Abstract
Purpose
To analyze the prognostic impact of Wilms’ tumor 1 (WT1) gene mutations in cytogenetically normal acute myeloid leukemia (CN-AML).
Patients and Methods
We studied 196 adults younger than 60 years with newly diagnosed primary CN-AML, who were treated similarly on Cancer and Leukemia Group B (CALGB) protocols 9621 and 19808, for WT1 mutations in exons 7 and 9. The patients also were assessed for the presence of FLT3 internal tandem duplications (FLT3-ITD), FLT3 tyrosine kinase domain mutations (FLT3-TKD), MLL partial tandem duplications (MLL-PTD), NPM1 and CEBPA mutations, and for the expression levels of ERG and BAALC.
Results
Twenty-one patients (10.7%) harbored WT1 mutations. Complete remission rates were not significantly different between patients with WT1 mutations and those with unmutated WT1 (P = .36; 76% v 84%). Patients with WT1 mutations had worse disease-free survival (DFS; P < .001; 3-year rates, 13% v 50%) and overall survival (OS; P < .001; 3-year rates, 10% v 56%) than patients with unmutated WT1. In multivariable analyses, WT1 mutations independently predicted worse DFS (P = .009; hazard ratio [HR] = 2.7) when controlling for CEBPA mutational status, ERG expression level, and FLT3-ITD/NPM1 molecular-risk group (ie, FLT3-ITDnegative/NPM1mutated as low risk v FLT3-ITDpositive and/or NPM1wild-type as high risk). WT1 mutations also independently predicted worse OS (P < .001; HR = 3.2) when controlling for CEBPA mutational status, FLT3-ITD/NPM1 molecular-risk group, and white blood cell count.
Conclusion
We report the first evidence that WT1 mutations independently predict extremely poor outcome in intensively treated, younger patients with CN-AML. Future trials should include testing for WT1 mutations as part of molecularly based risk assessment and risk-adapted treatment stratification of patients with CN-AML.
INTRODUCTION
Cytogenetically normal acute myeloid leukemia (CN-AML) is the largest cytogenetic subgroup of AML, representing approximately 45% of adult patients with AML who are younger than 60 years.1-3 During the last decade, CN-AML has been recognized as highly heterogeneous molecularly, because several abnormalities were discovered, including mutations in FLT3, NPM1, CEBPA, and MLL genes and aberrant expression of BAALC, ERG, and MN1 genes.4 These genetic alterations have been associated with treatment outcome and serve as a basis for molecularly guided risk assessment in CN-AML.4,5 However, discovery of novel genetic markers likely will improve molecular-risk stratification and will allow a more accurate prediction of response to current therapy.
The Wilms’ tumor 1 (WT1) gene, located on chromosome 11p13,6 encodes a transcriptional regulator that is capable of activating or repressing gene transcription, depending on the cell type, the WT1 protein isoform, and the target gene.7 Although initially considered a tumor suppressor gene,8 WT1 also has been demonstrated to act as an oncogene.7,9-11 The functional duality of WT1 as a tumor suppressor gene and an oncogene, however, is not well understood and appears to depend on the genomic and cellular milieu.7 Expression of the WT1 gene has been detected in 75% to 100% of patients with AML, but the results of studies that evaluated the impact of WT1 expression levels at diagnosis on clinical outcome have been inconsistent.12-17
Intragenic WT1 mutations are found in at least 5% of patients with sporadic Wilms’ tumors,18 which is one of the most common nonhematologic neoplasms in children.19 In addition, WT1 mutations have been found in rare congenital malformation syndromes with predisposition for the development of Wilms’ tumors20 and have been reported anecdotally in other cancers, including non–asbestos-related mesothelioma and juvenile granulosa cell tumor.18
In earlier studies of acute leukemias, WT1 mutations occurred in up to 15% of patients with AML,21-25 in 18% of patients with biphenotypic or undifferentiated acute leukemia,24 and rarely in those with acute lymphoblastic leukemia.24 To our knowledge, only two reports that address the prognostic relevance of WT1 mutations in AML have been published.24,26 One study that included 33 adult and childhood patients with AML who had various cytogenetic findings found that none of the four patients who harbored WT1 mutations achieved a complete remission (CR) and that their overall survival was worse than that of patients without WT1 mutations.24 A more recent study by Summers et al26 focused on CN-AML and found WT1 mutations, located primarily in exons 7 and 9, in seven (10%) of 70 patients with CN-AML.26 Of the six patients with WT1 mutations who received standard induction therapy, five did not achieve a CR. Interestingly, each of the five patients with WT1 mutations who failed induction therapy harbored simultaneously a FLT3-internal tandem duplication (FLT3-ITD).26 However, the analysis of other molecular markers with established prognostic relevance in CN-AML4 was not reported.26
The objectives of our study were to assess the frequency and prognostic value of WT1 mutations in the context of other prognostic molecular factors among younger patients with CN-AML who were treated similarly on Cancer and Leukemia Group B (CALGB) protocols that incorporated intensification therapy with autologous peripheral-blood stem-cell transplantation (APBSCT).
PATIENTS AND METHODS
Patients
We studied 196 adults who were younger than 60 years and who had untreated primary CN-AML. The diagnosis of CN-AML was based on standard cytogenetic analysis that was performed by CALGB-approved institutional cytogenetic laboratories as part of the cytogenetic companion study 8461.1 To be considered cytogenetically normal, at least 20 metaphase cells from diagnostic bone marrow (BM) had to be evaluated, and the karyotype had to be found normal in each patient. All cytogenetic results were confirmed by central karyotype review. All patients were enrolled on two similar CALGB treatment protocols (ie, 9621 or 19808). Institutional Review Board–approved informed consent for participation in the studies was obtained from all patients.
Treatment
Details of the treatment protocols have been previously reported.27,28 Briefly, patients on CALGB 9621 received induction chemotherapy with cytarabine, daunorubicin, and etoposide with (ADEP) or without (ADE) the multidrug resistance protein modulator PSC-833, also called valspodar.27 Patients who had CN-AML and who achieved a CR received high-dose cytarabine (HiDAC) and etoposide for stem-cell mobilization followed by myeloablative treatment with busulfan and etoposide supported by APBSCT. Patients unable to receive APBSCT received two additional cycles of HiDAC. Patients enrolled on CALGB 19808 were treated similarly28 to those on CALGB 9621. None of the patients received allogeneic stem-cell transplantation in first remission.
Mutational Analysis of WT1
Mononuclear cells from diagnostic BM and/or blood specimens were enriched by Ficoll density gradient centrifugation and were cryopreserved in liquid nitrogen until use. Genomic DNA was extracted from cryopreserved mononuclear cell preparations of diagnostic BM or blood by using the commercially available DNeasy Tissue Kit (Qiagen, Valencia, CA) or the Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturers’ instructions. DNA fragments that spanned the entire WT1 exons 7 and 9 were amplified by polymerase chain reaction (PCR) by using AmpliTaq Gold (Applied Biosystems, Foster City, CA). Intronic primers previously reported for WT1 exons 729 and 930 were used in the PCRs. PCR fragments of appropriate size were identified after agarose gel electrophoresis for all samples. Amplicons from each patient were pooled with unmutated reference amplicons, were denatured, and were cooled down slowly to 25°C. The reannealed DNA duplexes were analyzed for mutations by denaturing high-performance liquid chromatography (DHPLC)31 by using a WAVE 3500HT DNA Fragment Analysis System (Transgenomic Inc, Omaha, NE). The individual elution peaks were compared with the unmutated reference sample. Samples with elution peaks that differed from the unmutated reference were reamplified in an independent PCR and were assessed for variations in the coding DNA by direct sequencing in both directions. The results obtained by direct sequencing were confirmed by subcloning of mutated amplicons into the pCR2.1-TOPO vector (Invitrogen) and by sequencing of 18 or fewer independent clones.
Analyses of Other Molecular Markers
Other molecular markers (ie, FLT3-ITD,32 FLT3 tyrosine kinase domain mutation [FLT3-TKD],33-35 MLL partial tandem duplication [MLL-PTD],36,37 NPM138 and CEBPA39 mutations, and ERG5,40 and BAALC41 expression levels) were assessed as previously reported.
Definition of Clinical End Points
CR required an absolute neutrophil count ≥ 1,500/μL, a platelet count ≥ 100,000/μL, no leukemic blasts in the blood, BM cellularity greater than 20% with maturation of all cell lines, no Auer rods, less than 5% BM blast cells, and no evidence of extramedullary leukemia, all of which had persisted for at least 1 month. Relapse was defined by ≥ 5% BM blasts, circulating leukemic blasts, or the development of extramedullary leukemia.42 Disease-free survival (DFS) was measured from the date of CR until the date of relapse or death; patients alive and relapse-free at last follow-up were censored. Overall survival (OS) was measured from the date on study until the date of death, and patients alive at last follow-up were censored.
Statistical Analysis
Associations between patients with and without WT1 mutations and baseline demographic, clinical, and molecular features were described by using Fisher's exact and Wilcoxon rank sum tests for categoric and continuous variables, respectively. Estimated probabilities of DFS and OS were calculated using the Kaplan-Meier method, and the log-rank test evaluated differences between survival distributions. Proportional hazards models were constructed for DFS and OS to evaluate the impact of WT1 mutations when controlling for other prognostic variables. Variables other than WT1 mutational status that were considered for model inclusion were age, sex, race, hemoglobin, platelet count, log2(white blood cell count) [log2(WBC)], percentage of blasts in the blood and BM, ERG and BAALC expression levels (high v low), CEBPA status (mutated v wild-type), FLT3-TKD and MLL-PTD (present v absent), and molecular risk group (high v low) as defined by FLT3-ITD/NPM1 molecular features (ie, FLT3-ITDnegative/NPM1mutated as low risk and FLT3-ITDpositive and/or NPM1wild-type as high risk). Variables were entered by forward selection into the model by using the Wald test until no variable entered with P < .05. The proportional hazards assumption was checked for each variable individually. If the proportional hazards assumption was not met for a particular variable, then an artificial time-dependent covariate was included in all models that contained that variable.43 All analyses were performed by the CALGB Statistical Center.
RESULTS
Frequency and Types of WT1 Mutations
Twenty-one patients (10.7%) had at least one WT1 mutation (Table 1). Mutations in exon 7 were found in 16 patients. Among these 16 patients, two patients had two mutations in exon 7 simultaneously, and one patient had a mutation in exon 7 in addition to a mutation in exon 9. One WT1 mutation in exon 7 was a nonsense mutation; all others were frameshift mutations. The frameshift mutations were mainly caused by various small duplications; less frequent mutations were imperfect small repeats of exon 7 sequences or combined deletions/insertions. All WT1 mutations in exon 7 resulted in a premature truncation of the WT1 protein, with loss of the zinc finger region or truncation after the second zinc finger.
Table 1.
WT1 Mutations and Individual Outcome in 21 Patients With CN-AML
| Patient by Mutation | Age (years) | Sex | DNA Change*† | Protein Change†‡ | Individual Outcome |
|---|---|---|---|---|---|
| WT1mut7 | |||||
| 1 | 44 | F | 1301_1304dup | R302PfsX16 | Relapse/death |
| 2 | 25 | M | 1317_1318insCTGTGCCTGGAGT | A307CfsX14 | Relapse/death |
| 3 | 30 | F | 1317_1318insCTGTGCCTGGAGT | A307CfsX14 | Relapse/death |
| 4 | 52 | M | 1325_1331dup | V311DfsX8 | Relapse/death |
| 5 | 33 | F | 1328_1329dup | V311LfsX71§ | Relapse/death |
| 6 | 37 | F | 1334delinsGG | R312GfsX5 | Relapse/death |
| 7 | 51 | F | 1327_1334dup | R312LfsX72§ | Failure to achieve CR/death |
| 8 | 21 | F | 1324_1336dup | S313DfsX8 | Relapse/death |
| 9 | 38 | M | 1326_1335dup | A314CfsX6 | Relapse/death |
| 10 | 50 | F | 1329_1338dup | A314CfsX6 | Relapse/death |
| 11 | 51 | F | 1337_1340dup | A314VfsX4 | Relapse/death |
| 12 | 40 | M | 1338dupC | A314GfsX3 | Failure to achieve CR/death |
| 13 | 18 | M | 1335_1344dup | E316VfsX4 | Relapse/death |
| 14 | 58 | F | [1331_1337dup] + [1338C>A] | [S313CfsX6] + [S313X] | Failure to achieve CR/death |
| 15 | 43 | M | [1306dupT] + [1324_1333dup] | [V303CfsX14] + [R312DfsX8] | Achieved CR/alive‖ |
| WT1mut9 | |||||
| 16 | 57 | M | 1569_1570delinsCT | R390P | Relapse/death |
| 17 | 49 | F | 1581G>C | R394P | Failure to achieve CR/death |
| 18 | 56 | F | 1586G>C | D396H | Relapse/death |
| 19 | 37 | F | 1589C>G | H397D | Relapse/death |
| 20 | 41 | M | 1591C>A | H397Q | Failure to achieve CR/death |
| WT1mut7 and WT1mut9 | |||||
| 21 | 22 | M | [1330_1337dup(+)1581G>A] | [S313LfsX71(+)R394Q]§ | Achieved CR/alive |
Abbreviations: CN, cytogenetically normal; AML, acute myeloid leukemia; WT1mut7, WT1 mutations in exon 7; WT1mut9, WT1 mutations in exon 9; CR, complete response.
Nucleotide sequence numbering is according to Genebank accession number NM_024426.
The sequence variations are designated according to the current recommendations of the Human Genome Variation Society (http://www.hgvs.org/mutnomen/).
The protein changes are theoretically deduced.
The stop codons created by these WT1mut7 do not occur until the 5′ part of exon 9; DNA direct sequencing of exon 8 revealed a wild-type sequence.
During the preparation of the article, this patient suffered a relapse and was undergoing salvage therapy.
Mutations in exon 9 without accompanying mutations in exon 7 were found in five patients. All WT1 mutations in exon 9 resulted in single amino acid substitutions that affected the third zinc finger in the WT1 protein. All samples with WT1 mutations retained the wild-type sequence in addition to the mutated sequence, which suggested heterozygosity for the mutant allele.
Clinical Characteristics, Other Molecular Markers, and Treatment
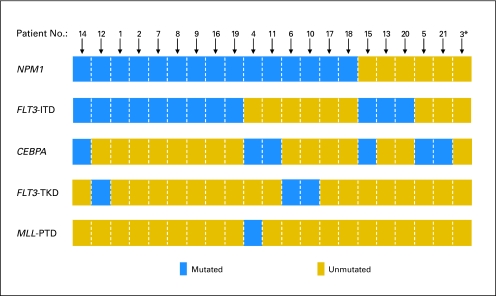
At diagnosis, patients with WT1 mutations had greater WBCs (P = .01), were more often high ERG (P = .01) and BAALC (P = .006) expressers, and tended to harbor FLT3-ITD more often (P = .06) than patients with unmutated WT1 (Table 2). The frequency of the FLT3-ITDnegative/NPM1mutated status was not significantly different between WT1-mutated and WT1-unmutated patient groups (P = .63; 29% v 37%), nor were the proportions of patients with a high (≥ 0.7) FLT3-ITD/FLT3–wild-type allele ratio (P = 1.00; 19% v 18%). Figure 1 shows the mutation status of the NPM1, FLT3 (ITD and TKD), CEBPA, and MLL (PTD) genes that were coexisting in individual patients with WT1 mutations. There was no significant difference between the WT1-mutated and WT1-unmutated patient groups with regard to the inclusion of PSC-833 (ie, valspodar) in induction (P = .63) or in the proportion of patients who received APBSCT for consolidation (P = 1.00).
Table 2.
Comparison of Pretreatment Clinical and Molecular Characteristics, Treatment and Outcomes of CN-AML Patients With (WT1mut) and Without (WT1wt) WT1 Mutations
| Characteristic | Mutation Status
|
P | |
|---|---|---|---|
| WT1mut (n = 21) | WT1wt (n = 175) | ||
| Age, years | .38 | ||
| Median | 41 | 46 | |
| Range | 18-58 | 19-59 | |
| Sex, male | .65 | ||
| No. | 9 | 86 | |
| % | 43 | 49 | |
| Race | .48 | ||
| White | |||
| No. | 20 | 151 | |
| % | 95 | 88 | |
| Nonwhite | |||
| No. | 1 | 21 | |
| % | 5 | 12 | |
| Hemoglobin, g/dL | .50 | ||
| Median | 9.4 | 9.4 | |
| Range | 4.9-12.1 | 4.6-13.6 | |
| Platelet count, × 109/L | .28 | ||
| Median | 54 | 62 | |
| Range | 7-235 | 8-466 | |
| WBC count, × 109/L | .01 | ||
| Median | 50.9 | 23.8 | |
| Range | 11.3-210.0 | 0.9-295.0 | |
| Blood blasts, % | .64 | ||
| Median | 61 | 59 | |
| Range | 10-93 | 0-97 | |
| Bone marrow blasts, % | .78 | ||
| Median | 70 | 65 | |
| Range | 32-93 | 10-99 | |
| FLT3-ITD | .06 | ||
| Absent | |||
| No. | 9 | 113 | |
| % | 43 | 65 | |
| Present | |||
| No. | 12 | 62 | |
| % | 57 | 35 | |
| FLT3-TKD | .42 | ||
| Absent | |||
| No. | 18 | 157 | |
| % | 86 | 91 | |
| Present | |||
| No. | 3 | 15 | |
| % | 14 | 9 | |
| NPM1 | .81 | ||
| Wild-type | |||
| No. | 6 | 60 | |
| % | 29 | 34 | |
| Mutated | |||
| No. | 15 | 115 | |
| % | 71 | 66 | |
| FLT3-ITD/NPM1 status* | .63 | ||
| Low-risk | |||
| No. | 6 | 64 | |
| % | 29 | 37 | |
| High-risk | |||
| No. | 15 | 111 | |
| % | 71 | 63 | |
| MLL-PTD | 1.00 | ||
| Absent | |||
| No. | 20 | 163 | |
| % | 95 | 93 | |
| Present | |||
| No. | 1 | 12 | |
| % | 5 | 7 | |
| CEBPA | .24 | ||
| Wild-type | |||
| No. | 15 | 121 | |
| % | 71 | 82 | |
| Mutated | |||
| No. | 6 | 26 | |
| % | 29 | 18 | |
| ERG expression† | .01 | ||
| Low | |||
| No. | 7 | 88 | |
| % | 37 | 68 | |
| High | |||
| No. | 12 | 41 | |
| % | 63 | 32 | |
| BAALC expression‡ | .006 | ||
| Low | |||
| No. | 4 | 74 | |
| % | 21 | 57 | |
| High | |||
| No. | 15 | 55 | |
| % | 79 | 43 | |
| Induction treatment | .63 | ||
| ADE | |||
| No. | 15 | 111 | |
| % | 71 | 63 | |
| ADEP | |||
| No. | 6 | 64 | |
| % | 29 | 37 | |
| Complete remission rate | .36 | ||
| No. | 16 | 147 | |
| % | 76 | 84 | |
| Receiving APBSCT for consolidation | 1.00 | ||
| No. | 11 | 101 | |
| % | 69 | 69 | |
| Disease-free survival, years | < .001 | ||
| Median | 0.6 | 3.4 | |
| Disease-free at 3 years | |||
| % | 13 | 50 | |
| 95% CI | 2 to 33 | 42 to 58 | |
| Overall survival, years | < .001 | ||
| Median | 0.8 | 4.5 | |
| Alive at 3 years | |||
| % | 10 | 56 | |
| 95% CI | 2 to 26 | 49 to 64 | |
Abbreviations: CN, cytogenetically normal; AML, acute myeloid leukemia; WT1mut, WT1 mutations; WT1wt, unmutated (wild-type) WT1 alleles; FLT3-ITD, internal tandem duplication of the FLT3 gene; FLT3-TKD, tyrosine kinase domain mutation of the FLT3 gene; MLL-PTD, partial tandem duplication of the MLL gene; ADE, cytarabine, daunorubicin, and etoposide; ADEP, cytarabine, daunorubicin, etoposide, and valspodar; APBSCT, autologous peripheral-blood stem-cell transplantation; CI, confidence interval.
FLT3-ITDnegative/NPM1mutated molecular status defines the low-risk group. FLT3-ITDpositive and/or NPM1wild-type molecular status defines the high-risk group.
In patients on protocol 9621, cut point was the same as in Marcucci et al.40 In patients on protocol 19808, cut point was the same as in Marcucci et al.5
In patients on protocol 9621, cut point was the same as in Baldus et al.41 In patients on protocol 19808, the median BAALC expression value was used for cut point.
Fig 1.
Coexistence of WT1 mutations with mutations in other genes, such as NPM1, FLT3 (ITD), CEBPA, FLT3 (TKD), and MLL (PTD) among 21 patients who harbored WT1 mutations. (*) Indicates the presence of the CEBPA polymorphism c.1175_1180dup in patient 3.
Prognostic Impact of WT1 Mutations
One-hundred sixty-three (83%) of 196 patients in this study achieved a CR, and the estimated DFS and OS rates at 3 years were 46% and 51%, respectively. The median follow-up for patients who remained alive was 4.2 years (range, 1.2 to 8.9 years).
The outcome results with respect to WT1 mutational status are summarized in Table 2. Among the 21 patients with WT1 mutations, 16 (76%) achieved a CR. This percentage was slightly lower, but not significantly different, than the 84% of patients with unmutated WT1 that achieved a CR (P = .36). All five patients with WT1 mutations who did not achieve a CR had resistant disease. Interestingly, four of them also harbored an NPM1 mutation, which in three of these patients coexisted with FLT3-ITD; the fifth patient had FLT3-ITD but no NPM1 mutation.
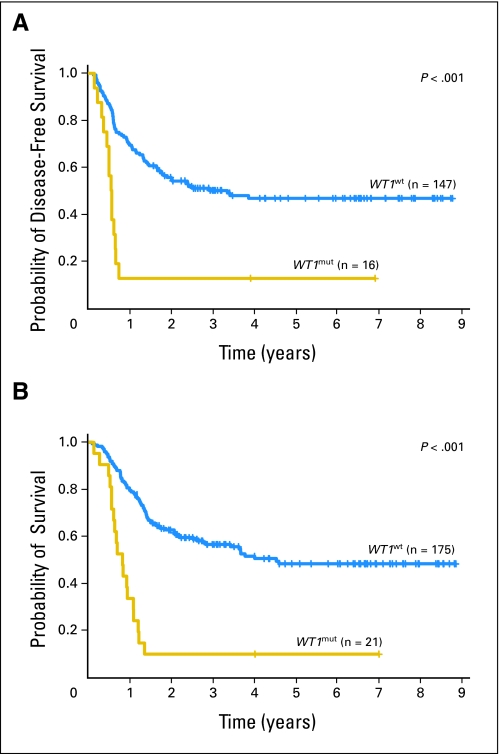
Of the patients who achieved a CR, those with WT1 mutations relapsed more frequently than those with unmutated WT1 (88% v 51%; P = .007). The estimated 3-year DFS rate was only 13% for patients with WT1 mutations compared with 50% for patients with unmutated WT1 (Fig 2A).
Fig 2.
(A) Disease-free and (B) overall survival of patients with CN-AML according to mutational WT1 status (ie, patients with WT1 mutations [WT1mut] v patients without WT1 mutations [WT1wt]).
In a multivariable analysis (Table 3), WT1 mutational status independently predicted worse DFS (P = .009) when controlling for CEBPA mutational status (P = .004), the FLT3-ITD/NPM1 molecular-risk group (P = .006), and ERG expression level (P = .04); the estimated risk of relapse or death was almost three times higher for patients who had WT1 mutations compared with patients who had unmutated WT1 (hazard ratio [HR] = 2.7; Table 3). At the time of the analysis, all relapses in the WT1-mutated patients occurred within 9 months of CR achievement; there were only two of the 16 patients with WT1 mutations who achieved a CR and had not relapsed—one at 4 years, and one at 7 years. However, during preparation of this manuscript, one of these patients experienced a relapse at 5 years after CR achievement. This patient currently is undergoing salvage therapy.
Table 3.
Multivariable Analyses for DFS and OS
| Endpoint | Variables in the Final Models | HR | 95% CI | P |
|---|---|---|---|---|
| DFS | WT1:WT1mutv WT1wt | 2.7 | 1.3 to 5.5 | .009 |
| FLT3-ITD/NPM1 status: high v low risk* | 2.5 | 1.3 to 4.7 | .006 | |
| CEBPA:CEBPAmutvCEBPAwt | 0.3 | 0.2 to 0.7 | .004 | |
| ERG expression: high v low† | 1.9 | 1.0 to 3.5 | .04 | |
| OS | WT1:WT1mutv WT1wt | 3.2 | 1.7 to 6.2 | < .001 |
| FLT3-ITD/NPM1 status: high v low risk* | 2.7 | 1.4 to 5.2 | .004 | |
| CEBPA:CEBPAmutvCEBPAwt | 0.4 | 0.2 to 0.8 | .02 | |
| Log2(WBC) two-fold increase‡ | 1.9 | 1.2 to 3.2 | .04 |
NOTE. Hazard ratios greater than or less than 1 indicate an increased or decreased risk, respectively, of an event for greater values of continuous variables and for the first category listed for dichotomous variables.
Abbreviations: DFS, disease-free survival; OS, overall survival; HR, hazard ratio; WT1mut, presence of WT1 mutations; WT1wt, absence of WT1 mutations; FLT3-ITD, internal tandem duplication of the FLT3 gene; CEBPAmut, presence of CEBPA mutations; CEBPAwt, absence of CEBPA mutations; Log2(WBC), log-transformed white blood cell count.
The high-risk molecular group is defined by the presence of FLT3-ITD and/or the absence of NPM1 mutations. The low-risk molecular group is defined by the absence of FLT3-ITD and by the simultaneous presence of an NPM1 mutation.
ERG expression did not meet the proportional hazards assumption. The P value corresponds to the Wald statistic of a two degree of freedom test that evaluated whether the coefficients for ERG expression and an artificial time-dependent covariate were equal to zero. The risk of an event was similar for high and low ERG expressers shortly after achieving complete remission, but the risk for high ERG expressers increased over time. The hazard ratio evaluated at 1 year is presented.
Log2(WBC) did not meet the proportional hazards assumption. The P value corresponds to the Wald statistic of a two degree of freedom test that evaluated whether the coefficients for log2(WBC) and an artificial time-dependent covariate were equal to zero. The risk of an event was larger for those with greater WBC counts soon after going on study, but this risk decreased over time. The hazard ratio presented is for a two-fold increase in WBC when evaluated at 3 months.
Likewise, patients with WT1 mutations had shorter OS than patients with unmutated WT1 (P < .001). The estimated OS rates at 3 years were 10% and 56% for patients with and without WT1 mutations, respectively (Fig 2B). WT1 mutations independently predicted a higher risk of death (P < .001; Table 3) when controlling for the FLT3-ITD/NPM1 molecular-risk group (P = .004), CEBPA mutational status (P = .02), and WBC (P = .04); the estimated risk of death was more than three times higher for patients who had WT1 mutations compared with patients who had unmutated WT1 (HR = 3.2; Table 3). Notably, none of the five patients with WT1 mutations who failed to achieve a CR and none of the 14 who experienced an early relapse had successful salvage treatment. All of these patients died within 17 months of study enrollment.
DISCUSSION
We present here a relatively large study that assessed the prognostic value of WT1 mutations in younger adults who had primary CN-AML and who received similar intensive treatment that did not include allogeneic stem-cell transplantation in first CR. We show that the presence of WT1 mutations at diagnosis is associated with an extremely poor outcome and that it independently predicts a higher risk of relapse and death when other molecular markers with established prognostic significance and clinical variables are taken into consideration.
Previous, relatively small studies on patients with AML who had diverse cytogenetic findings and/or secondary AML found WT1 mutations in ≤ 15% of the patients.21-25 To date, however, only Summers et al26 assessed the incidence and prognostic impact of WT1 mutations exclusively in CN-AML. This study included 70 patients with CN-AML, ranging in age between 19 and 78 years; seven (10%) of these patients had WT1 mutations, including five patients with heterozygous mutations in exon 7, one patient with concurrent mutations in exon 7 and exon 9, and one patient with a homozygous mutation in exon 9. Consistent with these results,26 we found WT1 mutations in 10.7% of patients with CN-AML, and mutations in exon 7 also were more frequent than those in exon 9. Although we found the CR rate of patients with WT1 mutations to be lower than that of patients with unmutated WT1, we did not observe a statistically significant difference. However, 14 (88%) of 16 patients who had WT1 mutations and who attained a CR relapsed within the first 9 months of CR achievement.
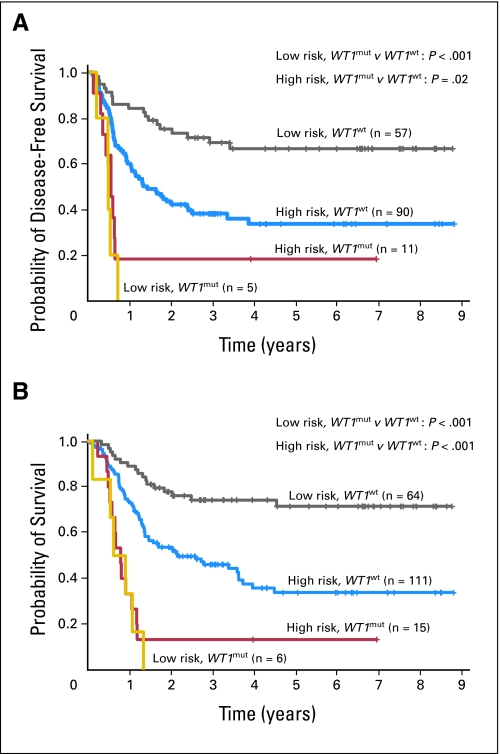
All but one patient with CN-AML who had WT1 mutations in our study had at least one additional mutation in the other genes analyzed (ie, NPM1, FLT3, CEBPA, and MLL; Fig 1). The most common among these were mutations in the NPM1 gene (71%), followed by FLT3-ITD (57%); other mutations were much less frequent. It is striking that four of five patients with WT1 mutations who did not achieve a CR also harbored an NPM1 mutation, which has been reported previously to impact favorably on the probability of CR achievement.44-46 Consistent with the study of Summers et al,26 patients who had WT1 mutations in our study tended to be FLT3-ITD–positive more often than patients with unmutated WT1. Thus, we performed multivariable analyses to assess whether the impact of WT1 mutations on outcome was independent from other established prognostic molecular markers and clinical characteristics. We show that mutations in the WT1 gene are indeed independent predictors for worse DFS and OS in younger patients who have primary CN-AML. Interestingly, all six patients with WT1 mutations who belonged to the low-risk molecular category by virtue of having FLT3-ITDnegative/NPM1mutated status died after they experienced a failure to achieve CR or relapse (Appendix Fig A1, online only), which suggests that the presence of WT1 mutations may be capable of overcoming the reported favorable prognostic impact of the coexistence of the NPM1 mutation with the lack of FLT3-ITD in this CN-AML subset.37,46,47
Patients with mutated WT1 were also high expressers of ERG and BAALC more frequently than patients with unmutated WT1. Overexpression of both the ERG5,40 and the BAALC41,48 genes has been associated with an adverse prognosis. In our study, WT1 mutations appeared to impact adversely on DFS and OS, regardless of the expression status of these genes.
The WT1 gene consists of 10 exons and encodes a transcriptional regulator that is characterized by two major functional domains—an N-terminal transcriptional regulatory domain and a C-terminal DNA and/or RNA binding domain that is composed of four zinc fingers.9,49 Mutational analyses in our study focused on WT1 exons 7 and 9, because these regions have been recognized previously as mutational hot spots in CN-AML.26 WT1 exons 7 and 9 encode the first and third zinc finger, respectively, in the WT1 protein.9,49 All mutations in exon 7 of the WT1 gene found in the present study led to a premature truncation of the protein and eliminated all or, less frequently, the last two zinc fingers. WT1 mutations in exon 9 led to single amino acid substitutions within the third zinc finger that affects residues expected to be crucial for the DNA binding ability.50 Thus, WT1 mutations would be expected to abolish, impair, or change the DNA binding ability of the WT1 protein to its target genes, including to those that encode proteins involved in the regulation of normal hematopoiesis (RARA, CSF1), apoptosis (BCL2, BCL2A1, BAK1), cell cycle (CCNE1, CDKN1A), gene transcription (MYC, PAX2, MYB, EGR1), and cell proliferation (TGFB1, PDGFA).9 Although preliminary in vitro10,51 and in vivo11 studies have implicated involvement of the WT1 protein in leukemogenesis, its role still is not understood fully,9 and the mechanisms by which WT1 mutations confer leukemic cell resistance to therapy remain to be elucidated.
In conclusion, we show for the first time that mutations in the WT1 gene represent a strong, independent predictor of poor outcome in intensively treated patients with CN-AML. On the basis of these results, we propose that upcoming clinical trials incorporate molecular testing for WT1 mutations in patients with CN-AML at diagnosis to prospectively confirm their prognostic significance, with the ultimate goals of improving current molecularly based risk stratification of CN-AML and of developing targeted therapies.
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Peter Paschka, Guido Marcucci, Clara D. Bloomfield
Financial support: Clara D. Bloomfield
Administrative support: Clara D. Bloomfield
Provision of study materials or patients: Bayard Powell, Maria R. Baer, Jonathan E. Kolitz, Richard A. Larson, Clara D. Bloomfield
Collection and assembly of data: Peter Paschka, Guido Marcucci, Amy S. Ruppert, Susan P. Whitman, Krzysztof Mrózek, Christian Langer, Claudia D. Baldus, Weiqiang Zhao, Andrew J. Carroll, Michael A. Caligiuri, Clara D. Bloomfield
Data analysis and interpretation: Peter Paschka, Guido Marcucci, Amy S. Ruppert, Krzysztof Mrózek, Kati Maharry, Clara D. Bloomfield
Manuscript writing: Peter Paschka, Guido Marcucci, Amy S. Ruppert, Krzysztof Mrózek, Clara D. Bloomfield
Final approval of manuscript: Peter Paschka, Guido Marcucci, Amy S. Ruppert, Susan P. Whitman, Krzysztof Mrózek, Kati Maharry, Christian Langer, Claudia D. Baldus, Weiqiang Zhao, Bayard Powell, Maria R. Baer, Andrew J. Carroll, Michael A. Caligiuri, Jonathan E. Kolitz, Richard A. Larson, Clara D. Bloomfield
Acknowledgments
We thank Jing Weng, Tamara Vukosavljevic, and Shunjun Liu, PhD, for their expert technical assistance and Donna Bucci for sample processing and storage services provided by the Cancer and Leukemia Group B Leukemia Tissue Bank at Ohio State University Comprehensive Cancer Center, Columbus, OH.
Appendix
The following Cancer and Leukemia Group B institutions, principal investigators, and cytogeneticists participated in this study:
The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield, Karl S. Theil, Diane Minka, and Nyla A. Heerema (Grant No. CA77658); Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, Wendy L. Flejter, and Mark J. Pettenati (Grant no. CA03927); North Shore University Hospital, Manhasset, NY: Daniel R. Budman and Prasad R.K. Koduru (grant No. CA35279); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine, and AnneMarie W. Block (Grant No. CA02599); University of Massachusetts Medical Center, Worcester, MA: William W. Walsh, Vikram Jaswaney, Kathleen Richkind, Michael J. Mitchell, and Patricia Miron (Grant No. CA37135); Dana-Farber Cancer Institute, Boston, MA: Eric P. Winer, Ramana Tantravahi, Paola Dal Cin, and Cynthia C. Morton (Grant No. CA32291); Washington University School of Medicine, St. Louis, MO: Nancy L. Bartlett, Michael S. Watson, and Jaime Garcia-Heras (Grant No. CA77440); University of North Carolina, Chapel Hill, NC: Thomas Shea and Kathleen W. Rao (Grant No. CA47559); Vermont Cancer Center, Burlington, VT: Hyman B. Muss, Elizabeth F. Allen, and Mary Tang (Grant No. CA77406); Dartmouth Medical School, Lebanon, NH: Marc S. Ernstoff and Thuluvancheri K. Mohandas (Grant No. CA04326); Duke University Medical Center, Durham, NC: Jeffrey Crawford and Mazin B. Qumsiyeh (Grant No. CA47577); University of Iowa Hospitals, Iowa City, IA: Gerald H. Clamon and Shivanand R. Patil (Grant No. CA47642); Christiana Care Health Services, Inc., Newark, DE: Stephen S. Grubbs, Digamber S. Borgaonkar, and Jeanne M. Meck (Grant No. CA45418); Massachusetts General Hospital, Boston, MA: Jeffrey W. Clark, Paola Dal Cin, and Cynthia C. Morton (Grant No. CA 12,449); Weill Medical College of Cornell University, New York, NY: John Leonard, Prasad R.K. Koduru, Andrew J. Carroll, and Susan Mathew (Grant No. CA07968); Eastern Maine Medical Center, Bangor, ME: Harvey M. Segal and Laurent J. Beauregard (Grant No. CA35406); University of Puerto Rico School of Medicine, San Juan, PR: Eileen I. Pacheco, Cynthia C. Morton, Paola Dal Cin, and Leonard L. Atkins; University of California at San Diego: Barbara A. Parker, Renée Bernstein, and Marie L. Dell'Aquila (Grant No. CA11789); Ft. Wayne Medical Oncology/Hematology, Ft. Wayne, IN: Sreenivasa Nattam and Patricia I. Bader; University of Chicago Medical Center, Chicago, IL: Gini Fleming, Diane Roulston, Yanming Zhang, and Michelle M. Le Beau (Grant No. CA41287); Western Pennsylvania Hospital, Pittsburgh, PA: Richard K. Shadduck and Gerard R. Diggans; Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (Grant No. CA04457); Rhode Island Hospital, Providence, RI: William Sikov, Shelly L. Kerman, and Aurelia Meloni-Ehrig (Grant No. CA08025); Southern Nevada Cancer Research Foundation CCOP, Las Vegas, NV: John Ellerton, E. Robert Wassman, Jr, and Marie L. Dell'Aquila (Grant No. CA35421); SUNY Upstate Medical University, Syracuse, NY: Stephen L. Graziano and Constance K. Stein (Grant No. CA21060); University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Michael C. Perry and Tim H. Huang (Grant No. CA12046); Virginia Commonwealth University MB CCOP, Richmond, VA: John D. Roberts and Colleen Jackson-Cook (Grant No. CA52784); Georgetown University Medical Center, Washington, DC: Minnetta C. Liu and Jeanne M. Meck (Grant No. CA77597); Long Island Jewish Medical Center, Lake Success, NY: Kanti R. Rai and Prasad R.K. Koduru (Grant No. CA11028); Medical University of South Carolina, Charleston, SC: Mark R. Green and G. Shashidhar Pai (Grant No. CA03927); Minneapolis VA Medical Center, Minneapolis, MN: Vicki A. Morrison and Sugandhi A. Tharapel (Grant No. CA47555); University of California at San Francisco: Charles J. Ryan and Kathleen E. Richkind (Grant No. CA60138); University of Illinois at Chicago: David J. Peace and Maureen M. McCorquodale (Grant No. CA74811); University of Minnesota, Minneapolis, MN: Bruce A. Peterson and Betsy A. Hirsch (Grant No. CA16450); University of Nebraska Medical Center, Omaha, NE: Anne Kessinger and Warren G. Sanger (Grant No. CA77298); Walter Reed Army Medical Center, Washington, DC: Thomas Reid and Digamber S. Borgaonkar (Grant No. CA26806).
Fig A1.
(A) Disease-free and (B) overall survival of patients with CN-AML according to the mutational WT1 status (ie, patients with WT1 mutations [WT1mut] v patients without WT1 mutations [WT1wt]) and the FLT3-ITD/NPM1 molecular-risk group (ie, high risk: patients with FLT3-ITD and/or wild-type NPM1; low risk: patients without FLT3-ITD and with NPM1 mutation).
published online ahead of print at www.jco.org on June 16, 2008.
Supported in part by Grants No. CA77658, CA101140, CA114725, CA31946, CA33601, and CA16058 from National Cancer Institute, Bethesda, MD and by the Coleman Leukemia Research Foundation.
Authors’ disclosures of potential conflicts of interest and author contributions are found at the end of this article.
REFERENCES
- 1.Byrd JC, Mrózek K, Dodge RK, et al: Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: Results from Cancer and Leukemia Group B (CALGB 8461). Blood 100:4325-4336, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Grimwade D, Walker H, Oliver F, et al: The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. Blood 92:2322-2333, 1998 [PubMed] [Google Scholar]
- 3.Slovak ML, Kopecky KJ, Cassileth PA, et al: Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group study. Blood 96:4075-4083, 2000 [PubMed] [Google Scholar]
- 4.Mrózek K, Marcucci G, Paschka P, et al: Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: Are we ready for a prognostically prioritized molecular classification? Blood 109:431-448, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcucci G, Maharry K, Whitman SP, et al: High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol 25:3337-3343, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Call KM, Glaser T, Ito CY, et al: Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell 60:509-520, 1990 [DOI] [PubMed] [Google Scholar]
- 7.Yang L, Han Y, Suarez Saiz F, et al: A tumor suppressor and oncogene: The WT1 story. Leukemia 21:868-876, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Haber DA, Buckler AJ, Glaser T, et al: An internal deletion within an 11p13 zinc finger gene contributes to the development of Wilms’ tumor. Cell 61:1257-1269, 1990 [DOI] [PubMed] [Google Scholar]
- 9.Ariyaratana S, Loeb DM: The role of the Wilms tumour gene (WT1) in normal and malignant haematopoiesis. Expert Rev Mol Med 14:1-17, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Yamagami T, Sugiyama H, Inoue K, et al: Growth inhibition of human leukemic cells by WT1 (Wilms tumor gene) antisense oligodeoxynucleotides: Implications for the involvement of WT1 in leukemogenesis. Blood 87:2878-2884, 1996 [PubMed] [Google Scholar]
- 11.Nishida S, Hosen N, Shirakata T, et al: AML1-ETO rapidly induces acute myeloblastic leukemia in cooperation with the Wilms tumor gene, WT1. Blood 107:3303-3312, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Bergmann L, Miething C, Maurer U, et al: High levels of Wilms’ tumor gene (wt1) mRNA in acute myeloid leukemias are associated with a worse long-term outcome. Blood 90:1217-1225, 1997 [PubMed] [Google Scholar]
- 13.Barragán E, Cervera J, Bolufer P, et al: Prognostic implications of Wilms’ tumor gene (WT1) expression in patients with de novo acute myeloid leukemia. Haematologica 89:926-933, 2004 [PubMed] [Google Scholar]
- 14.Yanada M, Terakura S, Yokozawa T, et al: Multiplex real-time RT-PCR for prospective evaluation of WT1 and fusion gene transcripts in newly diagnosed de novo acute myeloid leukemia. Leuk Lymphoma 45:1803-1808, 2004 [DOI] [PubMed] [Google Scholar]
- 15.Weisser M, Kern W, Rauhut S, et al: Prognostic impact of RT-PCR–based quantification of WT1 gene expression during MRD monitoring of acute myeloid leukemia. Leukemia 19:1416-1423, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Lapillonne H, Renneville A, Auvrignon A, et al: High WT1 expression after induction therapy predicts high risk of relapse and death in pediatric acute myeloid leukemia. J Clin Oncol 24:1507-1515, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues PC, Oliveira SN, Viana MB, et al: Prognostic significance of WT1 gene expression in pediatric acute myeloid leukemia. Pediatr Blood Cancer 49:133-138, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Little M, Wells C: A clinical overview of WT1 gene mutations. Hum Mutat 9:209-225, 1997 [DOI] [PubMed] [Google Scholar]
- 19.Brown KW, Malik KTA: The molecular biology of Wilms tumour. Expert Rev Mol Med 3:1-16, 2001 [DOI] [PubMed] [Google Scholar]
- 20.Scott RH, Stiller CA, Walker L, et al: Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J Med Genet 43:705-715, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mori N, Okada M, Motoji T, et al: Mutation of the WT1 gene in myelodysplastic syndrome and acute myeloid leukaemia post myelodysplastic syndrome. Br J Haematol 105:844-845, 1999 [DOI] [PubMed] [Google Scholar]
- 22.Takahashi T, Yamamoto R, Tanaka K, et al: Mutation analysis of the WT1 gene in secondary leukemia. Leukemia 14:1316-1317, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Hosoya N, Miyagawa K, Mitani K, et al: Mutation analysis of the WT1 gene in myelodysplastic syndromes. Jpn J Cancer Res 89:821-824, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King-Underwood L, Pritchard-Jones K: Wilms’ tumor (WT1) gene mutations occur mainly in acute myeloid leukemia and may confer drug resistance. Blood 91:2961-2968, 1998 [PubMed] [Google Scholar]
- 25.Miyagawa K, Hayashi Y, Fukuda T, et al: Mutations of the WT1 gene in childhood nonlymphoid hematological malignancies. Genes Chromosomes Cancer 25:176-183, 1999 [PubMed] [Google Scholar]
- 26.Summers K, Stevens J, Kakkas I, et al: Wilms’ tumour 1 mutations are associated with FLT3-ITD and failure of standard induction chemotherapy in patients with normal karyotype AML. Leukemia 21:550-551, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Kolitz JE, George SL, Dodge RK, et al: Dose escalation studies of cytarabine, daunorubicin, and etoposide with and without multidrug resistance modulation with PSC-833 in untreated adults with acute myeloid leukemia younger than 60 years: Final induction results of Cancer and Leukemia Group B study 9621. J Clin Oncol 22:4290-4301, 2004 [DOI] [PubMed] [Google Scholar]
- 28.Kolitz JE, George SL, Marcucci G, et al: A randomized comparison of induction therapy for untreated acute myeloid leukemia (AML) in patients < 60 years using P-glycoprotein (Pgp) modulation with valspodar (PSC833): Preliminary results of Cancer and Leukemia Group B study 19808. Blood 106:122a, 2005. (abstr 407) [Google Scholar]
- 29.Ruf RG, Schultheiss M, Lichtenberger A, et al: Prevalence of WT1 mutations in a large cohort of patients with steroid-resistant and steroid-sensitive nephrotic syndrome. Kidney Int 66:564-570, 2004 [DOI] [PubMed] [Google Scholar]
- 30.Aucella F, Bisceglia L, De Bonis P, et al: WT1 mutations in nephrotic syndrome revisited: High prevalence in young girls, associations and renal phenotypes. Pediatr Nephrol 21:1393-1398, 2006 [DOI] [PubMed] [Google Scholar]
- 31.Frueh FW, Noyer-Weidner M: The use of denaturing high-performance liquid chromatography (DHPLC) for the analysis of genetic variations: Impact for diagnostics and pharmacogenetics. Clin Chem Lab Med 41:452-461, 2003 [DOI] [PubMed] [Google Scholar]
- 32.Fröhling S, Schlenk RF, Breitruck J, et al: Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: A study of the AML Study Group Ulm. Blood 100:4372-4380, 2002 [DOI] [PubMed] [Google Scholar]
- 33.Thiede C, Steudel C, Mohr B, et al: Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood 99:4326-4335, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto Y, Kiyoi H, Nakano Y, et al: Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 97:2434-2439, 2001 [DOI] [PubMed] [Google Scholar]
- 35.Whitman SP, Ruppert AS, Radmacher MD, et al: FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood 111:1552-1559, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caligiuri MA, Strout MP, Lawrence D, et al: Rearrangement of ALL1 (MLL) in acute myeloid leukemia with normal cytogenetics. Cancer Res 58:55-59, 1998 [PubMed] [Google Scholar]
- 37.Whitman SP, Liu S, Vukosavljevic T, et al: The MLL partial tandem duplication: Evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy. Blood 106:345-352, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Döhner K, Schlenk RF, Habdank M, et al: Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood 106:3740-3746, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Fröhling S, Schlenk RF, Stolze I, et al: CEBPA mutations in younger adults with acute myeloid leukemia and normal cytogenetics: Prognostic relevance and analysis of cooperating mutations. J Clin Oncol 22:624-633, 2004 [DOI] [PubMed] [Google Scholar]
- 40.Marcucci G, Baldus CD, Ruppert AS, et al: Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: A Cancer and Leukemia Group B study. J Clin Oncol 23:9234-9242, 2005 [DOI] [PubMed] [Google Scholar]
- 41.Baldus CD, Tanner SM, Ruppert AS, et al: BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: A Cancer and Leukemia Group B Study. Blood 102:1613-1618, 2003 [DOI] [PubMed] [Google Scholar]
- 42.Cheson BD, Cassileth PA, Head DR, et al: Report of the National Cancer Institute–sponsored workshop on definitions of diagnosis and response in acute myeloid leukemia. J Clin Oncol 8:813-819, 1990 [DOI] [PubMed] [Google Scholar]
- 43.Klein JP, Moeschberger ML: Survival Analysis: Techniques for Censored and Truncated Data. New York, NY, Springer-Verlag, 1997
- 44.Falini B, Mecucci C, Tiacci E, et al: Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype [erratum appears in N Engl J Med 352:740, 2005]. N Engl J Med 352:254-266, 2005 [DOI] [PubMed] [Google Scholar]
- 45.Suzuki T, Kiyoi H, Ozeki K, et al: Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood 106:2854-2861, 2005 [DOI] [PubMed] [Google Scholar]
- 46.Schnittger S, Schoch C, Kern W, et al: Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood 106:3733-3739, 2005 [DOI] [PubMed] [Google Scholar]
- 47.Thiede C, Koch S, Creutzig E, et al: Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML). Blood 107:4011-4020, 2006 [DOI] [PubMed] [Google Scholar]
- 48.Baldus CD, Thiede C, Soucek S, et al: BAALC expression and FLT3 internal tandem duplication mutations in acute myeloid leukemia patients with normal cytogenetics: Prognostic implications. J Clin Oncol 24:790-797, 2006 [DOI] [PubMed] [Google Scholar]
- 49.Haber DA, Sohn RL, Buckler AJ, et al: Alternative splicing and genomic structure of the Wilms tumor gene WT1. Proc Natl Acad Sci U S A 88:9618-9622, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoll R, Lee BM, Debler EW, et al: Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA. J Mol Biol 372:1227-1245, 2007 [DOI] [PubMed] [Google Scholar]
- 51.Smith SI, Down M, Boyd AW, et al: Expression of the Wilms’ tumor suppressor gene, WT1, reduces the tumorigenicity of the leukemic cell line M1 in C.B-17 scid/scid mice. Cancer Res 60:808-814, 2000 [PubMed] [Google Scholar]