Abstract
Purpose
No biomarkers have been identified to predict outcome with the use of an antiangiogenesis agent for cancer. Vascular endothelial growth factor (VEGF) genetic variability has been associated with altered risk of breast cancer and variable promoter activity. Therefore, we evaluated the association of VEGF genotype with efficacy and toxicity in E2100, a phase III study comparing paclitaxel versus paclitaxel plus bevacizumab as initial chemotherapy for metastatic breast cancer.
Patients and Methods
DNA was extracted from tumor blocks of patients from E2100. Three hundred sixty-three samples were available to evaluate associations between genotype and outcome. Genotyping was performed for selected polymorphisms in VEGF and VEGF receptor 2. Testing for associations between each polymorphism and efficacy and toxicity was performed.
Results
The VEGF-2578 AA genotype was associated with a superior median overall survival (OS) in the combination arm when compared with the alternate genotypes combined (hazard ratio = 0.58; 95% CI, 0.36 to 0.93; P = .023). The VEGF-1154 A allele also demonstrated a superior median OS with an additive effect of each active allele in the combination arm but not the control arm (hazard ratio = 0.62; 95% CI, 0.46 to 0.83; P = .001). Two additional genotypes, VEGF-634 CC and VEGF-1498 TT, were associated with significantly less grade 3 or 4 hypertension in the combination arm when compared with the alternate genotypes combined (P = .005 and P = .022, respectively).
Conclusion
Our data support an association between VEGF genotype and median OS as well as grade 3 or 4 hypertension when using bevacizumab in metastatic breast cancer.
INTRODUCTION
Inhibition of angiogenesis has proven to be beneficial in multiple types of malignancies.1 Bevacizumab, a humanized monoclonal antibody against vascular endothelial growth factor (VEGF), is arguably the most clinically mature antiangiogenesis agent.2-5 Recently, E2100, a North American breast intergroup phase III trial, evaluated bevacizumab for patients receiving initial chemotherapy for metastatic, human epidermal growth factor receptor 2–negative breast cancer.6 Patients were randomly assigned to weekly paclitaxel alone or paclitaxel with bevacizumab. The addition of bevacizumab improved the response rate (RR) from 21.2% to 36.9% (P < .001) and the median progression-free survival (PFS) time from 5.9 months to 11.8 months (P < .001), but it did not improve median overall survival (OS). The addition of bevacizumab also increased grade 3 and 4 hypertension.
Bevacizumab, like conventional antitumor agents, has interindividual heterogeneity in efficacy and toxicity. Prior attempts to identify biomarkers for bevacizumab focused on tumor-derived features such as VEGF, k-raf, p53, and microvessel density, among others, and have been unsuccessful in predicting efficacy.7,8 This is not surprising because angiogenesis is a host-regulated process.
However, there is substantial inherited genetic variability within VEGF and its receptor, VEGF receptor 2 (VEGFR-2), including multiple single nucleotide polymorphisms (SNPs).9,10 Prior data have suggested that SNPs within VEGF have biologic importance in predicting risk and prognosis of breast cancer,10,11 although there are conflicting reports.12,13 The variability seen in common polymorphic sites from breast tumors seems to be a result of inherited variation rather than somatic mutations.14 In our prior work, we evaluated a common polymorphism in VEGF and two common polymorphisms in endothelial nitric oxide synthase and found 100% concordance in genotype when comparing DNA from primary breast tumors with germline DNA. Thus, genetic variability in VEGF and VEGFR-2 represents a logical candidate to study as a potential biomarker for bevacizumab.
The objective of this study was to test the hypothesis that, when patients are treated with bevacizumab, there exists an association between VEGF and VEGFR-2 candidate SNPs and efficacy and toxicity. In addition, this study planned to test the possibility of an association between candidate SNPs and protein expression assessed by immunohistochemistry (IHC) in the primary tumor. Finally, this study investigated for an association between VEGF and VEGFR-2 protein expression in the primary tumor and clinical outcomes.
PATIENTS AND METHODS
Samples
In the E2100 parent trial, there were 673 eligible patients with 623 disease progression events and 483 deaths as of November 13, 2007. Paraffin-embedded tumor blocks from E2100 were available from 363 eligible patients for genotyping with a median follow-up time of 43 months. One hundred eighty patients were from the experimental arm, and 183 patients were from the control arm. Three hundred seventy-seven eligible patients were available for VEGF IHC, and 341 eligible patients were available for VEGFR-2 IHC. All DNA specimens were provided to the investigators of this trial in a de-identified fashion. This retrospective trial was approved by the Institutional Review Board at Indiana University.
Candidate Polymorphisms
We selected genes and polymorphisms known to modulate angiogenesis (Table 1) using the following criteria: involved in the angiogenesis pathway; established genetic polymorphism; sufficient frequency that its impact on drug response at a population level would be meaningful; and/or polymorphism could alter the function of the gene in a biologically relevant manner.
Table 1.
Candidate Single Nucleotide Polymorphisms
| Gene and Single Nucleotide Polymorphism | Location | White: Frequency of Rare Allele10 | African American: Frequency of Rare Allele10 |
|---|---|---|---|
| VEGF | |||
| -2578 C/A | Promoter | A = 49% | A = 24% |
| -1498 C/T | Promoter | C = 49% | C = 33% |
| -1154 G/A | Promoter | A = 33% | A = 10% |
| -634 G/C | 5′ Untranslated region | C = 32% | C = 35% |
| 936 C/T | 3′ Untranslated region | T = 15% | T = 13% |
| VEGFR-2 | |||
| 889 G/A (V297I) | Exon 7 | A = 9% | A = 20% |
| 1416 A/T (Q472H) | Exon 11 | T = 25% | T = 10% |
Genotyping
DNA was extracted from 20-μm paraffin-embedded tissue sections using the DNeasy Tissue kit (Qiagen, Valencia, CA). Candidate SNPs were genotyped with Taqman-based real-time polymerase chain reaction. Details for genotyping of each SNP have been previously described.10 Overall, genotype was successfully determined in 88.2% of samples (range, 82% to 92%).
Assessment of Expression
For VEGF, slides were deparaffinized, rehydrated, and placed in a vegetable steamer with citrate buffer at pH of 6.0 for 30 minutes. After slides cooled to room temperature, they were washed in two changes of distilled water followed by two changes of phosphate-buffered saline with 0.05% Tween-20 (PBST; Fisher Scientific, Pittsburgh, PA). Slides were then placed on a Dako Autostainer (Dako Cytomation, Carpinteria, CA). Slides were incubated with peroxidase blocking solution (S2001; Dako) for 10 minutes followed by three changes of PBST for a minimum of 10 minutes total. Slides were then sequentially incubated with anti-VEGF antibody (VG1; Lab Vision, Freemont, CA) diluted at 1:100 for 60 minutes, Dako EnVision+ (K4001; Dako) for 60 minutes, and then diaminobenzidine Substrate-Chromogen System (K3466; Dako), with three changes of PBST between each step. Slides were counterstained with Harris hematoxylin (Fisher Scientific), dehydrated, and cleared and had a cover slip placed. A VEGF_inv score was calculated by estimating the percentage of invasive tumor cells with cytoplasmic VEGF staining from the entire slide.
For VEGFR-2 IHC, formalin-fixed paraffin-embedded breast tumor sections were first deparaffinized and rehydrated. Next, antigen retrieval was executed at 98°C for 20 minutes in Target Retrieval Solution at pH of 9.0 (S2367; Dako). Dual Endogenous Enzyme Block (K4065, EnVision+ Dual Link System-HRP; Dako) was then applied for 5 minutes at room temperature. Anti–VEGFR-2 clone 55B11 rabbit monoclonal antibody (#2479; Cell Signaling Technology, Danvers, MA) was administered at a dilution of 1:20 for 2 hours at room temperature. Signal development with diaminobenzidine was conducted by the protocol for the EnVision+ kit with minor modifications. Counterstaining was completed with Hematoxylin QS (H-3404; Vector, Burlingame, CA) followed by dehydration and cover slipping. Human placenta or liver sections were used as positive controls. Omission of the primary antibody and substitution with rabbit immunoglobulin G (X0936; Dako) served as negative controls. Scoring was conducted with the H-score method, which is calculated as follows:
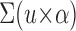 |
where u is the staining intensity (0 to 3+) and α is the percentage (0% to 100%) of tumor cells stained with each intensity.15,16
Statistics
Event-time distributions were estimated using the Kaplan-Meier method. Association of genotype with time to event outcome (median PFS and median OS) was evaluated using the Cox proportional hazards method. Planned pair-wise comparisons among the three genotypes were conducted for each polymorphism. A significance level of P = .017 corresponded to an overall type I error rate of 0.05 for each polymorphism based on Bonferroni correction for multiple comparisons. Given a 1.7% false-positive rate for each comparison, the probability that at least one false-positive result occurred among the 21 comparisons for all seven polymorphisms for PFS and OS was approximately 0.3, assuming that all of the comparisons were independent. Association of genotype with RR (defined as complete response/partial response v stable disease/progressive disease) and grade 3 or 4 hypertension was evaluated using Fisher's exact test with a significance level of P = .05. Association of genotype with expression was studied using the Kruskal-Wallis test. For RR and toxicity, given a 5% false-positive rate for each comparison, the probability that at least one false-positive result occurred among the seven comparisons was approximately 0.3, assuming that all of the comparisons were independent. Associations of expression with time to event outcome (median PFS and median OS) and RR were evaluated using the Cox proportional hazards method and Wilcoxon rank sum test, respectively. All P values were two sided.
RESULTS
Relationship of Genotype With Efficacy
All candidate genotypes were compared with efficacy in both treatment groups. The VEGF-2578 AA genotype and the VEGF-1154 AA genotype predicted a favorable median OS (Table 2; Fig 1) for patients in the combination arm but did not predict an improved median OS for patients in the control arm and did not predict a superior PFS or RR for either arm. There was a significant incremental benefit from each addition of the VEGF-1154 A allele.
Table 2.
Relationship of VEGF Genotype on OS in the Experimental Arm
| SNP and Genotype Comparison* | Hazard Ratio | 98.3% CI | P |
|---|---|---|---|
| VEGF-2578 | |||
| CA (24.4; 42.6%) v AA (37.0; 20.8%) | 1.78 | 0.96 to 3.32 | .026 |
| CC (22.2; 37.6%) v AA (37.0; 21%) | 1.70 | 0.91 to 3.17 | .043 |
| CC (22.2; 37.6%) v CA (24.4; 42.6%) | 0.99 | 0.62 to 1.58 | .95 |
| AA v CA + CC | 0.58 | 0.36 to 0.93† | .023 |
| VEGF-1154 | |||
| GG (22.3; 56.9%) v GA (29.8; 38.8%) | 1.60 | 0.98 to 2.60 | .022 |
| GG (22.3; 56.95) v AA (46.5; 9.4%) | 2.69 | 1.10 to 6.59 | .008 |
| GA (29.8; 38.8%) v AA (46.5; 9.4%) | 1.68 | 0.66 to 4.30 | .19 |
| AA v GA v GG | 0.62 | 0.46 to 0.83† | .001 |
Abbreviations: VEGF, vascular endothelial growth factor; OS, overall survival; SNP, single nucleotide polymorphism.
Median OS in months and percentage of patients are given in parentheses.
95% CI.
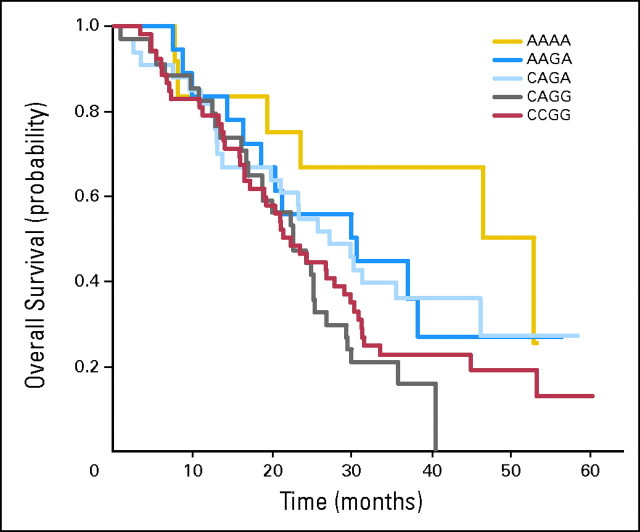
Fig 1.
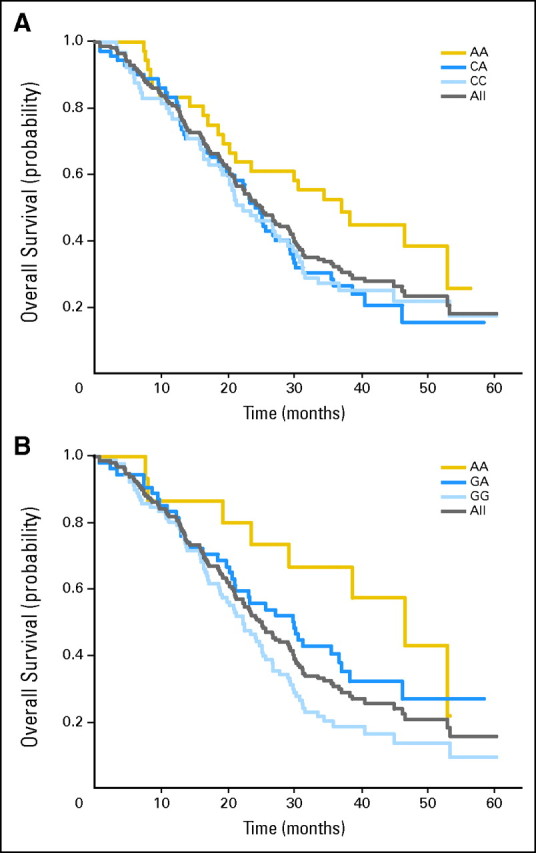
Kaplan-Meier curve for overall survival (OS) in experimental arm by genotype; (A) vascular endothelial growth factor (VEGF)-2578 C/A; (B) VEGF-1154 G/A.
We also combined all genotypes for VEGF-2578 and VEGF-1154 and evaluated for an association with median OS (Table 3; Appendix Fig A1, online only). There were nine possible combinations of which four groups had three or fewer samples (excluded from Fig A1). When comparing the VEGF-2578/-1154 AA/AA genotypes with all of the other genotypes, there was an improvement in median OS in the experimental arm (P = .041). When comparing the VEGF-2578/-1154 CA/GG genotypes with all of the other genotypes, there was an inferior median OS (P = .038).
Table 3.
Comparison of Combined VEGF Genotypes With Overall Survival in Experimental Arm
| VEGF-2578/-1154 | Median Overall Survival (months) | % of Patients | P (comparison with other genotypes combined) |
|---|---|---|---|
| AA/AA | 49.7 | 7.6 | .041 |
| AA/GA | 30.2 | 11.4 | .44 |
| CA/GA | 27.1 | 20.9 | .40 |
| CA/GG | 22.5 | 21.5 | .038 |
| CC/GG | 21.7 | 32.9 | .30 |
| Others | — | 5.7 | — |
Abbreviation: VEGF, vascular endothelial growth factor.
The median OS time was 25.2 months for the control arm (not subdivided by genotype) and 26.7 months for the experimental arm (not subdivided by genotype; P = .16).6 The median OS times for the subgroups in the VEGF-2578 AA and the VEGF-1154 AA genotypes in the experimental arm were significantly longer at 37.0 and 46.5 months, respectively, when compared with the control arm (not subdivided by genotype; P = .035 and P = .047, respectively; Fig 2). There were no other significant associations between genotypes and efficacy for the other VEGF and VEGFR-2 SNPs evaluated.
Fig 2.
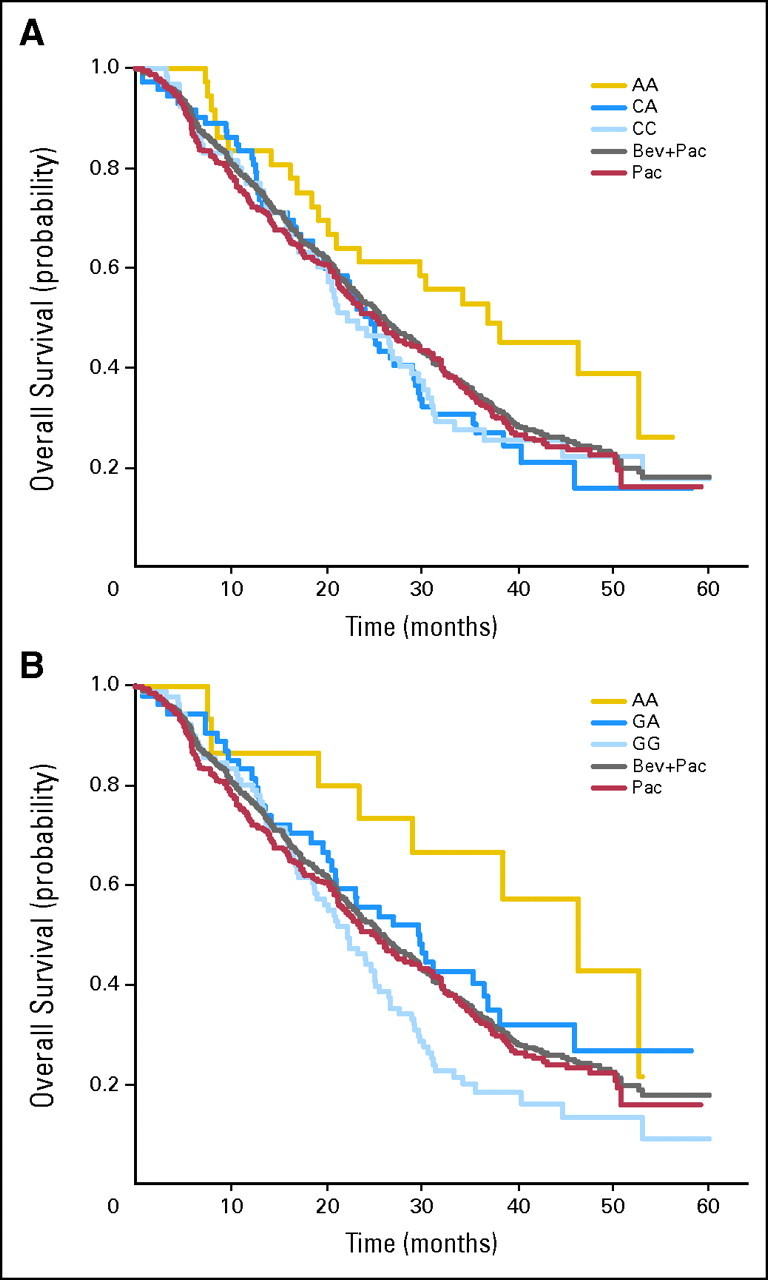
Kaplan-Meier curve for overall survival (OS) in the experimental arm (by vascular endothelial growth factor genotype) compared with the control and combination arms (not subdivided by genotype). Bev, bevacizumab; Pac, paclitaxel.
Relationship of Genotype With Grade 3 or 4 Hypertension
All candidate genotypes were compared with grade 3 or 4 hypertension (National Cancer Institute Common Toxicity Criteria version 2.0); 14.8% of all patients receiving bevacizumab in the parent trial experienced grade 3 or 4 hypertension compared with 0% of patients in the control arm (P < .001).6 The VEGF-634 CC and VEGF-1498 TT genotypes strongly correlated with less grade 3 or 4 hypertension (0% and 8%, respectively) when compared with the combined alternate genotypes (P = .005 and P = .022, respectively; Table 4). There were no other significant associations between genotypes and hypertension for the other VEGF and VEGFR-2 SNPs evaluated.
Table 4.
Relationship of VEGF Genotype With Grade 3 or 4 Hypertension
| Single Nucleotide Polymorphism | Patients
|
% of Patients With Grade 3 or 4 Hypertension | P | |
|---|---|---|---|---|
| No. | % | |||
| VEGF-634 | ||||
| CC | 27 | 15.3 | 0 | .013 |
| GC | 82 | 46.3 | 22 | |
| GG | 68 | 38.4 | 19 | |
| CC v GC + GG | .005 | |||
| VEGF-1498 | ||||
| TT | 60 | 33.9 | 8 | .056 |
| CT | 82 | 46.3 | 22 | |
| CC | 35 | 19.8 | 23 | |
| TT v CC + CT | .022 | |||
| VEGF-2578 | ||||
| AA | 36 | 20.8 | 22 | .32 |
| CA | 72 | 41.6 | 21 | |
| CC | 65 | 37.6 | 12 | |
| CC v CA + AA | .16 | |||
| VEGF-1154 | ||||
| AA | 15 | 9.4 | 27 | .29 |
| GA | 54 | 38.8 | 22 | |
| GG | 91 | 56.9 | 14 | |
| GG v GA + AA | .15 | |||
Abbreviation: VEGF, vascular endothelial growth factor.
Relationship of Hypertension With Efficacy
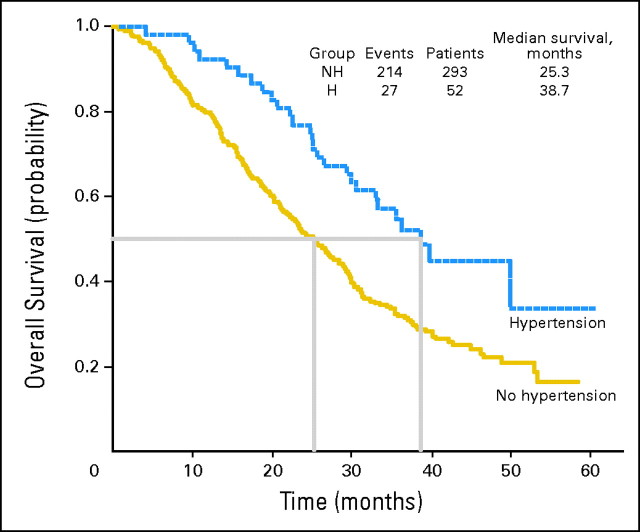
Although not a one-to-one relationship, the common haplotypes that contain the advantageous alleles for survival (-2578A and -1154A) never include the alleles that seem to protect against grade 3 or 4 hypertension (-634C and -1498T). Therefore, we assessed for an association between significant hypertension and median OS in the parent E2100 trial and found that patients with grade 3 or 4 hypertension had a superior median OS time compared with patients with no hypertension (38.7 v 25.3 months, respectively; P = .002; Appendix Fig A2, online only).
Relationship of Primary Tumor VEGF and VEGFR-2 Expression (IHC) With Clinical Outcome
An association between VEGF and VEGFR-2 and tumor expression was evaluated. The degree of VEGF expression was evaluated by the VEGF_inv score, which ranged from 0 to 100 (based on the percentage of invasive cells with cytoplasmic VEGF staining). The degree of VEGFR-2 expression was evaluated by an H-score, which could range from 0 (no detected expression) to 300 (100% of the cells had maximum 3+ expression). There was no statistically significant association between either VEGF or VEGFR-2 primary tumor expression and outcome.
Relationship of Genotype With Primary Tumor Expression of VEGF and VEGFR-2
All candidate genotypes were compared with primary tumor expression for both VEGF and VEGFR-2. The genotypes were compared with VEGF expression for the entire cohort, and there were no statistically significant associations determined. For the VEGF-2578 genotype, there was a trend for an association between genotype and VEGF inv_score. The median score for the AA genotype was lower (AA = 50) when compared with the alternate genotypes (CA = 60 and CC = 75), but this did not reach statistical significance (P = .08). The VEGF-1154 AA genotype also had a lower median expression (AA = 25) than the alternate genotypes (GA = 60 and GG = 70), but this also did not reach statistical significance (P = .13). No genotypes correlated with the expression of VEGFR-2.
DISCUSSION
Drugs that target VEGF and VEGFR-2 have made a major impact in cancer therapy.1,2,4,17-20 Recently, E2100 demonstrated an improvement in RR and PFS with the addition of bevacizumab to paclitaxel in the first-line metastatic setting of breast cancer.6 Although these drugs were largely touted as targeted therapy, we have had a difficult time identifying which patients will benefit most from them.21 These agents all demonstrate clear therapeutic heterogeneity in that they are active in some patients but inactive and toxic in others. A biomarker to predict which patients might experience the most activity and least toxicity would be of clinical and scientific value.
To our knowledge, these are the first data to describe biomarkers that seem to be associated with efficacy and toxicity for bevacizumab in cancer. Germline SNPs were logical candidates to study because angiogenesis is largely a host-mediated event, as opposed to a process mediated by somatic mutations in the tumor.22,23 This is critical in that these changes are not controlled by the tumor, but instead are an inherited predictive factor. From an analysis standpoint, SNPs are convenient because they can be determined at any time and are not affected by the status of the tumor.24 Much prior work has tested tumor-specific markers without success. Jubb et al7 found that testing for tumor VEGF expression, thrombospondin-2, and microvessal density did not predict benefit from bevacizumab in colorectal cancer. Ince et al25 have evaluated mutations in k-ras, k-raf, and p53, and these markers also did not predict benefit from bevacizumab.
It is not surprising that analysis of host-related variability might succeed where examination of tumor-related variability has failed. The heterogeneity in outcome seen with a therapy that targets a process (ie, angiogenesis) that is host mediated might best be explained by host-imprinted variability. There are multiple prior studies that suggest that SNPs within VEGF can impact outcome in conditions regulated by angiogenesis including cancer risk and prognosis,10,11,26-34 retinopathy,35-41 nephropathy,42-45 pre-eclampsia,46,47 recurrent pregnancy loss,48 and vasculopathy.49-51 Although the DNA evaluated here was derived from the primary tumor, we were attempting to evaluate germline variability. Although somatic mutations would not be unexpected, it seems that the majority of variability seen in the common polymorphic sites is inherited.14 Thus, the determination of germline variability in these specific sites can be assessed from primary tumor DNA, as was performed here.
A major strength of this study is that it was performed on a large, multicenter trial with a robust number of patients. These data suggest that patients who had the VEGF-2578 AA genotype and the VEGF-1154 AA genotype had a superior median OS compared with patients with alternative genotypes. Of the alleles that demonstrated a positive associations in this study, the minor allele frequency in a random white population is frequent and ranges from 33% to 49%.10 Thus, these findings are relevant to a large number of patients.
Another provocative finding was the prediction of clinically significant hypertension by VEGF genotype. Those with VEGF-1498 TT and VEGF-634 CC genotypes were largely protected from serious hypertension. The high blood pressure induced by bevacizumab can be a troubling toxicity, resulting in need for therapeutic intervention and even discontinuation for some. The ability to anticipate this toxicity could allow for either close monitoring with early intervention or even possibly prophylactic antihypertensive therapy. These finding may also lend insight into the fundamental biology of hypertension and provide clues for new therapeutic targets. The discovery of an association between those who experienced significant hypertension and an improved OS is also biologically provocative. On the basis of our haplotype data, we believe this represents an important biologic surrogate for those who may do well and not necessarily a toxicity that we should strive to achieve, as is common practice for conventional cytotoxic agents (ie, maximum-tolerated dose). Recently, it was found that patients who are extensive metabolizers of cytochrome P450 2D6 and take tamoxifen are more likely to gain the most benefit52,53 but are also most likely to have hot flashes and be less compliant.54 Similarly, the findings here suggest that those who experience significant hypertension comprise a subgroup that may have a greater potential to gain significant therapeutic benefit. This might provide motivation to aggressively treat drug-induced hypertension as opposed to abandoning therapy at the first sign of toxicity.
A weakness of this study was that blood was not collected. Thus, DNA had to be analyzed from paraffin-embedded tumor, which resulted in a genotype success rate of less than 100%. A limitation to the analysis of tumor protein VEGF/VEGFR-2 expression is that the expression was measured from the primary tumor. In contrast to SNP analysis, expression is likely to represent only a brief moment in the life of the malignancy. Because all of these patients had metastatic disease, it is possible that protein expression in the primary tumor may have been different from what the expression would have been at the time of therapy for metastatic disease. However, there was a trend for an association between the VEGF genotypes that had superior survival and lower VEGF expression in the primary tumor. This association may provide some mechanistic explanation to the genotype effect observed but can only be regarded as hypothesis generating at this time.
These findings pose a number of important questions. First, why was there a survival benefit, but only a hint of corresponding benefit in PFS and no RR advantage, detected by genotype? One possibility for this lack of concordance is that the genotype effect was a statistical artifact. For this reason, it is essential that these results be replicated. However, another possibility is that there is a complex interplay between the biology of angiogenesis and the use of an antibody that inhibits VEGF. The E2100 parent trial provides a hint that this interplay may be more complex than initially thought. Despite a doubling of RR and PFS, these dramatic differences did not translate into a statistically different OS. These findings would suggest that some of the patients in the experimental arm actually do worse after progression on an antiangiogenesis agent when compared with the control arm. This subgroup of patients may have an inherited predisposition for a rapid regrowth of blood vasculature either after loss of continued suppression from an antiangiogenesis agent or after becoming refractory to VEGF inhibition specifically. Recent data have demonstrated that an induction of tumor-independent circulating proangiogenic factors may occur after discontinuation of angiogenic blockade, possibly resulting in rapid tumor regrowth.55 Thus, it is plausible that a certain subgroup of patients with a specific genotype may derive a sustained benefit from VEGF inhibition with discontinuation at progression that translates into a significant improvement in OS compared with control, whereas others do not. Second, what potential biologic mechanisms underlie these findings? Further research to unravel why these variants in VEGF are associated with outcome has the potential to suggest new treatment modalities aimed at VEGF. Comprehensive studies aimed at understanding how these variants might alter the molecular biology of VEGF signaling are ongoing in this laboratory. Although there are many possible mechanistic explanations for these findings, it is clear that the next step should be validation of these findings in the treatment of breast cancer and other conditions.
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: Bryan P. Schneider, Genentech (C); George W. Sledge, Genentech (C); Maura Dickler, Genentech (C); Melody Cobleigh, Genentech (C); Kathy D. Miller, Genentech (C), Roche (C), Entremed (C), Pfizer Inc (C) Stock Ownership: None Honoraria: Julie Gralow, Genentech; Melody Cobleigh, Genentech; Kathy D. Miller, Roche Research Funding: Bryan P. Schneider, Genentech; Julie Gralow, Genentech; Maura Dickler, Genentech; Kathy D. Miller, Genentech, Roche Expert Testimony: None Other Remuneration: None
AUTHOR CONTRIBUTIONS
Conception and design: Bryan P. Schneider, Milan Radovich, George W. Sledge, David A. Flockhart, Kathy D. Miller
Financial support: Bryan P. Schneider, George W. Sledge, David A. Flockhart, Kathy D. Miller
Administrative support: Bryan P. Schneider, Milan Radovich, David A. Flockhart, Bradley Hancock, Kathy D. Miller
Provision of study materials or patients: Bryan P. Schneider, George W. Sledge, Sunil Badve, Nancy Davidson, Julie Gralow, Maura Dickler, Edith A. Perez, Melody Cobleigh, Tamara Shenkier, Kathy D. Miller
Collection and assembly of data: Bryan P. Schneider, Milan Radovich, Bradley Hancock, Edith A. Perez, Susan Edgerton
Data analysis and interpretation: Bryan P. Schneider, Molin Wang, Milan Radovich, Sunil Badve, Ann Thor, David A. Flockhart, Susan Edgerton
Manuscript writing: Bryan P. Schneider, Milan Radovich, George W. Sledge, Ann Thor, David A. Flockhart, Bradley Hancock, Nancy Davidson
Final approval of manuscript: Bryan P. Schneider, Milan Radovich, George W. Sledge, Sunil Badve, Ann Thor, David A. Flockhart, Bradley Hancock, Nancy Davidson, Julie Gralow, Maura Dickler, Edith A. Perez, Melody Cobleigh, Tamara Shenkier, Susan Edgerton, Kathy D. Miller
Appendix
Fig A1.
Relationship of combined vascular endothelial growth factor (VEGF)-2578C/A and VEGF-1154G/A genotypes with overall survival in the experimental arm.
Fig A2.
Superior median overall survival was seen for patients in E2100 who experienced grade 3 or 4 hypertension (H) or no hypertension (NH).
Supported by an American Society of Clinical Oncology Career Development Award, the Breast Cancer Research Foundation, and a grant from Genentech; also supported by funding for the Consortium on Breast Cancer Pharmacogenomics from the National Institute of General Medical Sciences Pharmacogenetics Research Network (Grant No. U01GM061373; D.A.F.).
Authors’ disclosures of potential conflicts of interest and author contributions are found at the end of this article.
REFERENCES
- 1.Schneider BP, Sledge GW Jr: Drug insight: VEGF as a therapeutic target for breast cancer. Nat Clin Pract Oncol 4:181-189, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Hurwitz HI, Fehrenbacher L, Hainsworth JD, et al: Bevacizumab in combination with fluorouracil and leucovorin: An active regimen for first-line metastatic colorectal cancer. J Clin Oncol 23:3502-3508, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Monk BJ, Han E, Josephs-Cowan CA, et al: Salvage bevacizumab (rhuMAB VEGF)-based therapy after multiple prior cytotoxic regimens in advanced refractory epithelial ovarian cancer. Gynecol Oncol 102:140-144, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Sandler A, Gray R, Brahmer J, et al: Randomized phase II/III trial of paclitaxel (P) plus carboplatin (C) with or without bevacizumab (NSC #704865) in patients with advanced non-squamous non-small cell lung cancer (NSCLC): An Eastern Cooperative Oncology Group (ECOG) Trial E4599. J Clin Oncol 23:2s, 2005. (suppl; abstr LBA4) [Google Scholar]
- 5.Yang JC, Haworth L, Sherry RM, et al: A randomized trial of bevacizumab, an anti-vascular endothelial growth factor antibody, for metastatic renal cancer. N Engl J Med 349:427-434, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller K, Wang M, Gralow J, et al: Paclitaxel plus bevacizumab versus paclitaxel alone for metastatic breast cancer. N Engl J Med 357:2666-2676, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Jubb AM, Hurwitz HI, Bai W, et al: Impact of vascular endothelial growth factor-A expression, thrombospondin-2 expression, and microvessel density on the treatment effect of bevacizumab in metastatic colorectal cancer. J Clin Oncol 24:217-227, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Miller K, Wang M, Gralow J, et al: A randomized phase III trial of paclitaxel versus paclitaxel plus bevacizumab as first-line therapy for locally recurrent or metastatic breast cancer: A trial coordinated by the Eastern Cooperative Oncology Group (E2100). Breast Cancer Res 94:S6, 2005. (suppl 1) [Google Scholar]
- 9.Stevens A, Soden J, Brenchley PE, et al: Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res 63:812-816, 2003 [PubMed] [Google Scholar]
- 10.Schneider BP, Radovich M, Sledge GW, et al: Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat [epub ahead of print on September 20, 2007] [DOI] [PubMed]
- 11.Krippl P, Langsenlehner U, Renner W, et al: A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer 106:468-471, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Jacobs EJ, Feigelson HS, Bain EB, et al: Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the Cancer Prevention Study II cohort. Breast Cancer Res 8:R22, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Q, Hemminki K, Enquist K, et al: Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin Cancer Res 11:3647-3653, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Schneider BP, Skaar TC, Sledge GW, et al: Analysis of angiogenesis genes from paraffin-embedded breast tumor and lymph nodes. Breast Cancer Res Treat 96:209-215, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Berger U, Wilson P, McClelland RA, et al: Correlation of immunocytochemically demonstrated estrogen receptor distribution and histopathologic features in primary breast cancer. Hum Pathol 18:1263-1267, 1987 [DOI] [PubMed] [Google Scholar]
- 16.Kinsel LB, Szabo E, Greene GL, et al: Immunocytochemical analysis of estrogen receptors as a predictor of prognosis in breast cancer patients: Comparison with quantitative biochemical methods. Cancer Res 49:1052-1056, 1989 [PubMed] [Google Scholar]
- 17.Demetri GD, van Oosterom AT, Garrett CR, et al: Efficacy and safety of sunitinib in patients with advanced gastrointestinal stromal tumour after failure of imatinib: A randomised controlled trial. Lancet 368:1329-1338, 2006 [DOI] [PubMed] [Google Scholar]
- 18.Escudier B, Eisen T, Stadler WM, et al: Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med 356:125-134, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Goodman VL, Rock EP, Dagher R, et al: Approval summary: Sunitinib for the treatment of imatinib refractory or intolerant gastrointestinal stromal tumors and advanced renal cell carcinoma. Clin Cancer Res 13:1367-1373, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Motzer RJ, Hutson TE, Tomczak P, et al: Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 356:115-124, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Jubb AM, Oates AJ, Holden S, et al: Predicting benefit from anti-angiogenic agents in malignancy. Nat Rev Cancer 6:626-635, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Folkman J: Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med 1:27-31, 1995 [DOI] [PubMed] [Google Scholar]
- 23.Ferrara N: Role of vascular endothelial growth factor in regulation of physiological angiogenesis. Am J Physiol Cell Physiol 280:C1358-C1366, 2001 [DOI] [PubMed] [Google Scholar]
- 24.Evans WE, McLeod HL: Pharmacogenomics: Drug disposition, drug targets, and side effects. N Engl J Med 348:538-549, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Ince WL, Jubb AM, Holden SN, et al: Association of k-ras, b-raf, and p53 status with the treatment effect of bevacizumab. J Natl Cancer Inst 97:981-989, 2005 [DOI] [PubMed] [Google Scholar]
- 26.Kim JG, Sohn SK, Chae YS, et al: Vascular endothelial growth factor gene polymorphisms associated with prognosis for patients with gastric cancer. Ann Oncol 18:1030-1036, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Park HM, Hong SH, Kim JW, et al: Gender-specific association of the VEGF-2578C > A polymorphism in Korean patients with colon cancer. Anticancer Res 27:2535-2539, 2007 [PubMed] [Google Scholar]
- 28.Chae YS, Kim JG, Sohn SK, et al: Investigation of vascular endothelial growth factor gene polymorphisms and its association with clinicopathologic characteristics in gastric cancer. Oncology 71:266-272, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Hefler LA, Mustea A, Konsgen D, et al: Vascular endothelial growth factor gene polymorphisms are associated with prognosis in ovarian cancer. Clin Cancer Res 13:898-901, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Closas M, Malats N, Real FX, et al: Large-scale evaluation of candidate genes identifies associations between VEGF polymorphisms and bladder cancer risk. PLoS Genet 3:e29, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzanakis N, Gazouli M, Rallis G, et al: Vascular endothelial growth factor polymorphisms in gastric cancer development, prognosis, and survival. J Surg Oncol 94:624-630, 2006 [DOI] [PubMed] [Google Scholar]
- 32.Sfar S, Hassen E, Saad H, et al: Association of VEGF genetic polymorphisms with prostate carcinoma risk and clinical outcome. Cytokine 35:21-28, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Balasubramanian SP, Brown NJ, Reed MW: Role of genetic polymorphisms in tumour angiogenesis. Br J Cancer 87:1057-1065, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarron SL, Edwards S, Evans PR, et al: Influence of cytokine gene polymorphisms on the development of prostate cancer. Cancer Res 62:3369-3372, 2002 [PubMed] [Google Scholar]
- 35.Al-Kateb H, Mirea L, Xie X, et al: Multiple variants in vascular endothelial growth factor (VEGFA) are risk factors for time to severe retinopathy in type 1 diabetes: The DCCT/EDIC genetics study. Diabetes 56:2161-2168, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Errera FI, Canani LH, Silva ME, et al: Functional vascular endothelial growth factor-634G>C SNP is associated with proliferative diabetic retinopathy: A case-control study in a Brazilian population of European ancestry. Diabetes Care 30:275-279, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Buraczynska M, Ksiazek P, Baranowicz-Gaszczyk I, et al: Association of the VEGF gene polymorphism with diabetic retinopathy in type 2 diabetes patients. Nephrol Dial Transplant 22:827-832, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Banyasz I, Bokodi G, Vannay A, et al: Genetic polymorphisms of vascular endothelial growth factor and angiopoietin 2 in retinopathy of prematurity. Curr Eye Res 31:685-690, 2006 [DOI] [PubMed] [Google Scholar]
- 39.Suganthalakshmi B, Anand R, Kim R, et al: Association of VEGF and eNOS gene polymorphisms in type 2 diabetic retinopathy. Mol Vis 12:336-341, 2006 [PubMed] [Google Scholar]
- 40.Ray D, Mishra M, Ralph S, et al: Association of the VEGF gene with proliferative diabetic retinopathy but not proteinuria in diabetes. Diabetes 53:861-864, 2004 [DOI] [PubMed] [Google Scholar]
- 41.Awata T, Inoue K, Kurihara S, et al: A common polymorphism in the 5′-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes 51:1635-1639, 2002 [DOI] [PubMed] [Google Scholar]
- 42.McKnight AJ, Maxwell AP, Patterson CC, et al: Association of VEGF-1499C–>T polymorphism with diabetic nephropathy in type 1 diabetes mellitus. J Diabetes Complications 21:242-245, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Chow KM, Szeto CC, Lai FM, et al: Genetic polymorphism of vascular endothelial growth factor: Impact on progression of IgA nephropathy. Ren Fail 28:15-20, 2006 [DOI] [PubMed] [Google Scholar]
- 44.Doi K, Noiri E, Nakao A, et al: Functional polymorphisms in the vascular endothelial growth factor gene are associated with development of end-stage renal disease in males. J Am Soc Nephrol 17:823-830, 2006 [DOI] [PubMed] [Google Scholar]
- 45.Summers AM, Coupes BM, Brennan MF, et al: VEGF-460 genotype plays an important role in progression to chronic kidney disease stage 5. Nephrol Dial Transplant 20:2427-2432, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Shim JY, Jun JK, Jung BK, et al: Vascular endothelial growth factor gene +936 C/T polymorphism is associated with preeclampsia in Korean women. Am J Obstet Gynecol 197:271.e1-e4, 2007 [DOI] [PubMed] [Google Scholar]
- 47.Banyasz I, Szabo S, Bokodi G, et al: Genetic polymorphisms of vascular endothelial growth factor in severe pre-eclampsia. Mol Hum Reprod 12:233-236, 2006 [DOI] [PubMed] [Google Scholar]
- 48.Papazoglou D, Galazios G, Papatheodorou K, et al: Vascular endothelial growth factor gene polymorphisms and idiopathic recurrent pregnancy loss. Fertil Steril 83:959-963, 2005 [DOI] [PubMed] [Google Scholar]
- 49.Petrovic D, Verhovec R, Globocnik Petrovic M, et al: Association of vascular endothelial growth factor gene polymorphism with myocardial infarction in patients with type 2 diabetes. Cardiology 107:291-295, 2007 [DOI] [PubMed] [Google Scholar]
- 50.Howell WM, Ali S, Rose-Zerilli MJ, et al: VEGF polymorphisms and severity of atherosclerosis. J Med Genet 42:485-490, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang B, Cross DF, Ollerenshaw M, et al: Polymorphisms of the vascular endothelial growth factor and susceptibility to diabetic microvascular complications in patients with type 1 diabetes mellitus. J Diabetes Complications 17:1-6, 2003 [DOI] [PubMed] [Google Scholar]
- 52.Goetz MP, Rae JM, Suman VJ, et al: Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 23:9312-9318, 2005 [DOI] [PubMed] [Google Scholar]
- 53.Jin Y, Desta Z, Stearns V, et al: CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst 97:30-39, 2005 [DOI] [PubMed] [Google Scholar]
- 54.Rae J, Sikora M, Henry N, et al: Cytochrome P450 2D6 activity predicts adherence to tamoxifen therapy. Breast Cancer Res Treat 106:S21, 2007. (suppl) [Google Scholar]
- 55.Ebos JM, Lee CR, Christensen JG, et al: Multiple circulating proangiogenic factors induced by sunitinib malate are tumor-independent and correlate with antitumor efficacy. Proc Natl Acad Sci U S A 104:17069-17074, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]