Abstract
A novel cancer targeted, internally cationic, and surface neutral Polyamidoamine (PAMAM) dendrimer was designed, synthesized, and evaluated as a nanocarrier for the targeted intracellular delivery of siRNA. The dendrimer contained a synthetic analog of Luteinizing Hormone-Releasing Hormone as cancer targeting moiety. The proposed delivery system possesses the following advantages: (1) internal cationic charges for complexation with siRNA and enhanced siRNA protection; (2) low cytotoxicity; (3) lesser degree of quaternization offering free tertiary amines for potential proton sponge effect; and (4) targeting specifically to cancer cells for enhancing siRNA uptake and efficiency and potential limitation of adverse side effects of chemotherapy on healthy organs. Both non-targeted and targeted dendrimer-siRNA complexes formed compact nanometer size spherical particles, exhibited very low cytotoxicity even at the higher concentration, and efficiently penetrated cancer cells in vitro. However, only the targeted dendrimer-siRNA complex was able to substantially decrease the expression of a targeted BCL2 gene.
Keywords: PAMAM dendrimer, Degree of quaternization, LHRH, siRNA, BCL2
Introduction
RNA interference is a natural process of sequence-specific, posttranscriptional gene silencing mediated by short double stranded RNA1. Given the ability to interfere with the disease-causing proteins at an early stage of gene expression, short interfering RNAs (siRNAs) have generated considerable attention as potential therapeutic agents for the treatment of cancer and other related diseases2-5. However, low resistance against enzymatic degradation, limited permeability across cell membranes, and substantial liver and renal clearance has restricted therapeutic applications of siRNA in vivo6. Consequently, the development of efficient delivery systems which will protect siRNA from the degradation during the voyage in the bloodstream and organs and facilitate its uptake by the targeted cells is one of the major challenges in the therapeutic application of siRNA. Recently, we reported novel, surface neutral, and internally cationic poly(amidoamine) (QPAMAM) generation four dendrimers for the efficient intracellular delivery of siRNA, including surface acetylated QPAMAM-NHAc and hydroxyl-terminated QPAMAM-OH dendrimers7. These dendrimers as nanocarriers possess the following advantages: (1) neutral surface of the dendrimer for low cytotoxicity; (2) existence of cationic charges inside the dendrimer (not on the outer surface) resulting in highly organized compact nanoparticles, which can potentially protect nucleic acids from degradation. Noteworthily, surface modified QPAMAM-NHAc dendrimer demonstrated enhanced cellular uptake of siRNA when compared with the internally cationic QPAMAM-OH dendrimer (degree of quaternization 97%). In the present study, to improve the siRNA delivery using the QPAMAM-OH dendrimer, we investigated two independent strategies: 1) degree of quaternization for proton sponge or buffering effect; and 2) targeting ligand as a penetration enhancer. Polyethyleneimine (PEI) and Polyamidoamine (PAMAM) dendrimers, exhibit high transfection efficiency due to buffering or the so-called proton sponge effect resulting from low pKa of tertiary amines8, 9. It was hypothesized that substitution of tertiary amines in polyamidoamine (PAMAM-OH) dendrimers with permanently charged quaternary amines may perhaps obstruct the transfection efficiency of siRNA10. We anticipated improved transfection efficiency by decreasing the degree of quaternization that would enable tertiary amines for buffering effect and quaternary amines for the complexation with siRNA.
Besides the proton sponge effect, conjugation of cell penetrating peptides to the macromolecular cargo represents another attractive method that is known to facilitate delivery of genetic materials across cell membranes with high efficiency11-13. In particular, cell penetrating peptides greatly improved internalization and effectiveness of antisense oligonucleotides into the mammalian cells14, 15. Cell penetrating peptides are positively charged short peptides that improve cellular uptake of different payloads including negatively charged chunks of DNA/RNA and their neutral complexes with macromolecules. While cell penetrating peptides possess distinct advantages in antisense oligonucleotides delivery, they were substantially less effective in enhancing the delivery of siRNA and siRNA-nanocarrier complexes16-18. Additionally, cell penetrating peptides enhance cellular uptake by virtually all cells in the body, not only by targeted cells, e.g. cancer cells in case of cancer treatment. Therefore, other penetration enhancers that are specific for targeted cells are required19.
Previously, we have successfully used a synthetic analog of Luteinizing Hormone-Releasing Hormone (LHRH peptide) for the effective targeting of anticancer drugs, therapeutic peptides, and different complex delivery systems to cancer cells in vitro and in vivo20-24. LHRH peptide is a targeting ligand to LHRH receptors that are over-expressed in the plasma membrane of several types of cancer cells and are not expressed detectably in normal visceral organs. Our previous findings in ovarian, breast, and prostate cancer cells provided the rationale of using LHRH peptide as a targeting moiety/penetration enhancer to target different drug delivery systems to tumors and facilitate their uptake by cancer cells. Resulting from this success, an introduction of a targeting LHRH peptide to the QPAMAM-OH dendrimer was envisaged to enhance the intracellular delivery of siRNA by a receptor mediated endocytosis pathway. The present study is aimed at designing, synthesizing, and validating different cancer-targeted QPAMAM-OH dendrimers as nanocarriers for the enhanced intracellular delivery of siRNA to cancer cells. The impact of two independent factors (degree of quaternization and LHRH targeting ligand) is also examined.
Material and Methods
Materials
Generation four PAMAM-OH dendrimer (Mw ∼ 14277 Da, 64 hydroxyl end groups, 1,2-diaminoethane dendrimer core), N,N’-dimethylformamide, succinic anhydride, 4-(methylamino)pyridine and methyl iodide were purchased from Sigma-Aldrich Co. (St. Louis, MO). Fluorescein isothiocyanate (FITC) and N-(3-dimethylaminopropyl)-N-ethylcarbodimide hydrochloride (EDC·HCl) were obtained from Fluka (Allentown, PA). Synthetic analog of LHRH, Lys6-des-Gly10-Pro9-ethylamide (Gln-His-Trp-Ser-Tyr-d-Lys-Leu-Arg-Pro-NH-Et), having a reactive amino group only on the side chain of the lysine at position 6 was synthesized according to our design by American Peptide Company, Inc. (Sunnyvale, CA). Spectra/Pore dialysis membranes with the molecular weight cutoff of 2000 and 500 Da were obtained from Spectrum Laboratories, Inc. (Rancho Dominguez, CA). Fluorescent RNA duplex - siRNA labeled with Pierce NuLight™ DY-547 fluorophores (siGLO Red Transfection Indicator, red fluorescence) was obtained from Applied Biosystems (Ambion, Inc., Foster City, CA). siRNA targeted to BCL2 mRNA was synthesized by Applied Biosystems (Ambion, Inc., Foster City, CA). The sequence of siRNA was 5′-GUGAAGUCAACAUGCCUGCTT-3′. All other chemicals were purchased from Fisher Scientific (Fairlawn, NJ).
Cell line
The human ovarian carcinoma A2780 cell line was obtained from Dr. T. C. Hamilton (Fox Chase Cancer Center). Cells were cultured in RPMI 1640 medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (Fisher Scientific, Fairlawn, NJ). Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 (v/v) in air. All experiments were performed on cells in the exponential growth phase.
Synthesis of internally quaternized QPAMAM-OH dendrimer
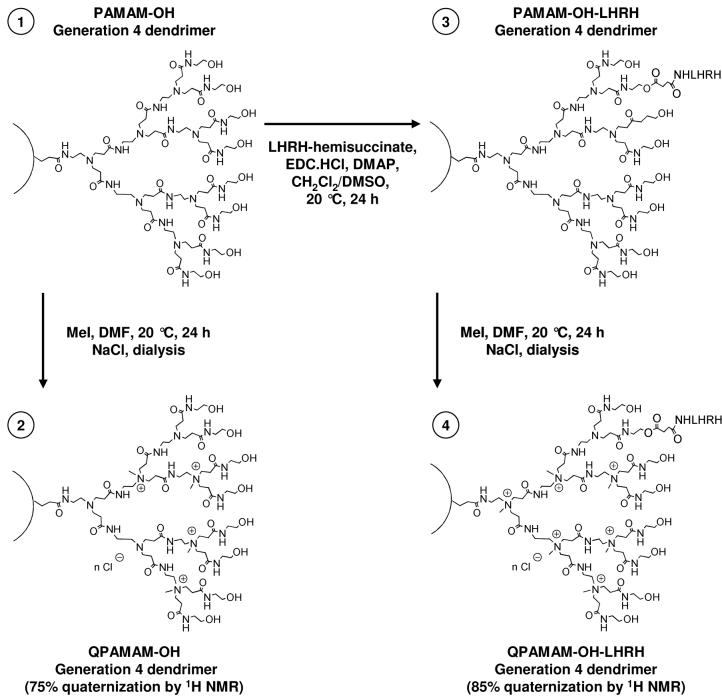
QPAMAM-OH dendrimer with 75% degree of quaternization was prepared using the previously described procedure7. Briefly, PAMAM-OH generation four (172 mg, 0.012 mmol, Fig. 1.1) was dissolved in N,N’-dimethylformamide (DMF, 1 mL) and methyl iodide (MeI, 50 μL) was added. The reaction mixture was sealed and stirred at room temperature for 24 h. The reaction mixture was then precipitated into diethyl ether to obtain solid, which was dried under a vacuum and redissolved in water (1 mL). The resulting solution was dialyzed against 2 M NaCl and deionizer water successively using a dialysis membrane (molecular mass cut off 2000 Da) and then lyophilized to afford the QPAMAM-OH dendrimer (Fig. 1.2) as white solid. Internally quaternized PAMAM dendrimers with various degree quaternization were synthesized by using appropriate mole equivalents of methyl iodide. To enforce a higher degree of quaternization, excess methyl iodide was used and the reaction was conducted at an elevated temperature.
Fig. 1.
Synthesis of internally quaternized non-targeted QPAMAM-OH and targeted QPAMAM-OH-LHRH dendrimers with 75% and 85% of quaternization.
Synthesis of PAMAM-OH-LHRH conjugate
Succinic anhydride (5 mg, 0.05 mmol) was added to a stirred solution of LHRH peptide (50 mg, 0.036 mmol) in anhydrous pyridine (1 mL). The reaction mixture was stirred at room temperature for 24 h. Evaporation of solvents under reduced pressure and purification by extensive dialysis against deionizer water using dialysis membrane (molecular mass cut off 500 Da) followed by freeze-drying afforded LHRH-hemisuccinate as white solid. The obtained conjugate was directly subjected to further reaction with the PAMAM-OH dendrimer. Briefly, PAMAM-OH generation four (86 mg, 0.006 mmol, Fig. 1.1) and LHRH-hemisuccinate (17.2 mg, 0.012 mmol) were dissolved in anhydrous dichloromethane (5 mL) and anhydrous dimethyl sulfoxide (5 mL). EDC·HCl (2.5 mg, 0.013 mmol) was added to the above solution as a condensing agent and DMAP (1 mg) was used as a catalyst and the reaction mixture was allowed to stir for 24 h. After evaporation of the solvent, the resulting conjugate was purified by extensive dialysis against deionizer water using dialysis membrane (molecular mass cut off 2000 Da) and passing through a Sephadex column and then lyophilized to afford PAMAM-OH-LHRH (Fig. 1.3) conjugate as pale yellow solid. One potential challenge of the design of our DDS is that the LHRH was conjugated to the dendrimer carrier via ester bond. Such a structure might be susceptible to esterases in circulation when applied in vivo. However our previous studies provided evidence that the conjugation of LHRH peptide through succinate ester to dendrimer or polymer did not affect biological efficiency in vivo20, 24, 25. Moreover, preliminary data showed that delivered by targeting DDS biologically active products, including anticancer drug, antisense oligonucleotides or siRNA, are substantially less effective if non-biodegradable amide bond is used to conjugate LHRH peptide to the carrier.
Synthesis of internally quaternized QPAMAM-OH-LHRH conjugate
Methyl iodide (0.5 mL) was added to a stirred solution of PAMAM-OH-LHRH conjugate (50 mg, Fig. 1.3) dissolved in N,N’-dimethylformamide (1 mL). The reaction mixture was sealed and stirred at room temperature for 24 h. The reaction mixture was then precipitated into diethyl ether to obtain solid, which was dried under a vacuum and redissolved in water (1 mL). The resulting solution was dialyzed against 2 M NaCl and deionizer water successively using dialysis membrane (molecular mass cut off 2000 Da) and then lyophilized to afford QPAMAM-OH-LHRH conjugate (Fig. 1.4).
Synthesis of fluorescein labeled internally quaternized QPAMAM-OH-FITC dendrimer
Fluorescein labeled PAMAM-OH dendrimer was prepared using our procedure that was previously reported26. The resulting PAMAM-OH-FITC was then internally quaternized using the protocol described above.
Proton nuclear magnetic resonance spectroscopy (1H NMR)
1H NMR was performed on a Varian VNMRS 400 MHz NMR spectrometer (Varian, Inc., Palo Alto, CA). The chemical shift was expressed as parts per million (ppm) and a solvent peak was used as reference (D2O, 4.8 ppm). The following abbreviations are used in the results section to identify multiplicities: s, singlet; m, multiplet; br, broad.
Atomic force microscopy
The samples of siRNA-dendrimer condensates were visualized with a tapping mode atomic force microscope (Nanoscope III A, Veeco Digital Instruments, Chadds Ford, PA). During imaging, a 125 μm long rectangular silicon cantilever/tip assembly was used with a spring constant of 40N/m, resonance frequency of 315-352 kHz and a tip radius of 5-10 nm. The images were generated by the change in amplitude of the free oscillation of the cantilever as it interacts with the sample. In order to image siRNA condensates, 5 μL of dendrimer-siRNA solutions were deposited on freshly cleaved mica. After 3-5 minutes of incubation, the mica surface was rinsed with 3 drops of deionized water 4 times and dried under a flow of nitrogen.
In Vitro Cytotoxicity
A modified MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was used to assess the cytotoxicity of the three dendrimers. To measure cytotoxicity, cells were separately incubated in a microtiter plate with different concentrations of QPAMAM-OH and QPAMAM-OH-LHRH dendrimers. Control cells received an equivalent volume of fresh medium. The duration of incubation was 24 h. On the basis of these measurements, cellular viability was calculated for each dendrimer concentration as previously described21, 27, 28. A decrease in the cellular viability indicated an increase in dendrimer toxicity.
Gel electrophoresis
The complexes of dendrimers (QPAMAM-OH and QPAMAM-OH-LHRH) and siRNA were prepared in water at N/P (Nitrogen to Phosphate, which reflects positive to negative charge ratio) ratios ranging from 0 to 3 relative units and incubated at room temperature for 30 min. The charge ratio was calculated by relating the number of positive charges on a dendrimer (quaternary amine groups) with the number of negatively charged phosphate groups of siRNA. Dendrimer-free siRNA was used as the control. The samples were further diluted with DPBS buffer and subjected to agarose gel electrophoresis and staining with ethidium bromide in 4% agarose gel at 100 V for 50 min in Tris-Borate-EDTA buffer. siRNA bands on the gel were visualized under ultraviolet light and digitally photographed.
Dynamic Light Scattering (DLS) analysis and zeta potential
Three types of siRNA complexes with N/P charge ratios of 1, 1.5 and 3 were prepared from dendrimers (QPAMAM-OH and QPAMAM-OH-LHRH) and siRNA in water. The resulting complexes were incubated for 30 min and the size was determined using the DynePro-MS800 dynamic light scattering/molecular sizing instrument with argon laser wavelength λ=830 nm, a detector angle 90°, and typical sample volume of 20 μL. Each light scattering experiment consisted of 20 or more independent readings, each 10 s in duration. Data analysis was conducted using DynaPro Instrument Control Software for molecular Research DYNAMICS (version 5.26.60). The obtained DLS data represents an average of three runs. Zeta potential was measured on PALS Zeta Potential Analyzer (Brookhaven Instruments Corp, New York, NY). Samples were taken as is and their volume was 1.5 mL. All measurements were carried out at room temperature. Each parameter was measured 5 times, and average values were calculated.
Cellular internalization
Two types of experiments were carried out to analyze cellular internalization and intracellular localization of siRNA. In the first experiment, fluorophore labeled siRNA (siGLO Red, red fluorescence) either free or complexed with dendrimers QPAMAM-OH and QPAMAM-OH-LHRH (N/P = 3) was incubated for 17 h with living A2780 human ovarian cancer cells. Fluorescence distribution within the cell from the top to the bottom cell surfaces (z-sections) was examined using a confocal microscope. In another series of experiments, cellular internalization of FITC-labeled dendrimer (green fluorescence) and its complex with fluorophore labeled siRNA (siGLO Red, red fluorescence) were studied in living A2780 human ovarian cancer cells by confocal microscopy. Cells were incubated with non-targeted and targeted dendrimer-siRNA conjugates within 100 min and images were taken with 25 min intervals. Superposition of green (dendrimer) and red (siRNA) fluorescence images allows for detecting co-localization of siRNA with the dendrimer resulting in yellow color.
Gene Expression
siRNA targeted to the BCL2 mRNA was used to study the gene silencing efficacy of siRNA delivered by the dendrimers. In these experiments, the final siRNA concentration in the complexes was 1 μM. A2780 cells were incubated with dendrimer-siRNA complexes and appropriate controls within 24 h and total cellular RNA was isolated using an RNeasy kit (Qiagen, Valencia, CA). The Reverse Transcription-Polymerase Chain Reaction (RT-PCR) procedure was used for the analysis of gene expression as previously described22. First-strand cDNA was synthesized by Ready-To-Go You-Prime First-Strand Beads (Amersham Biosciences, Piscataway, NJ) with 2 μg of total cellular RNA and 100 ng of random hexadeoxynucleotide primer (Amersham Biosciences). After synthesis, the reaction mixture was immediately subjected to PCR, which was carried out using GenAmp PCR System 2400 (Perkin-Elmer, Shelton, CT). β2-microglobulin was used as an internal standard. The following pairs of primers were used: BCL2: 5′-GGA TTG TGG CCT TCT TTG AG-3′ (sense), 5′-CCA AAC TGA GCA GAG TCT TC-3′ (antisense); β2-microglobulin (β2-m,) -ACC CCC ACT GAA AAA GAT GA (sense), ATC TTC AAA CCT CCA TGA TG (antisense). PCR regimen was as follows: 94°C for 5 minutes; 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1 minute for 41 cycles; and 60°C for 10 minutes. PCR products were separated in 4% NuSieve 3:1 Reliant-agarose gels (Lonza, Basel, Switzerland) in 1x Tris-borate EDTA buffer (0.089 mol/L Tris-borate, 0.002 mol/L EDTA, pH 8.3; Research Organics Inc., Cleveland, OH) by submarine electrophoresis. The gels were stained with ethidium bromide and digitally photographed.
Results
Synthesis and characterization of internally quaternized PAMAM dendrimers (QPAMAM-OH)
The internal quaternization of a PAMAM-OH dendrimer was carried out according to the described procedure (Fig. 1). Analysis of the dendrimer by 1H NMR (400 MHz, D2O) revealed the following major peaks (Fig. 2, A): δ 2.46-2.56 (br m, COCH2CH2N), 2.66-2.74 (br m, CONHCH2CH2N) 2.83-2.98 (br m, COCH2CH2N+), 3.16 (br s, N+CH3), 3.32-3.38 (br m, CH2CH2OH), 3.46-3.57 (br m, CONHCH2 CH2N+N), 3.62-3.78 (br m, N+CH2CH2CONH and CH2OH). The degree of quaternization was confirmed by 1H NMR spectroscopy. Proton peaks arising from the (-CH2CH2N-) methylene group adjacent to tertiary nitrogen indicated downfield chemical shift. Furthermore, the exact degree of quaternization (75%) was determined by comparing the integrated peak area of newly introduced methyl group (N+-CH3) at δ 3.16 to that of the unmodified methylene protons (-CH2CH2OH) at δ 3.32-3.38.
Fig. 2.
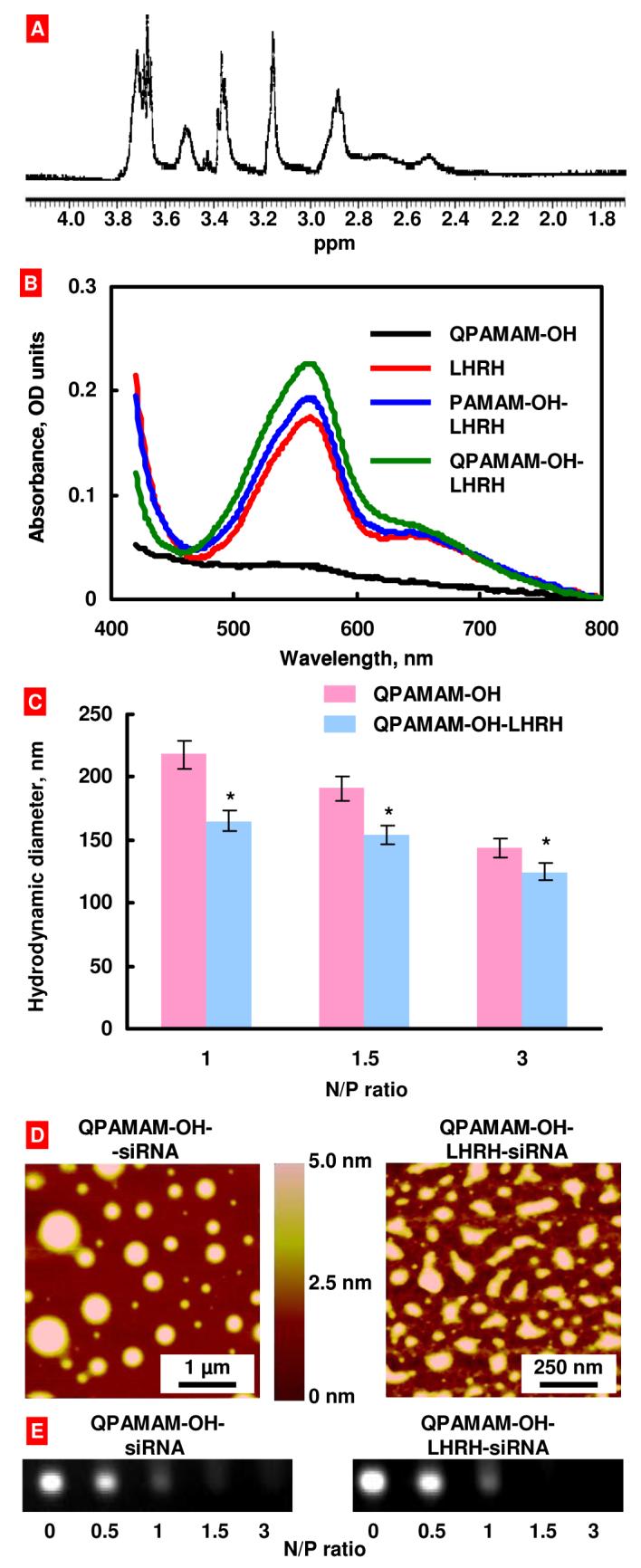
Characterization of synthesized dendrimers. (A) 1H-NMR chart for the QPAMAM-OH dendrimer. (B) UV-visible spectra of different dendrimers. (C) Average particle size of different siRNA-dendrimer complexes determined by dynamic light scattering. (D) Atomic force microscopy images of dendrimer-siRNA complexes. The height differences on the surface are indicated by the color code: lighter regions indicate higher heights. (E) Agarose gel electrophoresis of dendrimer-siRNA complexes.
Synthesis of targeting ligand (LHRH peptide) conjugated and internally quaternized PAMAM dendrimer (QPAMAM-OH-LHRH)
The targeted and internally quaternized PAMAM dendrimer (QPAMAM-OH-LHRH) was synthesized in three simple steps (Fig. 1). During the first step, LHRH analog, Lys6-des-Gly10-Pro9-ethylamide (Gln-His-Trp-Ser-Tyr-D-Lys[D-Cys]-Leu-Arg-Pro-NH-Et), having a reactive primary amine group only on the side chain of the lysine at position 6 was reacted with succinic anhydride to form a LHRH-hemisuccinate. In the second step, PAMAM-OH-LHRH conjugate was synthesized by reacting the PAMAM-OH dendrimer with LHRH-hemisuccinate using EDC·HCl as coupling agent. Finally, the internal quaternization of the PAMAM-OH-LHRH dendrimer was carried out according to the previously described procedure to yield QPAMAM-OH-LHRH conjugate. 1H NMR (400 MHz, D2O) spectral data for this conjugate was similar to that of described for QPAMAM-OH and the exact degree of quaternization was determined to be 85%. Since it was difficult to detect LHRH peptide by proton NMR due to low concentration, a well known BCA protein assay method was used. The presence and concentration of LHRH peptide in targeted dendrimers was detected by a colorimetric method using Pierce Bicinchoninic Acid (BCA) protein assay29 (Thermo Fisher Scientific Inc., Rockford, IL) according to manufacturer recommendations. Typical UV-visible spectra of colored reaction product are presented in Fig. 2, B. The spectra of the product corresponding to free LHRH and all the conjugates containing LHRH have well defined absorbance maximum around 560 nm corresponding to the absorbance of the BCA/copper complex formed as a result of the reaction of BCA reagent with the cuprous cation produced from the reduction of Cu2+ to Cu1+ by the LHRH peptide. This maximum is absent in the assay spectra of the QPAMAM-OH dendrimer that does not contain LHRH. The average estimated concentration of LHRH peptide in working PAMAM-OH-LHRH and QPAMAM-OH-LHRH dendrimer solution (50 μM) was about 50 μg/ml (38 μM). Based on this value one can estimate that the targeted DDS contained an average one LHRH peptide per one dendrimer molecule.
Dynamic Light Scattering, zeta potential and Atomic Force Microscopy
The particle size of each dendrimer-siRNA complex was determined by dynamic light scattering at charge ratio ranging from 1 to 3 relative units as shown in Fig. 2, C. Similar to our previous studies, the complexes formed by using QPAMAM-OH and QPAMAM-OH-LHRH dendrimers exhibited a decrease in particle size as the charge ratio was increased7. Furthermore, atomic force microscopy at N/P ratio 3 relative units revealed formation of well-condensed spherical particles (Fig. 2, D). The average potential of dendrimer-siRNA complexes was 0.11±0.88 mV. Therefore, the dendrimer-siRNA complexes can be considered neutral.
In Vitro Cytotoxicity
In general the cytotoxicity of a dendrimer is greatly influenced by the surface charge of the dendrimer and cytotoxicity increases with an increase in surface charge. The analysis of cytotoxicity of each dendrimer at various concentrations by the MTT assay revealed that the surface neutral and internally charged dendrimers QPAMAM-OH and QPAMAM-OH-LHRH does not lead to the death of more than 5-10% of cells even at relatively high concentrations up to 12.5 μM.
Analysis of dendrimer/siRNA complex formation by agarose gel electrophoresis
Dendrimers QPAMAM-OH and QPAMAM-OH-LHRH, were mixed with siRNA in water at various N/P ratios and were subjected to electrophoresis in agarose gel (Fig. 2, E). The complex formation was observed for both the non-targeted QPAMAM-OH and targeted QPAMAM-OH-LHRH dendrimers as evidenced by oligonucleotide bands disappearance from agarose gels. It should be stressed that at N/P ratios higher than 1 relative unit siRNA band completely disappeared.
Cellular Uptake of siRNA
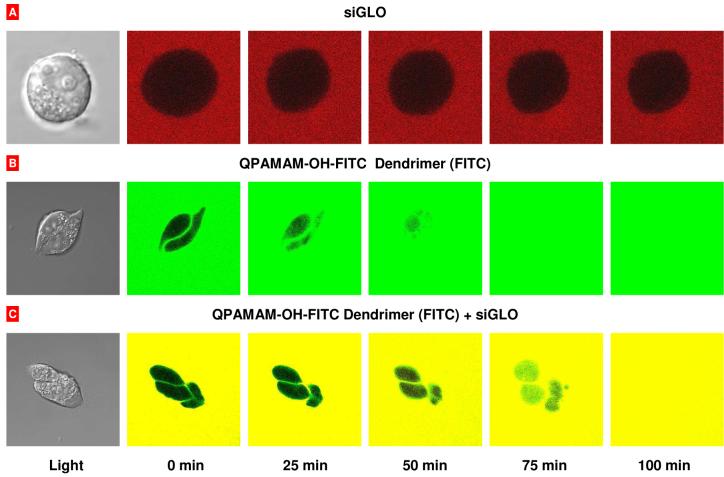
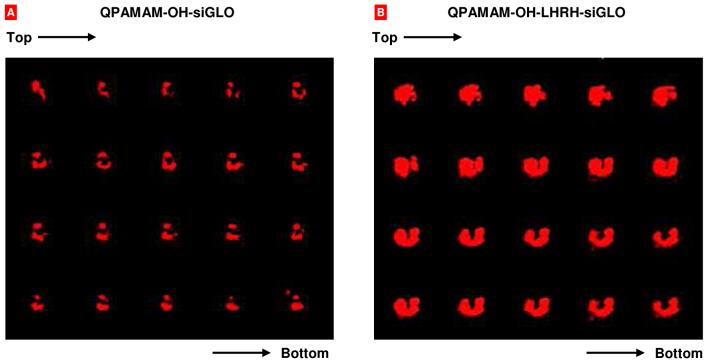
Two series of experiments were carried out to analyze cellular internalization and intracellular localization of free siRNA and siRNA-dendrimer complexes. In the first series, living A2780 cells were separately incubated with free and complexed with dendrimers fluorophore labeled siRNA (siGLO Red, red fluorescence) within 100 min. The dendrimers were labeled with FITC (green fluorescence). The fluorescence of each fluorophore (green and red) was registered with a confocal microscope every 25 min and digitally photographed. Green and red fluorescence images of a dendrimer and siRNA respectively were then digitally overlaid. The results of these experiments are shown in Fig. 3. The data obtained showed that naked siRNA failed to penetrate cancer cells. In contrast, both the dendrimer and dendrimer-siRNA complex were internalized by cancer cells and distributed uniformly in cellular cytoplasm and nucleus resulting in homogenous yellow color on overlaid pictures (Fig. 3, bottom panel). The comparison of cellular internalization of siRNA delivered by dendrimers with different degree of quaternization showed that dendrimers with lower degree of quaternization (20-30%) delivered siRNA more efficiently when compared with those with higher degree (70-85%) of quaternization (data not shown). One can speculate that a uniform distribution of fluorescence on two-dimensional fluorescent images may not reflect a distribution of a fluorescence substance within the cell. Theoretically, a dendrimer and/or siRNA might stick to the surface of the cells and create the impression that these substances are internalized by the cells and distributed through the cytoplasm and nuclei. In order to reject such an assumption and to study the distribution of siRNA delivered by dendrimers in the third dimension (from the top to the bottom of the cell), a special king of measurement, so-called “z-sections”, were performed by a confocal microscope using cells incubated with dendrimers-siRNA complexes and washed out from the excess of unbound substances by free media. In this series, A2780 human ovarian cancer cells were incubated 17 h with dendrimer-siRNA complexes, washed out by fresh media and subjected to confocal microscopy. Data showed that siRNA delivered by both QPAMAM-OH and QPAMAM-OH-LHRH dendrimers were distributed from the top to the bottom of the cell (Fig. 4). A comparison of the left and right panels on Fig. 4 clearly shows that targeting the dendrimer to cancer cells by LHRH peptide substantially enhanced internalization of siRNA conjugated with the QPAMAM-OH-LHRH dendrimer leading to a considerably higher concentration of delivered siRNA inside cancer cells.
Fig. 3.
Cellular internalization of free siRNA and siRNA delivered by the internally quaternized dendrimer. Typical confocal microscopy images of living A2780 cells incubated with fluorophore labeled siRNA (siGLO Red, red fluorescence, A), a dendrimer labeled with FITC (green fluorescence, B) and a dendrimer-siRNA complex (C). Superposition of green and red fluorescence images (C) allows for detecting of co-localization of the dendrimer and siRNA in the solution and inside cells (yellow color).
Fig. 4.
Confocal microscopy images of human A2780 ovarian carcinoma cells incubated for 17 hours with non-targeted (A) and cancer targeted (B) dendrimer-siGLO complexes (z-series, from the top of the cell to the bottom).
Gene Expression
The gene knockdown efficiency of siRNA delivered by non-targeted QPAMAM-OH and targeted QPAMAM-OH-LHRH dendrimer was investigated using quantitative RT-PCR. We selected BCL2 protein responsible for cellular antiapoptotic defense as a target for siRNA. The results of these experiments are shown in Fig. 5. It was found that siRNA delivered by all non-targeted QPAMAM-OH dendrimers with different degree of quaternization slightly, but statistically significantly, lowered the expression of the targeted gene (Fig. 5, bars 2-4). Targeted to cancer cells QPAMAM-OH-LHRH-siRNA complex led to a significant suppression of the expression of the BCL2 gene.
Fig. 5.
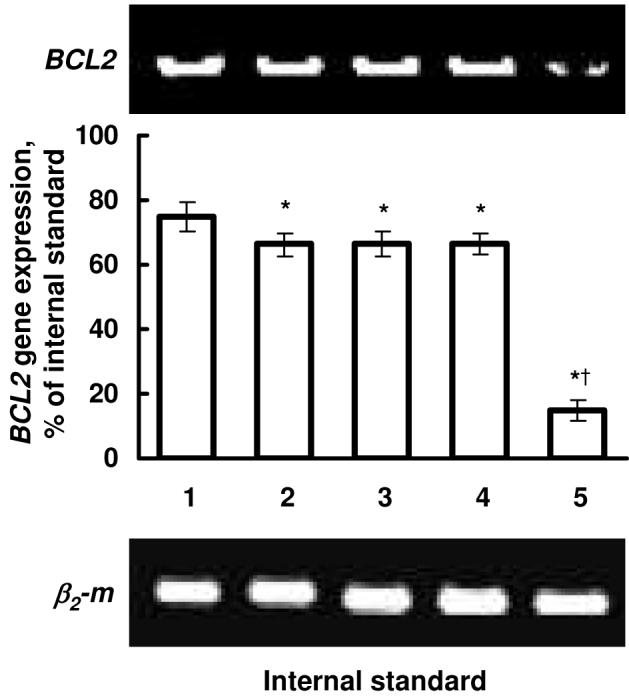
Suppression of the expression of the BCL2 gene by complexes of siRNA with dendrimers of different degree of quaternization and N/P ratio equal to 3 relative units. Typical images of RT-PCR products of genes encoding BCL2 protein and β2-microglobulin (β2-m, internal standard) in human ovarian cancer cells. 1 - Control (cells incubated with fresh media); 2 - Cells incubated with QPAMAM-OH-0.20-siRNA (20% of quaternization); 3 - Cells incubated with QPAMAM-OH-0.70-siRNA (70% of quaternization); 4 - Cells incubated with QPAMAM-OH-siRNA (97% of quaternization); 5 - Cells incubated with QPAMAM-OH-LHRH-siRNA (85% of quaternization).
Discussion
In recent years, dendrimers have emerged as novel cationic nanocarriers for exogenous gene transfer into mammalian cells mainly due to their well-defined structure, nanometer size, and relatively easy structural modifications30. Among them, different generations of polyamidoamine dendrimers have been successfully tested as promising nanocarriers for the delivery of plasmid DNA and antisense oligonucleotides that exhibited high transfection efficiency31-33. However, PAMAM dendrimers offered only a limited success in the delivery of short oligonucleotides (siRNA)34. Recently, we developed, designed, and evaluated a novel surface modified and internally cationic PAMAM dendrimer (QPAMAM-NHAc) with a high degree of quaternization (∼90%)7. This dendrimer efficiently delivered siRNA into cancer cells. In contrast, unmodified PAMAM-NH2 as well as internally cationic, hydroxyl terminated and almost completely quaternized dendrimers (QPAMAM-OH) failed to internalize siRNA into the ovarian cancer cells. Interestingly, both internally quaternized dendrimers QPAMAM-NHAc and QPAMAM-OH formed compact and spheroid nanoparticles with siRNA, while the PAMAM-NH2 dendrimer exhibited nanofiber shaped structures. These results clearly indicated the importance of surface modification and internal quaternization for intracellular delivery of siRNA. We figured out that the size and morphology of the PAMAM-NH2-siRNA complex and neutral surface of the QPAMAM-OH dendrimer with a high degree of quaternization are the major obstacles for the cellular uptake and efficiency of siRNA.
The mechanism by which dendrimers efficiently deliver genes has been extensively investigated31-33. A PAMAM dendrimer possess several internal tertiary amine groups, which are believed to play a critical role in the suppression of lowering the pH in endosomes and lysosomes. These tertiary amine groups are known to induce osmotic swelling of the endosome due to endosomal buffering that leads to rupture of endocytic vesicles and subsequent release of their payload. Such an effect of tertiary amine in a dendrimer or polymer is called a proton sponge effect. We envisioned that almost complete quaternization of tertiary amine groups in a QPAMAM-OH dendrimer could hamper the proton sponge effect and might be one of the reasons for the low efficacy of siRNA. Therefore, in order to increase the transfection efficiency of siRNA, a partially quaternized PAMAM dendrimer (QPAMAM-OH) with approximately 75% of quaternization was synthesized and evaluated as a nanocarrier for siRNA delivery. The evaluation of cellular uptake and transfection efficacy of siRNA delivered by this dendrimer showed the following. With the decrease in the degree of quaternization enhanced cellular uptake of the QPAMAM-OH-siRNA complex, the resulting efficacy of the delivered siRNA in terms of the suppression of the expression of targeted gene was very low. Based on these results one can suggest that despite the decrease in the quaternization resulting in the increase in cellular uptake of an entire dendrimer-siRNA complex, the aforementioned suppression of the proton sponge effect was still sufficient to prevent the release of free siRNA from the endosome.
To further increase the cellular uptake of dendrimer-siRNA complexes, we added a cancer cell targeting moiety to the dendrimeric delivery system. A synthetic analog of natural LHRH peptide conjugated and cancer-targeted internally quaternized PAMAM dendrimer (QPAMAM-OH-LHRH) was synthesized for this purpose. The rationale for using LHRH peptide was based on the following main considerations23: 1) The receptors for this peptide are over-expressed in several types of cancer cells, including ovarian, breast, endometrial, and prostate cancers, 2) The receptors are less expressed in normal cells, 3) LHRH peptide targets the conjugated nanocarrier specifically to cancer cells and facilitates cellular uptake using over-expressed LHRH receptors through receptor-mediated endocytosis. 4) LHRH-conjugated nanocarriers preferentially accumulated in tumors, limiting side effects on healthy organs. In our previous experiments, we extensively studied targeting mechanisms of LHRH peptide in cancer cells, including experiments that involve competitive binding35. It was clearly shown that LHRH peptide works through specific LHRH receptors which are overexpressed in many types of cancer cells and specific targeting is due to LHRH molecules.
The surface neutral and internally cationic QPAMAM-OH dendrimer was prepared by a known method using controlled internal quaternization of a PAMAM dendrimer. A two step synthetic protocol was used for the synthesis of the QPAMAM-OH-LHRH dendrimer. In the first step, the PAMAM-OH dendrimer was reacted with LHRH-hemisuccinate to produce PAMAM-OH-LHRH conjugate. During the second step, PAMAM-OH-LHRH was internally quaternized to afford the QPAMAM-OH-LHRH dendrimer. The degree of quaternization in both of the dendrimers was estimated by 1H-NMR spectroscopy. Unfortunately, 1H-NMR was not useful in this case to detect the presence of LHRH peptide due to its low concentration in the conjugate. Consequently, the presence of LHRH peptide in QPAMAM-OH-LHRH was confirmed by BCA protein assay. Furthermore, the concentration of LHRH peptide in the QPAMAM-OH-LHRH conjugate was estimated to be around 50 μg/ml based on the calibration curve for standard LHRH peptide using BCA protein assay.
Among several advantages, the low cytotoxicity of internally quaternized dendrimer makes them most suitable nanocarriers for the safe delivery of genes and other therapeutic drugs. In the present study, both the dendrimers, QPAMAM-OH and QPAMAM-OH-LHRH, showed low toxicity even at higher concentrations. Low cytotoxicity is particularly important when the high loading of nanocarrier is required and it has been shown that the higher dendrimer-siRNA ratio results in smaller particles for relatively easy internalization7, 26. It has been documented that a higher dendrimer-siRNA (N/P) ratio also gives superior results in the in vitro gene expression studies36.
The oligonucleotide band disappearance in agarose gel electrophoresis studies suggested that both the dendrimers had almost identical ability in forming a complex with siRNA and a stable complex was formed at N/P ratio 1 and above.
Dynamic Light Scattering analysis was in good agreement with our previous studies and the particle size decreased with an increase in the N/P (Nitrogen to Phosphate, positive to negative charge ratio) ratio7. However, the particle size in partially quaternized dendrimers was larger when compared to that previously obtained for a completely quaternized PAMAM dendrimer7. Atomic Force Microscope studies revealed compact and spherical particles for QPAMAM-OH-siRNA complex and the presence of LHRH marginally influenced the morphology of QPAMAM-OH-LHRH-siRNA complex. The size of dendrimer-siRNA complexes used in the present study varied between different dendrimers and was in most cases relatively higher when compared with that registered by other investigators36-38. It is known that the size of dendrimer-RNA complexes, their stability and uniformity critically depend on the size of the RNA molecule, the dendrimer generation, and the charge ratio between the dendrimer and the RNA39. Larger RNA molecules, higher generations of dendrimers, and larger dendrimer-to-RNA charge ratios usually form stable, uniform nanoscale RNA/dendrimer complexes. Previously, we reported that the complexation of PAMAM generation 4 dendrimer with siRNA produced very large size nanofibers7. It has been shown that dendrimer generation greatly affects the particle size and a higher generation dendrimer could lead to smaller particles. The formation of relatively large dendrimer-siRNA complexes in the present study could most likely be explained by their low resulting positive charge where cationic charges of dendrimers were hindered by dendrimer surface. This probably leads to the formation of less condensed complexes. An increase in N/P ratio led to the condensation of complexes and decrease in their diameter. At N/P ratio about 3 relative units, atomic force microscopy revealed formation of well condensed nanoparticles along with a small amount of larger particles. Such a broad size diversity of dendrimer-siRNA complexes have previously been registered in independent studies 7, 26, 36, 39. It should also be stressed that AFM images have a tendency to overestimate the size particles28. The average size of complexes at N/P ratio equal to 3 relative units (the ratio which we used for cellular uptake study) determined by dynamic light scattering technique was about 150 nm. Our previous investigations clearly showed that delivery systems of such size with a low surface charge provide for an effective intracellular internalization of anticancer drugs, antisense oligonucleotides, and siRNA. Moreover, targeting of DDS carriers specific to cancer cells substantially enhanced this internalization23-25. Our new data support the previous findings and show that cancer targeted QPAMAM-OH-LHRH-siRNA with 85% of quaternization and N/P ratio equal to 3 relative units provide for an enhanced cytoplasmic delivery of siRNA, uniform distribution of delivered siRNA within the cytoplasm, and led to the effective suppression of the targeting gene.
Next, the role of degree of quaternization and targeting ligand as a penetration enhancer was investigated in the cellular uptake of siRNA by human ovarian cancer A2780 cells. Indeed, the degree of quaternization influenced the intracellular uptake of siRNA and relatively less quaternized dendrimer QPAMAM-OH improved the internalization of siRNA when compared with the previously studied QPAMAM-OH dendrimer with almost complete quaternization7. The cellular uptake of siRNA was significantly improved when the targeting ligand was conjugated to the dendrimer, even though the degree of quaternization of this dendrimer was slightly higher than QPAMAM-OH (85% vs. 75%). These studies clearly indicate the importance of the degree of quaternization and targeting ligand LHRH for the enhanced intracellular delivery of siRNA using nontoxic nanocarriers. Although the exact reason for improved cellular internalization of siRNA using QPAMAM-OH is unknown, two possible contributing factors are suggested (1) more efficient interaction of free tertiary amines with negatively charged cell membrane than permanently charged quaternary amines, and (2) tertiary amines assisted the endosomal escape of siRNA to cytoplasm by the so-called proton sponge effect. On the other hand, the receptor mediated endocytosis pathway was considered a reason for the higher uptake of siRNA by the QPAMAM-OH-LHRH dendrimer. A series of our previous experiments clearly demonstrated such a phenomenon for the enhancement of other various drug delivery systems when LHRH peptide was used as targeting ligand20, 22-25.
In order to exploit the effect of lower degree of quaternization for effective siRNA delivery, a QPAMAM-OH dendrimer with approximately 30% of quaternization and its complex with siRNA were prepared. The preliminary testing showed that such a complex possesses good cell penetrating activity (data not shown). Unfortunately, the less quaternized dendrimers degraded in water over a period of time presumably through Hofmann elimination or a retro Michael type of reaction40, which precluded their further use in the siRNA delivery.
Having been encouraged by the higher cellular uptake of siRNA by QPAMAM-OH and QPAMAM-LHRH-OH dendrimers with relatively low degree of quaternization, we further compared gene silencing efficiency for BCL2 gene of siRNA complexes with dendrimers of different degree of quaternization. The delivery of siRNA by the non-targeted QPAMAM-OH dendrimers with different degrees of quaternization led to only a mild decrease in the expression of the targeted gene. In contrast, targeted QPAMAM-OH-LHRH-siRNA complex significantly suppressed the expression of the BCL2 gene in cancer cells. The result suggests that degree of quaternization to some extent is important for the cellular uptake but not sufficient to achieve a gene silencing effect. In contrast, delivery of siRNA by a cancer-targeted dendrimer substantially enhances its gene silencing effectiveness. These data show that an effective intracellular delivery of siRNA by dendrimers does not guarantee its high gene silencing activity. In order to successfully decrease the expression of targeted mRNA, siRNA should be delivered into the cellular cytoplasm, released from the delivery system with preserved silencing activity, and enter the RNA interference pathway. For instance, it was found that poor endosomal escape of the carrier and inefficient cytoplasmic decoupling of the complexed nucleic acid may be a critical step in limiting gene silencing activity of siRNA delivery systems41. Recently, Hollins et al42 reported that PAMAM dendrimeric drug delivery systems, differing only in their structural architecture, elicit opposing effects on the expression of the gene targeted for silencing by siRNA. Despite providing similar improvements in siRNA uptake, dendrimer formulations with comparable efficiency of cytoplasmic delivery led to an approximately 10-fold variation in the expression of targeted mRNA. A relative independence of the efficiency of cellular uptake of different DDS and their gene silencing activities was reported in other independent studies41, 43-45. In addition, it was found that some dendrimers without siRNA can influence gene expression45. Similarly, in the present study, three non-targeted QPAMAM-OH-siRNA complexes induced a comparable decrease in the expression of BCL2 mRNA despite different efficiency in delivering of siRNA in the cytoplasm of cancer cells. Although mechanisms of such phenomenon require more detailed study, one can suggest that intracellular internalization of a dendrimer-siRNA complex by receptor-mediated endocytosis provides an effective delivery to the place of action and allows for preserving the activity of complexated siRNA. The present data show that cancer-targeted delivery system down regulated the expression of targeted mRNA 4-4.5 times more effectively when compared with non-targeted dendrimers.
The present experimental data support our previous conclusion that targeting of nanocarriers to cancer minimized the influence of the architecture, composition, size, and molecular mass of nanocarriers on the efficacy of their payload. Previously, we experimentally supported this important statement for the efficacy of the delivered anticancer drug25. In the present publication, we provide experimental support of the phenomenon for siRNA delivered by internally quaternized dendrimers. This finding shows that the observation probably has general character, which in turn can potentially produce a high impact on nanocarrier-based drug delivery of cancer therapeutics. This conception implies that one can design nanocarrier architecture with specific composition, size, molecular mass, and other characteristics based solely on the effective encapsulation of active ingredient(s), desired their release profile, intracellular distribution, cost, and other factors ensuring that the high specific efficacy of the delivered agent(s) could be achieved automatically by cancer targeting.
Conclusion
Two internally cationic polyamidoamine dendrimers were designed and evaluated for siRNA delivery into cancer cells. The result suggested that a lesser degree of quaternization improved the cellular uptake of siRNA but did not considerably increase its gene silencing activity. In contrast, targeting of the dendrimer specifically to the plasma membrane of cancer cells by LHRH peptide further improved internalization of siRNA by cancer cells and significantly enhanced its intracellular activity leading to a substantial suppression of the expression of a targeted gene. Data obtained show the high potential of targeted internally cationic dendrimers as nanocarriers for efficient delivery of siRNA to cancer cells and their possible use in cancer chemotherapy.
Acknowledgements
The research was supported in part by NIH CA100098 and CA111766 grants from the National Cancer Institute.
References
- (1).Caplen NJ, Mousses S. Ann N Y Acad Sci. 2003;1002:56–62. doi: 10.1196/annals.1281.007. [DOI] [PubMed] [Google Scholar]
- (2).Dave RS, Pomerantz RJ. Rev Med Virol. 2003;13:373–385. doi: 10.1002/rmv.407. [DOI] [PubMed] [Google Scholar]
- (3).Sontheimer EJ. Nat Rev Mol Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- (4).Mello CC, Conte D., Jr. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- (5).Riddihough G. Science. 2005;309:1507–1533. [Google Scholar]
- (6).Paroo Z, Corey DR. Trends Biotechnol. 2004;22:390–394. doi: 10.1016/j.tibtech.2004.06.004. [DOI] [PubMed] [Google Scholar]
- (7).Patil ML, Zhang M, Betigeri S, Taratula O, He H, Minko T. Bioconjug Chem. 2008;19:1396–1403. doi: 10.1021/bc8000722. [DOI] [PubMed] [Google Scholar]
- (8).Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Sonawane ND, Szoka FC, Jr., Verkman AS. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- (10).Lee J, Lim Y-B, Choi J, Choi M-U, Yang C-H, Park J-S. Bull. Korean Chem. Soc. 2003;24:1637–1640. [Google Scholar]
- (11).Futaki S. Adv Drug Deliv Rev. 2005;57:547–558. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- (12).Snyder EL, Dowdy SF. Pharm Res. 2004;21:389–393. doi: 10.1023/B:PHAM.0000019289.61978.f5. [DOI] [PubMed] [Google Scholar]
- (13).Juliano RL. Ann N Y Acad Sci. 2006;1082:18–26. doi: 10.1196/annals.1348.011. [DOI] [PubMed] [Google Scholar]
- (14).Astriab-Fisher A, Sergueev DS, Fisher M, Shaw BR, Juliano RL. Biochem Pharmacol. 2000;60:83–90. doi: 10.1016/s0006-2952(00)00310-5. [DOI] [PubMed] [Google Scholar]
- (15).Tung CH, Stein S. Bioconjug Chem. 2000;11:605–618. doi: 10.1021/bc0000334. [DOI] [PubMed] [Google Scholar]
- (16).Abes R, Arzumanov AA, Moulton HM, Abes S, Ivanova GD, Iversen PL, Gait MJ, Lebleu B. Biochem Soc Trans. 2007;35:775–779. doi: 10.1042/BST0350775. [DOI] [PubMed] [Google Scholar]
- (17).Kang H, DeLong R, Fisher MH, Juliano RL. Pharm Res. 2005;22:2099–2106. doi: 10.1007/s11095-005-8330-5. [DOI] [PubMed] [Google Scholar]
- (18).Moschos SA, Williams AE, Lindsay MA. Biochem Soc Trans. 2007;35:807–810. doi: 10.1042/BST0350807. [DOI] [PubMed] [Google Scholar]
- (19).Jayant S, Khandare JJ, Wang Y, Singh AP, Vorsa N, Minko T. Pharm Res. 2007;24:2120–2130. doi: 10.1007/s11095-007-9406-1. [DOI] [PubMed] [Google Scholar]
- (20).Chandna P, Saad M, Wang Y, Ber E, Khandare J, Vetcher AA, Soldatenkov VA, Minko T. Mol Pharm. 2007;4:668–678. doi: 10.1021/mp070053o. [DOI] [PubMed] [Google Scholar]
- (21).Dharap SS, Minko T. Pharm Res. 2003;20:889–896. doi: 10.1023/a:1023839319950. [DOI] [PubMed] [Google Scholar]
- (22).Dharap SS, Qiu B, Williams GC, Sinko P, Stein S, Minko T. J Control Release. 2003;91:61–73. doi: 10.1016/s0168-3659(03)00209-8. [DOI] [PubMed] [Google Scholar]
- (23).Dharap SS, Wang Y, Chandna P, Khandare JJ, Qiu B, Gunaseelan S, Sinko PJ, Stein S, Farmanfarmaian A, Minko T. Proc Natl Acad Sci U S A. 2005;102:12962–12967. doi: 10.1073/pnas.0504274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Khandare JJ, Chandna P, Wang Y, Pozharov VP, Minko T. J Pharmacol Exp Ther. 2006;317:929–937. doi: 10.1124/jpet.105.098855. [DOI] [PubMed] [Google Scholar]
- (25).Saad M, Garbuzenko OB, Ber E, Chandna P, Khandare JJ, Pozharov VP, Minko T. J Control Release. 2008;130:107–114. doi: 10.1016/j.jconrel.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Khandare JJ, Jayant S, Singh A, Chandna P, Wang Y, Vorsa N, Minko T. Bioconjug Chem. 2006;17:1464–1472. doi: 10.1021/bc060240p. [DOI] [PubMed] [Google Scholar]
- (27).Betigeri S, Pakunlu RI, Wang Y, Khandare JJ, Minko T. Mol Pharm. 2006;3:424–430. doi: 10.1021/mp060014x. [DOI] [PubMed] [Google Scholar]
- (28).Yang D-Q, Xiong Y-Q, Guo Y, Da D-A, Lu W-G. J Mater Sci. 2001;36:263–267. [Google Scholar]
- (29).Stoscheck CM. Methods Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- (30).Paleos CM, Tsiourvas D, Sideratou Z. Mol Pharm. 2007;4:169–188. doi: 10.1021/mp060076n. [DOI] [PubMed] [Google Scholar]
- (31).Braun CS, Vetro JA, Tomalia DA, Koe GS, Koe JG, Middaugh CR. J Pharm Sci. 2005;94:423–436. doi: 10.1002/jps.20251. [DOI] [PubMed] [Google Scholar]
- (32).Eichman JD, Bielinska AU, Kukowska-Latallo JF, Baker JR., Jr. Pharm Sci Technolo Today. 2000;3:232–245. doi: 10.1016/s1461-5347(00)00273-x. [DOI] [PubMed] [Google Scholar]
- (33).Kubasiak LA, Tomalia DA. Cationic Dendrimers as Gene Transfection Vectors: Dendri-Poly(amidoamines) and Dendri-Poly(propylenimines), Chapter 9. CRC Press; Boca Raton: 2004. pp. 133–157. [Google Scholar]
- (34).Kang H, DeLong R, Fisher MH, Juliano RL. Pharm Res. 2005;22:2099–2106. doi: 10.1007/s11095-005-8330-5. [DOI] [PubMed] [Google Scholar]
- (35).Dharap SS, Minko T. Pharm Res. 2003;20:889–896. doi: 10.1023/a:1023839319950. [DOI] [PubMed] [Google Scholar]
- (36).Zhou J, Wu J, Hafdi N, Behr JP, Erbacher P, Peng L. Chem Commun (Camb) 2006:2362–2364. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- (37).Shen XC, Zhou J, Liu X, Wu J, Qu F, Zhang ZL, Pang DW, Quelever G, Zhang CC, Peng L. Org Biomol Chem. 2007;5:3674–3681. doi: 10.1039/b711242d. [DOI] [PubMed] [Google Scholar]
- (38).Weber N, Ortega P, Clemente MI, Shcharbin D, Bryszewska M, de la Mata FJ, Gomez R, Munoz-Fernandez MA. J Control Release. 2008;132:54–64. doi: 10.1016/j.jconrel.2008.07.035. [DOI] [PubMed] [Google Scholar]
- (39).Shen XC, Zhou J, Liu X, Wu J, Qu F, Zhang ZL, Pang DW, Quelever G, Zhang CC, Peng L. Org Biomol Chem. 2007;5:3674–3681. doi: 10.1039/b711242d. [DOI] [PubMed] [Google Scholar]
- (40).March J, Smith MB. 6 ed. Wiley & Sons Inc; New York, NY: 2007. p. 2384. [Google Scholar]
- (41).Ghosn B, Kasturi SP, Roy K. Curr Top Med Chem. 2008;8:331–340. doi: 10.2174/156802608783790947. [DOI] [PubMed] [Google Scholar]
- (42).Hollins AJ, Omidi Y, Benter IF, Akhtar S. J Drug Target. 2007;15:83–88. doi: 10.1080/10611860601151860. [DOI] [PubMed] [Google Scholar]
- (43).Akhtar S, Benter I. Adv Drug Deliv Rev. 2007;59:164–182. doi: 10.1016/j.addr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- (44).Moriguchi R, Kogure K, Akita H, Futaki S, Miyagishi M, Taira K, Harashima H. Int J Pharm. 2005;301:277–285. doi: 10.1016/j.ijpharm.2005.05.021. [DOI] [PubMed] [Google Scholar]
- (45).Omidi Y, Hollins AJ, Drayton RM, Akhtar S. J Drug Target. 2005;13:431–443. doi: 10.1080/10611860500418881. [DOI] [PubMed] [Google Scholar]