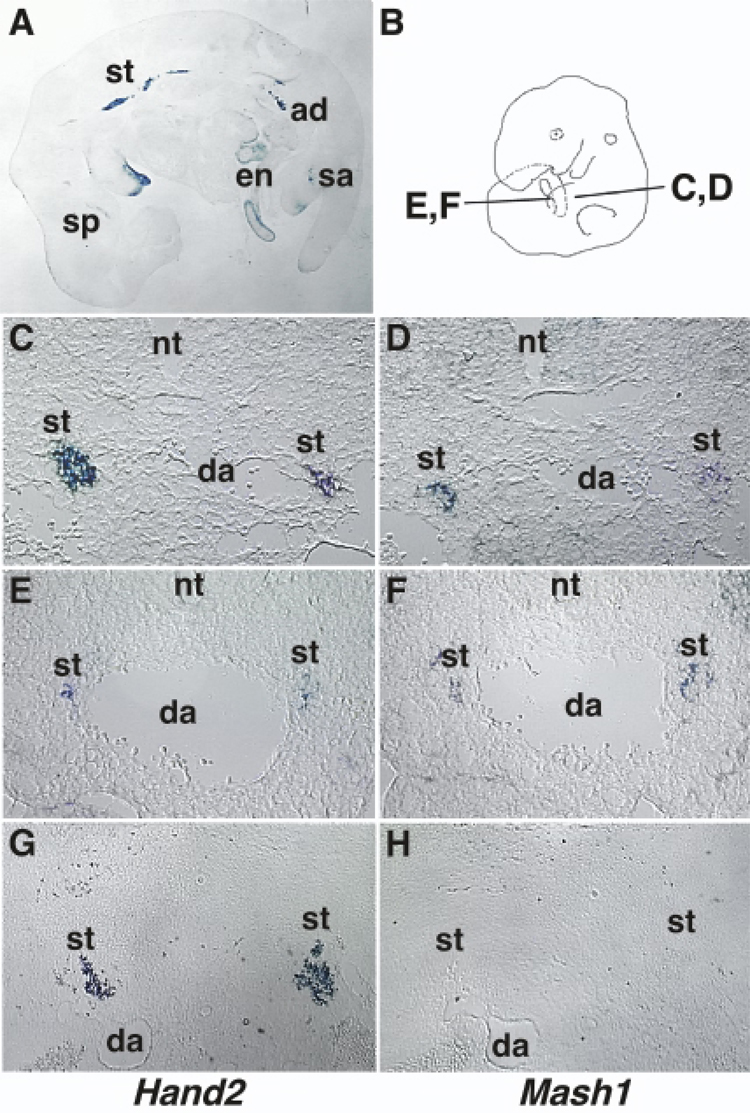
Fig. 1. Expression of Hand2 and Mash1 during PNS development.
Hand2 and Mash1 gene expression was examined by in situ hybridization using digoxigenin-labeled anti-sense cRNAs. A: In situ hybridization of a 12.5 dpc mouse embryo with Hand2. Hand2 is expressed throughout the length of the sympathetic trunk, in the coalescing adrenal gland, and in the parasympathetic and enteric nervous systems. B–H: Comparison of Hand2 (C, E, G) and Mash1 (D, F, H) expression in the SNS of 10.5 dpc (C–F) and 14.5 dpc (G–H) mouse embryos by in situ hybridization using alternate 10 µm cryosections. The regions of the embryo where sections were obtained for plates C–F are shown in plate B. Hand2 and Mash1 are co-expressed during early SNS development (C–F) but Hand2 expression continues after down regulation of Mash1 at 14.5 dpc (G–H). ad: adrenal gland; da: dorsal aorta; en: enteric plexus; nt: neural tube; sa: sacral parasympathetic plexus; sp: sphenopalatine ganglia; st: sympathetic trunk.
To determine the exact temporal relationship between Hand2 and Mash1, we focused our studies on the SNS. Since SNS development proceeds from rostral to caudal, we sectioned through the length of a 10.5 dpc mouse embryo obtaining sections containing SNS ganglia at all stages of differentiation. Alternate serial sections were analyzed for Hand2 and Mash1 expression by in situ hybridization (Fig. 1C–F). The cervical region of the embryo contains a large population of sympathetic cells expressing both Hand2 and Mash1 transcripts (Fig. 1C, D). Sections through more caudal regions contain progressively fewer Hand2 and Mash1 expressing cells (Fig. 1E, F). The analysis showed that the initiation of Hand2 and Mash1 expression in the SNS occurs concurrently suggesting they are activated in parallel.
To determine if Hand2 expression is dependent on continued Mash1 expression, we examined the expression of Hand2 in later stage embryos. Mash1 is expressed at high levels until 13 dpc followed by a rapid down regulation at 14.5 dpc (Guillemot and Joyner, 1993). At 14.5 dpc of development, Hand2 expression remains high (Fig. 1G) while Mash1 transcripts were undetectable (Fig. 1H). To determine if Hand2 expression is maintained in the adult SNS and if Mash1 expression is reactivated, we examined the levels of Hand2 and Mash1 transcripts by semi-quantitative RT-PCR. Hand2 is detected in the adult SNS while Mash1 expression was not detected (data not shown). The continued expression of Hand2 after cessation of Mash1 suggests that Hand2 expression in not dependent of Mash1.
Hand2 expression is not regulated by Mash1
Although our expression results show that continued expression of Hand2 is independent of Mash1, it is possible that initiation of Hand2 expression is Mash1 dependent. To test this, we examined Hand2 expression in embryos lacking the Mash1 gene (Fig. 2). Mash1+/+ and Mash1−/− embryos were analyzed by in situ hybridization using Hand2 probes to examine Hand2 expression and Phox2b probes to identify SNS neuroblasts. Sagittal (Fig. 2A, B) analysis showed that at 12.5 dpc, Hand2 was expressed in the sympathetic trunk in Mash1+/+ embryos although fewer Hand2 expressing cells were observed. At 10.5 dpc, a half day after the initiation of Mash1 expression, Hand2 expression was observed in the sympathetic trunk of Mash1−/− embryos (data not shown). These data suggest that activation and continued expression of the Hand2 gene are independent on Mash1.
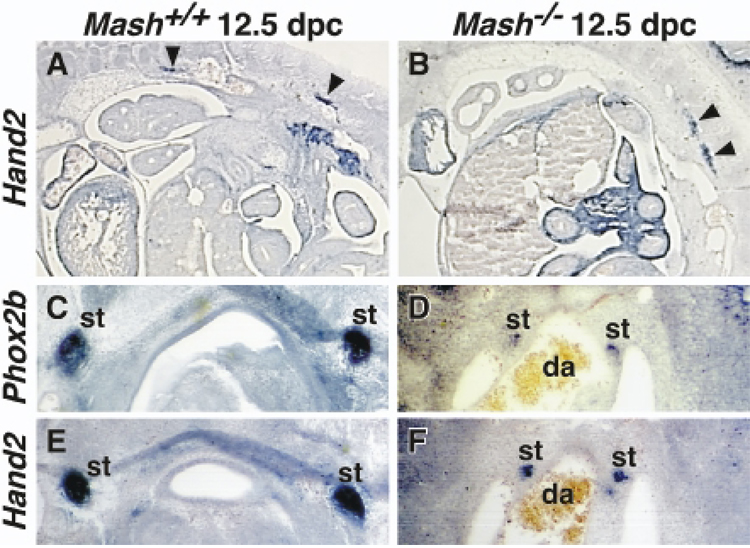
Fig. 2. Hand2 expression is not dependent on Mash1.
Expression of Hand2 and Phox2b genes was examined in 12.5 dpc Mash+/+ (A, C, E) and Mash1−/− (B, D, F) littermates using in situ hybridization. In situ hybridization was performed using digoxigenin -labeled anti-sense probes on 10 µm sagittal and 25 µm paraffin embedded cross sections. A, B: Sagittal sections stained with Hand2 probe. Hand2 is expressed in the developing SNS of both Mash+/+ and Mash1−/− embryos including the trunk (arrowheads). C–F: Cross sections hybridized with Phox2b (C, D) and Hand2 (E, F) on alternate sections. Hand2 and Phox2b expression overlaps in the sympathetic trunk of both Mash+/+ and Mash1−/− embryos. The decreased level of transcript in the Mash1−/− embryos reflects a decreased cell number in the forming SNS ganglia. da: dorsal aorta; st: sympathetic trunk.
The smaller number of Hand2 expressing cells in Mash1−/− embryos could be due either to a dependence on Mash1 expression in a subset of cells or to the loss of cells during development (Guillemot and Joyner, 1993; Sommer et al., 1995). To determine if the decrease in Hand2 expressing cells in the sympathetic trunk was due to Mash1 dependence or to the presence of fewer cells (neuroblasts), alternate thin sections from Mash1+/+ and Mash1−/− embryos were hybridized with Hand2 and a marker for SNS neuroblasts and neurons, Phox2b. More cells in the sympathetic trunk express Phox2b and Hand2 in Mash1+/+ (Fig. 2C, E) than in Mash1−/− (Fig. 2D, F) embryos. A comparison of the number of Hand2 and Phox2b expressing SNS cells in Mash1−/− embryos was determined in a series of cross sections along the embryo. An equivalent number of cells express Hand2 and Phox2b. This suggests that the reduced Hand2 expression in Mash1−/− embryos is due to the loss of developing sympathetic neurons. These results further demonstrate that Hand2 expression is not dependent on Mash1.
Hand2 activates the ANS developmental program
Previous studies have shown that Ectopic expression of Hand2 in primary NC stem cell cultures enhances the differentiation of catecholaminergic neurons (Howard and Cserjesi, 1996). However, in the culture conditions used, the NC cells spontaneously differentiated into catecholaminergic neurons suggesting that Hand2 may not be activating the SNS developmental program but may instead promote their terminal differentiation. To investigate if Hand2 can activate the ANS developmental program, we ectopically expressed Hand2 in P19 embryonic carcinoma (P19-EC) cells and examined the ability of Hand2 to activate the neuronal program and more specifically, the SNS developmental program.
P19-EC cells are pleuripotent cells that can be induced to differentiate into CNS neurons when aggregated into embryoid bodies in the presence of retinoic acid (RA) (Bain et al., 1994). As reported previously, transient Hand2 expression or RA addition to monolayer cultures was not sufficient to induce neuronal differentiation (Farah et al., 2000). To determine if Hand2 can activate neuronal development in P19-EC cells after embryoid body formation, we generated cell lines stably expressing Hand2 (P19-H2). When cultured as a monolayer, P19-H2 cell lines are morphologically indistinguishable from control cells and express the inner cell mass antigen SSEA1 indicating they remain pleuripotent (data not shown).
To determine if Hand2 has the ability to activate the neuronal program after embryoid body formation, cells were aggregated, replated, and examined for morphological changes and the expression of neural markers (Fig. 3). P19-EC cells that were aggregated for 4 days and replated did not develop neuronal projections, express the panneuronal marker TuJ1 or the PNS marker peripherin (Fig. 3A, B). We next examined if the neuronal inductive agent RA could activate the PNS developmental program in P19-EC cells. Embryoid bodies were formed and treated with 0.3 µm RA for 4 days, replated, and examined for expression of TuJ1 or peripherin. P19-EC cells express TuJ1 but not peripherin (Fig. 3C, D). When P19-H2 cells were aggregated and replated in the absence of RA, the cultures developed extensive neuronal projections, and cells expressed TuJ1 and peripherin (Fig. 3E, F). The expression of peripherin in the P19-H2 cultures suggests that Hand2 can activate formation of PNS neurons.
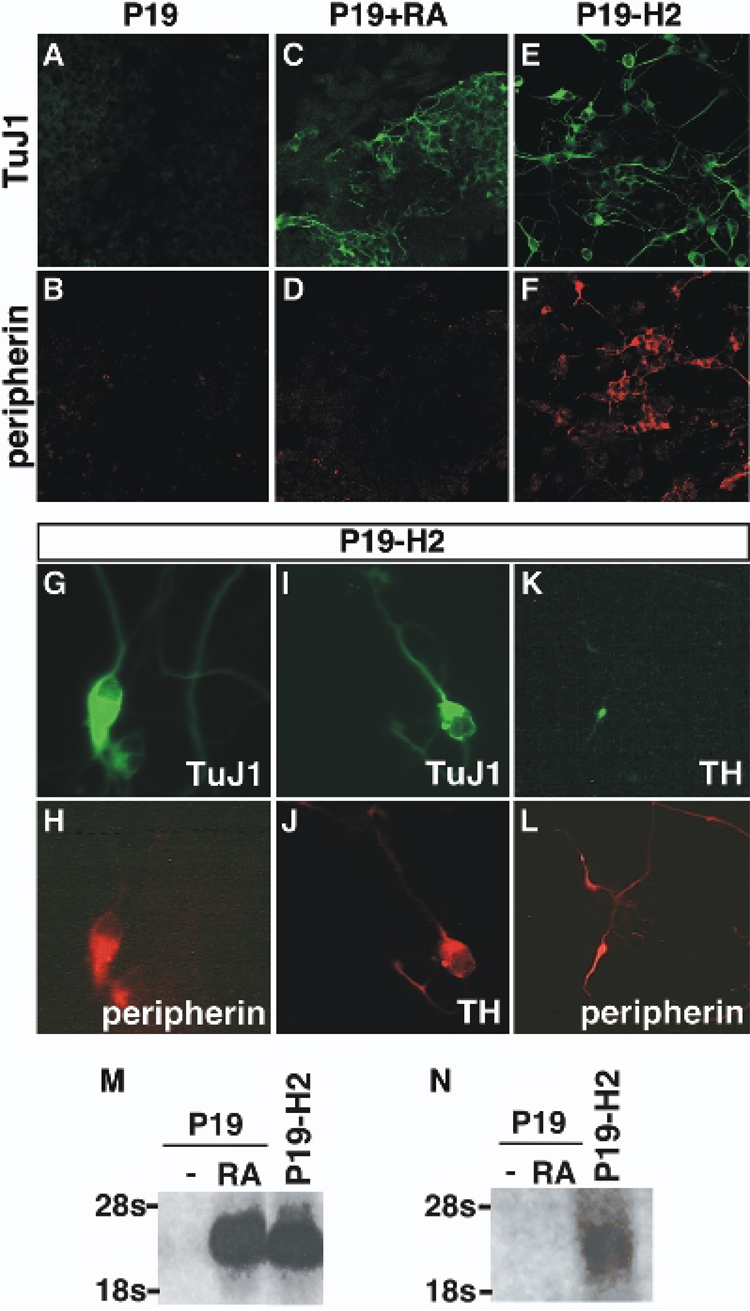
Fig. 3. Hand2 activates neuronal development in P19-EC cells.
The ability of Hand2 to activate the neuronal program was examined by constitutively expressing Hand2 in P19-EC cells. P19-EC and P19-H2 cells were aggregated to form embryoid bodies, allowed to reattach as a monolayer and then examined for expression of neuronal markers. A–F: Cells were cultured for 4 days after reattachment and analyzed for expression of the pan-neuronal neuronal marker TuJ1 or the peripheral neuronal marker peripherin. P19-EC untreated (A, B), P19-EC treated with RA (C, D) and P19-H2 untreated (E, F) cells were examined for expression of TuJ1 (A, C, E) and peripherin (B, D, F). G–L: Untreated P19-H2 embryoid bodies were dissociated, re-plated and cultured for 12 days prior to analysis. P19-H2 cultures were examined for the co-expression of the pan-neural marker TuJ1 (G) and peripherin (H), co-expression of TuJ1 (I) and TH (J), and for TH (K) and peripherin (L). M–N: Northern analysis of Mash1 (M) and endogenous Hand2 (N) gene expression in P19-EC and P19-H2 cultures was performed 4 days after replating embryoid bodies.
In the developing nervous system, Hand2 expression is restricted to the peripheral autonomic lineage suggesting a role in the development of peripheral neurons. To determine if the neurons that form in the P19-H2 cultures are all peripheral neurons or a mixture of CNS and PNS neurons, we examined if all TuJ1 expressing cells also co-express the peripheral neuronal marker peripherin (Fig. 3G, H). We found that all neurons that express the pan-neuronal marker TuJ1 co-express peripherin. Because Hand2 has been shown to enhance SNS development in cultured neural crest cells (Howard et al., 1999), we asked if Hand2 also activates the SNS differentiation program in P19-EC cells. We found that a subset of Hand2 induced neurons, which express the pan-neuronal marker TuJ1, also co-express TH (Fig. 3I, J). To confirm that these catecholaminergic cells are indeed SNS neurons, we examined the co-localization of peripherin and TH (Fig. 3K, L). All Hand2 induced TH expressing neurons also expressed peripherin showing that expression of Hand2 is able to activate the SNS developmental program in P19-EC cells.
The SNS developmental program involves activation of a number of transcription factors including Mash1 and Hand2. To determine if the Hand2 induced neuronal development in P19-EC cells involves activation of these bHLH factors, we examined their expression by Northern analysis. The generation of embryoid bodies from P19-EC cells is not sufficient to induce neuronal differentiation or the expression of Mash1 while RA treatment robustly activated both (Fig. 3M). When P19-H2 cells are aggregated and replated in the absence of RA, Hand2 was able to activate Mash1 expression. To further investigate the gene hierarchy induced by Hand2 expression in P19-EC cells, we examined the ability of Hand2 to activate expression of the endogenous Hand2 gene. Aggregation of P19-EC cultures and activation of the neuronal program with RA is insufficient to activate the endogenous Hand2 gene (Fig. 3N). However, aggregation of P19-H2 cultures is sufficient to activate endogenous Hand2 expression (Fig. 3N). These data suggests that Hand2 regulates the ANS developmental program by auto and cross regulation of the ANS transcriptional network.
Hand2 activates expression of Mash1 by a p300-dependent mechanism
Hand2 has been shown to act through recruitment of the histone acetyltransferase p300 during cardiac gene regulation (Dai et al., 2002). To determine if Hand2’s function in the activation of neuronal development also depends on p300 recruitment, we examined Hand2’s dependence on p300 in the activation of Mash1 (Fig. 4). Hand2 was expressed in the mouse neuroblastoma cell line, neuro-2a, a cell line that does not normally express Mash1. Neuro2a cells were transfected with the dual promoter construct pEGFPJH-H2 that expresses Hand2 and EGFP from independent promoters. Transfection was monitored by EGFP expression (Fig. 4A) and Mash1 expression was determined by immunostaining with Mash1 monoclonal antibody (Fig. 4B). In Hand2 expressing cells, over 80% of cells co-express Mash1. Although Hand2 activated Mash1 in neuro-2a cells, it was unable to promote terminal differentiation, which may reflect the transformed nature of this cell line.
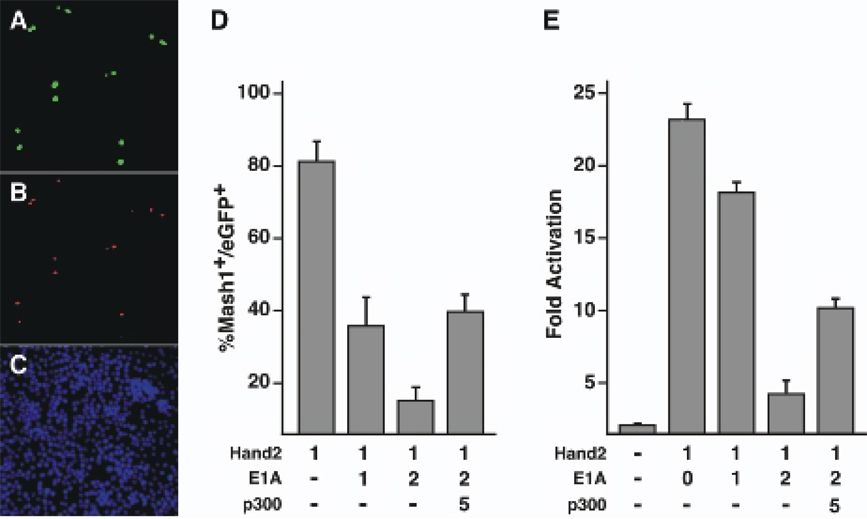
Fig. 4. Hand2 activation of Mash1 and transcriptional activity is dependent on p300/CBP.
The role of p300/CBP in Hand2 function was examined by inhibition of p300/CBP function with E1A. (A–C) Neuro2a cells were transfected with pEGFP-HAND2 and monitored for co-expression of Hand2 by GFP fluorescence (A), expression of Mash1 with a Mash1 specific monoclonal antibody (B) and cell number with DAPI nuclear staining (C). Mash1 expression was activated in over 80% of cells co-expressing Hand2 and eGFP. (D) Hand2 was co-expressed with the p300/CBP inhibitor protein E1A (µg of plasmid transfected). Hand2 activation of Mash1 expression was inhibited in a dose dependent manner by E1A. Co-expression of p300 rescues the inhibitory effect of E1A suggesting E1A inhibition targets the Hand2-p300/CBP complex. (E) Expression of E1A repressed the transcriptional activity of Hand2 in a dose dependent manner that was overcome by co-expression of p300.
To determine if p300 or the functionally redundant factor CBP, was an essential co-factor for Hand2 dependent Mash1 activation, Hand2 was co-expressed with the p300/CBP inhibitory protein E1A (Fig. 4D). The activation of Mash1 by Hand2 was repressed by E1A in a dose dependent manner. To determine if the effect of E1A was due to an inhibition of p300 or by non-specific repression, p300 was co-expressed with Hand2 and E1A to determine if p300 could rescue E1A repression of Mash1. E1A inhibition of Mash1 expression was rescued by p300 (Fig. 4D) suggests that Hand2 activation of Mash1 requires the recruitment of p300/CBP.
To analyze the mechanism by which p300 regulates Hand2 function, we examined its ability to regulate the transcriptional activity of Hand2 using a mammalian one-hybrid assay (Fig. 4E). We fused Hand2 with the yeast Gal4 minimal DNA binding domain to create a chimeric protein dependent on Hand2 for transcriptional activity but not DNA binding. The Gal4-Hand2 fusion plasmid and a CAT reporter plasmid containing five Gal4 DNA binding sites upstream of the E1b basal promoter was transfected into neuro-2a cells. Hand2 activated transcription by 23 fold. Co-transfection of E1A with Gal4-Hand2 reduced transcriptional activity in a dose-dependent manner. E1A repression of Hand2 transcription is rescued by co-transfection with p300 showing that E1A repression is acting through p300/CBP. Taken together, our results suggest that Hand2 promotes neuronal development by functioning in a transcriptional complex that includes p300/CBP.
DISCUSSION
Our understanding of the transcriptional network regulating ANS development remains incomplete. Only two transcription factors, Phox2b and Mash1, have been assigned essential early roles in ANS development. Phox2b is expressed soon after NC cells have reached the dorsal aorta and have become specified to form the SNS suggesting it may function as an early determination factor. However, genetic analysis has shown it is not required for the initial formation of the SNS, based upon the transient expression of Mash1 and β-galactosidase knock into the Phox2b gene (Pattyn et al., 1999). Phox2b is required for the continued expression of Mash1, a bHLH transcription factor that is required for differentiation and survival of the SNS during terminal differentiation but not its initial formation (Guillemot et al., 1993; Sommer et al., 1995; Pattyn et al., 1999). Genetic analysis in mice suggests that these two transcription factors are not essential for commitment of NC cells to an ANS lineage but are essential for their survival. In addition, both factors are expressed in a number of neuronal lineages other than the ANS including the CNS, further suggesting that these factors, whether acting alone or in concert, are insufficient to specifically activate the ANS developmental program. These data suggest that other transcription factors must be involved in the specification and differentiation of the ANS.
The work presented here demonstrates that Hand2 is a member of the gene regulatory network that controls ANS development. Unlike other neuronal transcription factors, Hand2 is expressed exclusively in the ANS lineage, indicating that it plays a unique role in its development. We have shown that the expression of Hand2 and Mash1 begins concurrently, or within a short time of each other. The close timing of expression suggests that Hand2 is regulated in parallel with Mash1 and expression of these two bHLH transcription factors may initially be co-regulated by an upstream factor. However, it has been reported that Hand2 expression is dependent on Mash1 (Anderson and Jan, 1998; Goridis and Rohrer, 2002) suggesting a linear relationship where Hand2 is downstream and regulated by Mash1. Our analysis of Mash1 null mice shows that Hand2 expression is independent of Mash1 during SNS differentiation. Although there is reduced Hand2 expression in the SNS of Mash1 null embryos, this is due to a reduced cell number, not a dependence on Mash1. After Mash1 is no longer expressed, Hand2 continues to be expressed during development and in the adult SNS, indicating that Mash1 does not play a role in regulating Hand2. However, our finding that Hand2 is expressed in the adult SNS does support a role in maintaining the noradrenergic phenotype by regulating expression of the noradrenalin biosynthetic enzymes dopamine-ß-hydroxylase (DBH) gene in combination with Phox2 (Kim et al., 1998; Yang et al., 1998; Rychlik et al., 2003; Xu et al., 2003).
When ectopically expressed in NC derived cells, Hand2 has been shown to activate the SNS developmental program (Howard et al., 1999; Howard et al., 2000). Whether this activity is permissive or instructive is unclear. We have extended these results by demonstrating that Hand2 activates the ANS neuronal developmental program in P19 EC cells, a cell line not predisposed to form ANS neurons. This supports an instructive role for Hand2 in neuronal specification. The ability of Hand2 to activate the ANS program and the expression of Mash1 also suggests it acts upstream of Mash1 and functions as a proneural gene. However, in the embryo, Hand2 expression begins after Phox2b, a marker for NC cells committed to ANS lineage (Howard et al., 2000). Since expression of Hand2 begins after ANS specification in vivo, Hand2 does not initiate the ANS program. The proneural activity of Hand2 in vitro most likely reflects an ability to activate neuronal development through a cross-regulatory mechanism where Hand2 activates other members of the transcriptional network that act concurrently during ANS development. This supports a parallel transcriptional regulatory model for ANS development in which Hand2, Mash1 and Phox2 genes act in parallel to promote differentiation (Fig. 5). The genes act in concert during neuroblast to neuron transition with Hand2 and Phox2 continuing to function in the maintenance of the catecholaminergic phenotype by directly regulating expression of the DBH gene (Rychlik et al., 2003; Xu et al., 2003). However, the ability of Hand2 to activate the ANS-specific developmental program in non-NC cells appears to be a property unique to Hand2.
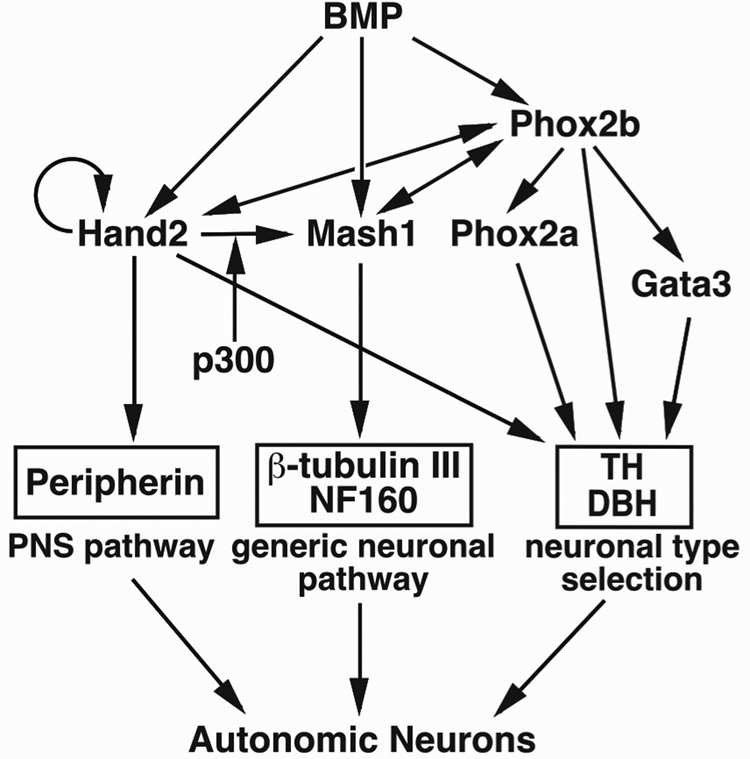
Fig. 5. Transcriptional regulatory network during development of the ANS.
During ANS development, transcription factors act in a network to induce other transcription factors. BMP is an inductive cue for the SNS lineage and activates the expression of the transcription factors Phox2b (Lo et al., 1998; Schneider et al., 1999), Mash1 (Shah et al., 1996; Lo et al., 1998) and Hand2 (Howard et al., 2000). The bHLH factor Mash1, is able to cross-activate the paired homeodomain transcription factor Phox2b (Lo et al., 1998; Lo et al., 1999) which is required for maintenance of Mash1 expression (Pattyn et al., 1999) and for expression of another bHLH factor Hand2 (Goridis and Rohrer, 2002). Hand2 activates Phox2b (Howard et al., 2000), itself and Mash1 (this study). These three factors together activate the expression of the neuronal structural genes. Each factor activates different pathways that converge during autonomic neuronal development.Generic neuronal pathway: Expression of Mash1 is sufficient for the expression of generic neuronal structural genes such as TuJ1 and NF160 (Farah et al., 2000). Mash1 is required for the expression of the noradrenergic biosynthetic genes TH, DBH (Guillemot et al., 1993) and peripherin (Sommer et al., 1995) but transcription regulation of these genes must be indirect since Mash1 expression stops at mid-gestation (Guillemot and Joyner, 1993). Neuronal type selection: Phox2b regulates expression of the zinc finger factor GATA3 (Tsarovina et al., 2004) and Phox2a (Pattyn et al., 1999). Phox2a/b and Gata3 directly regulate the expression of noradrenergic biosynthetic enzymes TH and DBH in sympathetic neurons (Kim et al., 1998; Yang et al., 1998; Lim et al., 2000) and may regulate other neuronal genes in parasympathetic and enteric neurons. Peripheral pathway: Hand2 is only expressed in the ANS lineages of the nervous system suggesting a role in its specification (Srivastava et al., 1995; Dai et al., 2004). Ectopic expression of Hand2 activates the PNS program and regulates the expression of TH in the SNS. The ability of Hand2 to activate Mash1 is p300/CBP dependent.
The ability of Hand2 to activate other members of the transcriptional network regulating ANS development is through recruitment of p300. An interaction between p300 and Hand2 has also been show to occur during regulation of the ANF gene (Dai et al., 2002). During ANF regulation, the interaction between Hand2 and p300 is direct and forms a complex that includes the zinc finger transcription factor GATA4 (Dai et al., 2002). The formation of a transcriptional complex between a bHLH factor and p300 is not unique to Hand2. Other members of the Twist family (Hamamori et al., 1999; Huang et al., 1999) and the atonal and achaete-scute families of neuronal bHLH factors (Sun et al., 2001; Vojtek et al., 2003) also require formation of a transcriptional complex with p300/CBP for their function. Further support of the requirement for Hand2 to act through the formation of multi-protein complexes to regulate developmental events is its ability to regulate limb development in the absence of a DNA binding domain (McFadden et al., 2002). The ability of Hand2 to function without direct DNA binding suggests it can function through protein-protein interactions in vivo.
Because the p300/CBP proteins bind a large number of transcription factors (Reviewed in (Goodman and Smolik, 2000), the number of factors modulating Hand2 function during ANS development may also be large and dynamic. The requirement for Hand2 to bind p300/CBP may also explain the apparent contradictory role of Hand2 regulation of Mash1. Hand2 activates Mash1 yet continues to be expressed in SNS neurons after cessation of Mash1 expression. During early SNS development, Hand2 can recruit p300/CBP to a transcriptional complex that activates Mash1 transcription, as development proceeds, this ability is lost. This dynamic interaction between Hand2 and p300/CBP may occur through the extensive post-translational modifications that have been reported previously to affect the transcriptional activity of Hand2 (Firulli et al., 2005).
ACKNOWLEDMENTS
The Mash1 mutant mice were a gift from David Anderson (California Institute of Technology, Pasadena). The Mash1 monoclonal antibody was a gift from Jane Johnson (University of Texas, Southwestern, Dallas) and peripherin antibody from Ron Liem (Columbia University). We are grateful to Judith Venuti and Mark Alliegro for critical reading of the manuscript and wish to thank Taneasha Washington and Ralston Barnes for their technical assistance.
This work was supported by an NIH training grant (5T32 NS07062) to K. B. and by support to P. C. from the American Cancer Society, Whitehall Foundation, NSF (IBN-0345924), and NIH (2RO1NS015547-22).
REFERENCES
- Anderson DJ, Jan YN. The determination of the neuronal phenotype. In: Cowan WM, Jessell TM, Zimursky SL, editors. Molecular and Cellular Approaches to Neural Development. Oxford, UK: Oxford University Press; 1998. pp. 26–63. [Google Scholar]
- Bain G, Ray WJ, Yao M, Gottlieb DI. From embryonal carcinoma cells to neurons: the P19 pathway. Bioessays. 1994;16:343–348. doi: 10.1002/bies.950160509. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Dai YS, Cserjesi P. The basic helix-loop-helix factor, HAND2, functions as a transcriptional activator by binding to E-boxes as a heterodimer. J Biol Chem. 2002;277:12604–12612. doi: 10.1074/jbc.M200283200. [DOI] [PubMed] [Google Scholar]
- Dai YS, Cserjesi P, Markham BE, Molkentin JD. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J Biol Chem. 2002;277:24390–24398. doi: 10.1074/jbc.M202490200. [DOI] [PubMed] [Google Scholar]
- Dai YS, Hao J, Bonin C, Morikawa Y, Cserjesi P. JAB1 enhances HAND2 transcriptional activity by regulating HAND2 DNA binding. J Neurosci Res. 2004;76:613–622. doi: 10.1002/jnr.20105. [DOI] [PubMed] [Google Scholar]
- Eckner R, Yao TP, Oldread E, Livingston DM. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Gene Develop. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Firulli BA, Krawchuk D, Centonze VE, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. Phosphorylation regulates Twist1 and Hand2 dimerization and implicates a mechanism for Saethre-Chotzen Syndrome. Nat Genet. 2005 doi: 10.1038/ng1525. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Joyner AL. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech Dev. 1993;42:171–185. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L. Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell. 1999;96:405–413. doi: 10.1016/s0092-8674(00)80553-x. [DOI] [PubMed] [Google Scholar]
- Howard M, Foster DN, Cserjesi P. Expression of HAND gene products may be sufficient for the differentiation of avian neural crest-derived cells into catecholaminergic neurons in culture. Dev Biol. 1999;215:62–77. doi: 10.1006/dbio.1999.9450. [DOI] [PubMed] [Google Scholar]
- Howard MJ, Cserjesi P. Society for Neuroscience. Washington: 1996. Chicken eHAND and dHAND influence neural crest cell differentiation; p. 7. [Google Scholar]
- Howard MJ, Stanke M, Schneider C, Wu X, Rohrer H. The transcription factor dHAND is a downstream effector of BMPs in sympathetic neuron specification. Development. 2000;127:4073–4081. doi: 10.1242/dev.127.18.4073. [DOI] [PubMed] [Google Scholar]
- Huang S, Qiu Y, Stein RW, Brandt SJ. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene. 1999;18:4958–4967. doi: 10.1038/sj.onc.1202889. [DOI] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Genetic control of cell fate specification in Drosophila peripheral nervous system. Annu Rev Genet. 1994;28:373–393. doi: 10.1146/annurev.ge.28.120194.002105. [DOI] [PubMed] [Google Scholar]
- Kim HS, Seo H, Yang C, Brunet JF, Kim KS. Noradrenergic-specific transcription of the dopamine beta-hydroxylase gene requires synergy of multiple cis-acting elements including at least two Phox2a-binding sites. J Neurosci. 1998;18:8247–8260. doi: 10.1523/JNEUROSCI.18-20-08247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet. 2000;25:209–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- Lo L, Morin X, Brunet JF, Anderson DJ. Specification of neurotransmitter identity by Phox2 proteins in neural crest stem cells. Neuron. 1999;22:693–705. doi: 10.1016/s0896-6273(00)80729-1. [DOI] [PubMed] [Google Scholar]
- Lo L, Tiveron MC, Anderson DJ. MASH1 activates expression of the paired homeodomain transcription factor Phox2a, and couples pan-neuronal and subtype-specific components of autonomic neuronal identity. Development. 1998;125:609–620. doi: 10.1242/dev.125.4.609. [DOI] [PubMed] [Google Scholar]
- McFadden DG, McAnally J, Richardson JA, Charite J, Olson EN. Misexpression of dHAND induces ectopic digits in the developing limb bud in the absence of direct DNA binding. Development. 2002;129:3077–3088. doi: 10.1242/dev.129.13.3077. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Extra-embryonic vasculature development is regulated by the transcription factor HAND1. Development. 2004;131:2195–2204. doi: 10.1242/dev.01091. [DOI] [PubMed] [Google Scholar]
- Morin X, Cremer H, Hirsch MR, Kapur RP, Goridis C, Brunet JF. Defects in sensory and autonomic ganglia and absence of locus coeruleus in mice deficient for the homeobox gene Phox2a. Neuron. 1997;18:411–423. doi: 10.1016/s0896-6273(00)81242-8. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- Reissmann E, Ernsberger U, Francis-West PH, Rueger D, Brickell PM, Rohrer H. Involvement of bone morphogenetic protein-4 and bone morphogenetic protein-7 in the differentiation of the adrenergic phenotype in developing sympathetic neurons. Development. 1996;122:2079–2088. doi: 10.1242/dev.122.7.2079. [DOI] [PubMed] [Google Scholar]
- Rychlik JL, Gerbasi V, Lewis EJ. The interaction between dHAND and Arix at the dopamine beta-hydroxylase promoter region is independent of direct dHAND binding to DNA. J Biol Chem. 2003;278:49652–49660. doi: 10.1074/jbc.M308577200. [DOI] [PubMed] [Google Scholar]
- Schneider C, Wicht H, Enderich J, Wegner M, Rohrer H. Bone morphogenetic proteins are required in vivo for the generation of sympathetic neurons. Neuron. 1999;24:861–870. doi: 10.1016/s0896-6273(00)81033-8. [DOI] [PubMed] [Google Scholar]
- Shah NM, Groves AK, Anderson DJ. Alternative neural crest cell fates are instructively promoted by TGFbeta superfamily members. Cell. 1996;85:331–343. doi: 10.1016/s0092-8674(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Sommer L, Shah N, Rao M, Anderson DJ. The cellular function of MASH1 in autonomic neurogenesis. Neuron. 1995;15:1245–1258. doi: 10.1016/0896-6273(95)90005-5. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Stanke M, Junghans D, Geissen M, Goridis C, Ernsberger U, Rohrer H. The Phox2 homeodomain proteins are sufficient to promote the development of sympathetic neurons. Development. 1999;126:4087–4094. doi: 10.1242/dev.126.18.4087. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Tiveron MC, Hirsch MR, Brunet JF. The expression pattern of the transcription factor Phox2 delineates synaptic pathways of the autonomic nervous system. J Neurosci. 1996;16:7649–7660. doi: 10.1523/JNEUROSCI.16-23-07649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Moriyoshi K, Nakanishi S, Guillemot F, Kageyama R. Mammalian achaete-scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. Embo J. 2000;19:5460–5472. doi: 10.1093/emboj/19.20.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsarovina K, Pattyn A, Stubbusch J, Muller F, Van Der Wees J, Schneider C, Brunet JF, Rohrer H. Essential role of Gata transcription factors in sympathetic neuron development. Development. 2004;131:4775–4786. doi: 10.1242/dev.01370. [DOI] [PubMed] [Google Scholar]
- Varley JE, Maxwell GD. BMP-2 and BMP-4, but not BMP-6, increase the number of adrenergic cells which develop in quail trunk neural crest cultures. Exp Neurol. 1996;140:84–94. doi: 10.1006/exnr.1996.0118. [DOI] [PubMed] [Google Scholar]
- Varley JE, McPherson CE, Zou H, Niswander L, Maxwell GD. Expression of a constitutively active type I BMP receptor using a retroviral vector promotes the development of adrenergic cells in neural crest cultures. Dev Biol. 1998;196:107–118. doi: 10.1006/dbio.1998.8853. [DOI] [PubMed] [Google Scholar]
- Vojtek AB, Taylor J, DeRuiter SL, Yu JY, Figueroa C, Kwok RP, Turner DL. Akt regulates basic helix-loop-helix transcription factor-coactivator complex formation and activity during neuronal differentiation. Mol Cell Biol. 2003;23:4417–4427. doi: 10.1128/MCB.23.13.4417-4427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Firulli AB, Zhang X, Howard MJ. HAND2 synergistically enhances transcription of dopamine-beta-hydroxylase in the presence of Phox2a. Dev Biol. 2003;262:183–193. doi: 10.1016/s0012-1606(03)00361-0. [DOI] [PubMed] [Google Scholar]
- Yang C, Kim HS, Seo H, Kim CH, Brunet JF, Kim KS. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for noradrenergic cell-specific transcription of the dopamine beta-hydroxylase gene. J Neurochem. 1998;71:1813–1826. doi: 10.1046/j.1471-4159.1998.71051813.x. [DOI] [PubMed] [Google Scholar]