Abstract
Purpose
Primary CNS lymphoma (PCNSL) is an aggressive lymphoma but clinically validated biologic markers that can predict natural history to tailor treatment according to risk are lacking. Several genetic changes including BCL6 rearrangements and deletion of 6q22, containing the putative tumor suppressor gene PTPRK, are potential risk predictors. Herein we determined the prevalence and survival impact of del(6)(q22) and BCL6, immunoglobulin heavy chain (IGH), and MYC gene rearrangements in a large PCNSL cohort treated in a single center.
Patients and Methods
Interphase fluorescence in situ hybridization was performed using two-color probes for BCL6, MYC, IGH-BCL6, and del(6)(q22) on thin sections of 75 paraffin-embedded samples from 75 HIV-negative, immunocompetent patients newly diagnosed with PCNSL. Survival data were analyzed using Kaplan-Meier survival curves, log-rank tests, and proportional hazards regression adjusting for age, deep structure involvement, and high-dose methotrexate (HDMTX) treatment.
Results
The prevalence of del(6)(q22) and BCL6, IGH, and MYC translocations was 45%,17%, 13%, and 3%, respectively. The presence of del(6)(q22) and/or a BCL6 translocation was associated with inferior overall survival (OS; P = .0097). The presence of either del(6)(q22) alone or a BCL6 translocation alone was also associated with inferior OS (P = .0087). Univariable results held after adjusting for age, deep structure involvement, and HDMTX.
Conclusion
Del (6)(q22) and BCL6 rearrangements are common in PCNSL and predict for decreased OS independent of deep structure involvement and HDMTX. Unlike systemic diffuse large B-cell lymphoma, del(6)(q22) is common and IGH translocations are infrequent and usually involve BCL6 rather than BCL2, suggesting a distinct pathogenesis.
INTRODUCTION
Primary CNS lymphoma (PCNSL) is an aggressive non-Hodgkin's lymphoma (NHL) that is confined to the CNS. In immunocompetent patients PCNSL is an uncommon tumor, accounting for approximately 1% of NHL and 5% of primary brain tumors.1,2 PCNSL typically shows the morphologic and immunophenotypic features of diffuse large B-cell lymphoma (DLBCL); however, extranodal marginal zone B-cell lymphoma of mucosa associated lymphoid tissue (MALT) lymphoma, peripheral T-cell lymphoma, and Hodgkin's lymphoma may rarely show isolated CNS involvement.2-7 Because PCNSL has heterogeneous clinical behavior despite its relative morphologic homogeneity, investigators have tried to identify clinically relevant prognostic biomarkers in order to individually tailor treatment and minimize toxicity. So far these attempts have met with limited success and clinical parameters, such as age and performance status, are the only consistently identified independent prognostic variables.8-11 Therefore, identification of new prognostic biomarkers and treatment targets remains a high clinical priority.
Only a limited number of genetic studies have been performed in PCNSL, partly due to lack of available tissue specimens. The few karyotypes obtained from PCNSL have shown no recurrent abnormalities. The frequency of chromosomal translocations common to systemic DLBCL has recently been assessed by interphase fluorescence in situ hybridization (FISH) in two small series of PCNSL.12,13 These studies suggested that chromosomal translocations that involve the BCL6 gene are relatively common (23% to 37%) but other abnormalities characteristic of systemic DLBCL, such as IGH-BCL2, appear to be rare. Interestingly, most of the BCL6 translocations in PCNSL involve nonimmunoglobulin partners unlike systemic DLBCL, in which BCL6 is frequently juxtaposed to immunoglobulin genes. Translocations involving MYC at 8q24, which have been associated with poorer survival in systemic DLBCL, are thought to be rare in PCNSL but very few cases have been studied.12
Another abnormality reported to be common in PCNSL is deletion of the long arm of chromosome 6, primarily involving 6q21-23.14-18 The genes involved in this region are not known. Loss of heterozygosity studies in PCNSL have demonstrated loss of one or more loci at 6q22-23 in 66% of cases in a small series.16 More precise mapping has implicated the putative tumor suppressor gene PTPRK within the 140 kb common minimally deleted region in these cases. Deletion of this region was often associated with loss of expression of PTPRK, and both loss of heterozygosity at the PTPRK locus as well as lack of PTPRK protein expression was associated with a poorer prognosis.
BCL6 translocations and 6q22-23/PTPRK deletions may be important in the pathogenesis of PCNSL and may have prognostic significance, but the prevalence and survival impact of these markers have not been well studied. The aim of this study is to determine the prevalence and survival impact of del(6)(q22), BCL6, MYC, and IGH gene rearrangements as well as prevalence of Epstein-Barr virus (EBV) infection in PCNSL in immunocompetent patients.
PATIENTS AND METHODS
Patient Characteristics
The cohort comprised 75 formalin-fixed, paraffin-embedded (FFPE) specimens from 75 HIV-negative, immunocompetent patients with PCNSL newly diagnosed and treated at Mayo Clinic between 1985 and 2006. Clinical information including age, sex, and therapy and imaging records (in order to determine unifocal v multifocal involvement and presence or absence of deep structure involvement) were available on all 75 patients. Performance score (PS) was available for only four patients. All cases were classified as DLBCL according to the WHO classification.19 All patients consented to research use of their tissue. The study was approved by Mayo Foundation institutional review board.
FISH Probes
Interphase FISH was performed on thin sections of the FFPE tumor samples as described previously.20 All cases were screened using a homebrew two-color probe for del(6)(q22), consisting of a SpectrumGreen labeled control probe at 6p24 and an Alexa-594-labeled probe at 6q22-23 (RP11-151E20 and CTD-2378A7). All cases were also screened for BCL6 and MYC translocations. For BCL6, a two-color breakapart probe (BAP; Vysis Inc, Downers Grove, IL) and a homebrew two-color dual fusion (D-FISH) IGH-BCL6 probe consisting of a SpectrumGreen labeled probe and a SpectrumOrange-labeled probe labeled at 3q27.3 (BACS RP11-88P6, RP11-211G3, and CTD2522K3) were used. Two-color IGH BAP (Vysis Inc) probes were also used in cases showing extra FISH signals without fusion using the IGH-BCL6 probe. For MYC, a two-color BAP probe (Vysis Inc) was used.
A minimum of 50 tumor cells were scored in each sample. For BAP and D-FISH probes, a minimum of 20 cells with a recognized abnormal signal pattern were required to deem the cytogenetic abnormality present.21 For the del(6)(q22) probe, a cohesive group of at least 20 cells, of which at least 80% were abnormal was required for that sample to be considered abnormal.22
In Situ Hybridization
In situ hybridization was performed using probes that recognize EBV-encoded RNA on FFPE tumor sections in 75 cases. Any nuclear staining in tumor cells was considered positive.
Statistical Analysis
Survival data were analyzed for all patients, with overall survival (OS) being calculated from the date of tissue diagnosis to date of death or last contact. Survival curves were estimated using the Kaplan-Meier method. The log-rank test was used to compare survival across groups. Proportional hazards regression was used to evaluate the association of genetic abnormalities with survival adjusting for age, deep structure involvement, and high-dose methotrexate (HDMTX). Two-tailed P values less than .05 were considered statistically significant.
RESULTS
The cohort comprised 43 men and 32 women. Mean and median age at diagnosis were 63.5 and 67.0 years, respectively, with a range of 26 to 87 years. There were four patients with a recorded PS; two stuporous patients had a PS of 4 and two intact patients who presented only with seizures had a PS of 0. Sixteen patients (21%) had unifocal disease. Fifty-four patients (72%) had involvement of deep structures (periventricular areas, corpus callosum, basal ganglia, brainstem, and cerebellum). Twenty-eight patients (37%) received HDMTX as initial therapy.
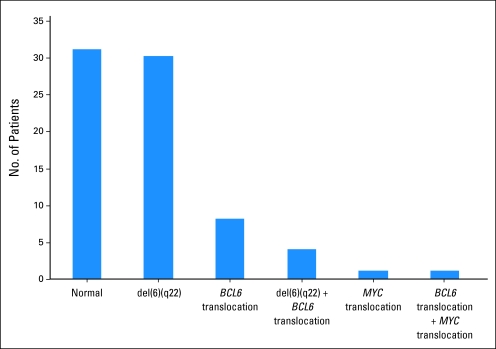
BCL6 translocations were identified in 13 (17%) of 75 PCNSL cases. Eight cases showed IGH-BCL6 fusion and in the remaining five cases the BCL6 translocation partner was unknown. Del(6)(q22) was present in 34 cases (45%), with a spectrum of homozygous (n = 9), heterozygous (n = 4), and mixed homozygous/heterozygous (n = 21) del(6)(q22) patterns. Four cases (5%) possessed both a BCL6 translocation and del(6)(q22). Translocations involving MYC were present in two cases; one also had IGH-BCL6 fusion while the other lacked a BCL6 translocation or del(6)(q22) (Fig 1).
Fig 1.
Prevalence of del(6)(q22), BCL6 translocations, and MYC translocations in primary CNS lymphoma.
Using the IGH-BCL6 D-FISH probe, in addition to the eight cases showing IGH-BCL6 fusion described earlier, 65 cases lacked an abnormality involving IGH and three cases had an extra IGH signal in the absence of IGH-BCL6. An IGH BAP FISH probe used on the latter three cases showed that two had a 1R1G1F pattern indicating IGH translocation to an unknown gene partner (one also had del(6)(q22) while the other also had a BCL6 translocation to an unknown gene partner) while the third showed three intact fusion signals indicating trisomy 14. All cases were negative for EBV-encoded RNA.
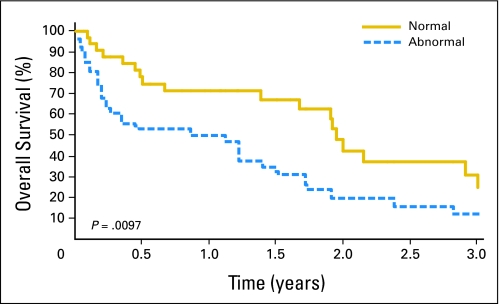
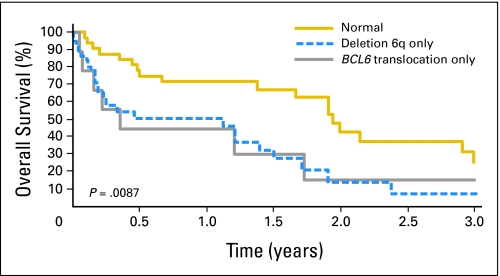
There were 51 deaths and the median follow-up time for surviving patients was 399 days (range, 0 to 2,520 days). Patients who had a BCL6 translocation and/or del(6)(q22) had a median OS of 316 days, compared with those who lacked a BCL6 translocation or del(6)(q22), whose median OS was 713 days (P = .0097; Fig 2). Differences in OS were also statistically significant between patients with a BCL6 translocation only (median OS, 129 days), patients with del(6)(q22) only (median OS, 412 days) and patients who lacked a BCL6 rearrangement or del(6)(q22) (median OS, 713 days, P = .0087; Fig 3). Proportional hazards regression was used to adjust for age, HDMTX, and deep structure involvement. Similar results were observed after adjusting for the above variables (Table 1). We were unable to adjust for performance score, lactate dehydrogenase, or CSF protein due to limited data for these variables.
Fig 2.
Kaplan-Meier estimate of overall survival (OS) for all patients. OS according to the presence or absence of del(6)(q22) and/or a BCL6 translocation.
Fig 3.
Kaplan-Meier estimate of overall survival (OS) for all patients. OS according to the presence of del(6)(q22) only, a BCL6 translocation only, or neither of these abnormalities.
Table 1.
Association of Genetic Abnormalities With Survival
| Parameter | Univariable
|
Adjusted*
|
||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | P† | Hazard Ratio | 95% CI | P† | |
| BCL6 translocation and/or del (6)(q22) | 2.08 | 1.18 to 3.70 | .012 | 2.40 | 1.30 to 4.42 | .005 |
| Normal | 1.0 | — | 1.0 | — | ||
| Del (6)(q22) | 2.55 | 1.36 to 4.77 | .012 | 2.62 | 1.36 to 5.05 | .011 |
| BCL6 translocation | 2.08 | 0.86 to 4.99 | 2.62 | 1.0 to 6.88 | ||
| Normal | 1.0 | — | 1.0 | — | ||
| Del (6)(q22) | 1.83 | 1.04 to 3.20 | .035 | 1.87 | 1.05 to 3.33 | .034 |
| Normal | 1.0 | — | 1.0 | — | ||
Adjusted for age, high-dose methotrexate use, and deep structure involvement.
Proportional hazards regression.
Only four patients had both a BCL6 translocation and del(6)(q22); these abnormalities did not appear to have an additive deleterious effect on survival. Younger patients (age < 60 years) showed a trend toward a more favorable OS than older patients (age > 60 years; OS 713 days v 412 days), but this did not reach statistical significance (P = .113).
DISCUSSION
PCNSL is a rare extranodal lymphoma with aggressive clinical behavior. Although there are effective treatment modalities none are reliably curative and treatment-associated neuro-toxicity is common. Patient-specific therapy is difficult to achieve as there are no good biologic markers that predict the natural history of the disease because little is known regarding the molecular pathogenesis of PCNSL. In this study, we have demonstrated that the prevalence of BCL6 translocations and del(6)(q22) in PCNSL is 17% and 45%, respectively, and both are associated with diminished OS.
The prevalence of several specific cytogenetic abnormalities in our PCNSL cohort is different than that previously reported in systemic DLBCL. Specifically, although the prevalence of BCL6 rearrangements in PCNSL (17%) is comparable with that of systemic DLBCL (19% to 36%),23-27 del(6)(q22) is more common in PCNSL (45%) than in systemic DLBCL (25%)28 and IGH translocations are less common in PCNSL (13%) than in systemic DLBCL (45% to 51%).29,30 Furthermore, the most common IGH translocation partner in PCNSL is BCL6 (80%) while in systemic DLBCL IGH is more frequently juxtaposed to BCL2 (12% to 20%)29-32 than to BCL6 (6.5% to 8%).29,30
In our study, the prevalence of both BCL6 rearrangements (17%) and del(6)(q22) (45%) is comparable to that previously reported in PCNSL (23% to 37%12,13 and 47% to 75%,14-18 respectively), although in the prior PCNSL studies smaller cohorts were evaluated. The prevalence of MYC translocations in our study was 3%, which is similar to that of systemic DLBCL.33 Interestingly, recent gene expression profiling studies have demonstrated that PCNSL shows higher expression of MYC than nodal DLBCL.34 Our data suggest that a mechanism other than a MYC translocation may be responsible for the MYC overexpression in PCNSL. In addition, all cases lacked EBV-encoded RNA. These data are in contrast to post-transplant lymphoproliferative disorders arising in immunosuppressed patients, in which both MYC translocations and EBV positivity are common, and suggest that both MYC translocation and EBV status are less critical in the routine diagnostic evaluation of lymphoproliferative lesions involving the CNS of immunocompetent patients.
The discrepancies in translocation frequency between our PCNSL data and the accumulated systemic DLBCL data suggest distinct pathogeneses for these two disorders. Although PCNSL tumor cells are morphologically and immunohistochemically identical to malignant lymphocytes of systemic DLBCL, PCNSL differs from systemic DLBCL in several important ways. First, PCNSL by definition arises only in the CNS and relapses locally/regionally; systemic relapse is rare. Second, although PCNSL is highly responsive to HDMTX, it is rarely curable. Third, although gene expression profiling studies have demonstrated that both PCNSL and systemic DLBCL can be subcategorized into activated B cell (ABC), germinal center B-cell (GBC), and non-ABC, non-GBC forms,34,35 unlike systemic DLBCL, a subset of PCNSL cases show concurrent expression of both activation genes, such as cyclin D2, and germinal center genes, such as BCL6,34 suggesting a unique molecular origin. It is interesting to note that a subset of PCNSL of both the ABC and GBC subtypes show high expression of BCL6, raising the question of whether this may be associated with a BCL6 translocation regardless of subtype.
When analyzed separately or grouped together, both del(6)(q22) and BCL6 rearrangements showed a negative survival impact in our immunocompetent PCNSL cohort. Deletion of 6q has been previously associated with diminished survival in immunocompetent PCNSL patients.16,17 Because R-PTP-κ (PTPRK) is within a common minimally deleted region at 6q22 and its loss has been associated with loss of PTPRK expression in PCNSL,16 PTPRK is a candidate tumor suppressor gene in PCNSL. PTPRK has also been shown to be a functional tumor suppressor in Hodgkin's lymphoma.36 It is thought to be a regulator of growth factor receptor mediated phosphorylation and thus may be a key mechanism for inhibition/control of cell proliferation. PTPRK belongs to the protein tyrosine phosphatase superfamily of enzymes.37-39 In the context of antigen receptor-mediated signaling in lymphocytes protein tyrosine phosphatases tend to have a primarily inhibitory role,38 and by controlling proliferation and survival signals generated by protein tyrosine kinases, such as SRC or SYK, have emerged as a new generation of candidate tumor suppressor genes and potential therapy targets.
The International Prognostic Index (IPI) was recently adapted to include PCNSL. Based on five measures—lactate dehydrogenase, CSF total protein, involvement of deep structures (periventricular areas, corpus callosum, basal ganglia, brainstem, and cerebellum), age, and PS—three prognostic tiers were proposed.10 These results have been validated and the IPI-PCNSL is now widely used. In our study, too few patients had elevated lactate dehydrogenase (n = 7), elevated CSF protein (n = 20), and a PS (n = 4) in their initial episode of care and thus an IPI-PCNSL score could not be determined. Although there have been attempts to retrospectively assess stroke score,40,41 these focused on neurologic examination findings and did not assess performance as defined by Karnofsky or Zubrod/Eastern Cooperative Oncology Group systems. We could find no report supporting the retrospective assignment of PS.
There were sufficient cases to assess the influence of involvement of deep structures on survival. In this analysis the significance of both del(6)(q22) and a BCL6 rearrangement was maintained. Similarly, in our series, the use of HDMTX in newly diagnosed patients did not seem to correct for the presumed loss of the tumor suppressor effect of del(6)(q22). This is not surprising. Although HDMTX is considered the standard regimen for newly diagnosed PCNSL patients,42 the improved survival may be as much a function of salvage therapies as of the HDMTX.43 A population-based analysis did not confirm that OS improved consistently over the past three decades despite the introduction of HDMTX and the impressive clinical trials results.44
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Francois M. Cady, Brian P. O'Neill, Mark E. Law, Paul A. Decker, David M. Kurtz, Caterina Giannini, Alyx B. Porter, Paul J. Kurtin, Patrick B. Johnston, Ahmet Dogan, Ellen D. Remstein
Financial support: Brian P. O'Neill, Ellen D. Remstein
Provision of study materials or patients: Francois M. Cady, Brian P. O'Neill, Mark E. Law, Paul A. Decker, David M. Kurtz, Alyx B. Porter, Patrick B. Johnston, Ahmet Dogan, Ellen D. Remstein
Collection and assembly of data: Francois M. Cady, Brian P. O'Neill, Mark E. Law, Paul A. Decker, David M. Kurtz, Caterina Giannini, Alyx B. Porter, Patrick B. Johnston, Ahmet Dogan, Ellen D. Remstein
Data analysis and interpretation: Francois M. Cady, Brian P. O'Neill, Mark E. Law, Paul A. Decker, David M. Kurtz, Caterina Giannini, Paul J. Kurtin, Patrick B. Johnston, Ahmet Dogan, Ellen D. Remstein
Manuscript writing: Francois M. Cady, Brian P. O'Neill, Mark E. Law, Paul A. Decker, David M. Kurtz, Caterina Giannini, Alyx B. Porter, Patrick B. Johnston, Ahmet Dogan, Ellen D. Remstein
Final approval of manuscript: Francois M. Cady, Brian P. O'Neill, Mark E. Law, Paul A. Decker, David M. Kurtz, Caterina Giannini, Alyx B. Porter, Paul J. Kurtin, Patrick B. Johnston, Ahmet Dogan, Ellen D. Remstein
Acknowledgments
We thank Leslie Ottjes for expert secretarial and administrative assistance.
published online ahead of print at www.jco.org on July 21, 2008.
Supported in part by the University of Iowa/Mayo Clinic Lymphoma Specialized Program of Research Excellence (SPORE; P50 CA97274), and by the Mayo SPORE in Brain Cancer (P50 CA108961); cores and shared resources were supported by the Cancer Center Support Grant No. P30 CA15083 to Mayo Clinic Cancer Center, a National Cancer Institute–designated Comprehensive Cancer Center.
Presented at the Annual Meetings of the United States and Canadian Academy of Pathology (March 24-30, 2007, San Diego, CA) and the American Society for Hematology (December 8-11, 2007, Atlanta, GA).
Authors’ disclosures of potential conflicts of interest and author contributions are found at the end of this article.
REFERENCES
- 1.Behin A, Hoang-Xuan K, Carpentier AF, et al: Primary brain tumours in adults. Lancet 361:323-331, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Miller DC, Hochberg FH, Harris NL, et al: Pathology with clinical correlations of primary central nervous system non-Hodgkin's lymphoma: The Massachusetts General Hospital experience 1958-1989. Cancer 74:1383-1397, 1994 [DOI] [PubMed] [Google Scholar]
- 3.George DH, Scheithauer BW, Aker FV, et al: Primary anaplastic large cell lymphoma of the central nervous system: Prognostic effect of ALK-1 expression. Am J Surg Pathol 27:487-493, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Bataille B, Delwail V, Menet E, et al: Primary intracerebral malignant lymphoma: Report of 248 cases. J Neurosurg 92:261-266, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Tu PH, Giannini C, Judkins AR, et al: Clinicopathologic and genetic profile of intracranial marginal zone lymphoma: A primary low-grade CNS lymphoma that mimics meningioma. J Clin Oncol 23:5718-5727, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Johnson MD, Kinney MC, Scheithauer BW, et al: Primary intracerebral Hodgkin's disease mimicking meningioma: Case report. Neurosurgery 47:454-456, 2000; discussion 456-457, 2000 [DOI] [PubMed] [Google Scholar]
- 7.Shenkier TN, Blay JY, O'Neill BP, et al: Primary CNS lymphoma of T-cell origin: A descriptive analysis from the international primary CNS lymphoma collaborative group. J Clin Oncol 23:2233-2239, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Abrey LE, Yahalom J, DeAngelis LM: Treatment for primary CNS lymphoma: The next step. J Clin Oncol 18:3144-3150, 2000 [DOI] [PubMed] [Google Scholar]
- 9.Corry J, Smith JG, Wirth A, et al: Primary central nervous system lymphoma: Age and performance status are more important than treatment modality. Int J Radiat Oncol Biol Phys 41:615-620, 1998 [DOI] [PubMed] [Google Scholar]
- 10.Ferreri AJ, Blay JY, Reni M, et al: Prognostic scoring system for primary CNS lymphomas: The International Extranodal Lymphoma Study Group experience. J Clin Oncol 21:266-272, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Abrey LE, Ben-Porat L, Panageas KS, et al: Primary central nervous system lymphoma: The Memorial Sloan-Kettering Cancer Center prognostic model. J Clin Oncol 24:5711-5715, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Montesinos-Rongen M, Zuhlke-Jenisch R, Gesk S, et al: Interphase cytogenetic analysis of lymphoma-associated chromosomal breakpoints in primary diffuse large B-cell lymphomas of the central nervous system. J Neuropathol Exp Neurol 61:926-933, 2002 [DOI] [PubMed] [Google Scholar]
- 13.Schwindt H, Akasaka T, Zuhlke-Jenisch R, et al: Chromosomal translocations fusing the BCL6 gene to different partner loci are recurrent in primary central nervous system lymphoma and may be associated with aberrant somatic hypermutation or defective class switch recombination. J Neuropathol Exp Neurol 65:776-782, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Boonstra R, Koning A, Mastik M, et al: Analysis of chromosomal copy number changes and oncoprotein expression in primary central nervous system lymphomas: Frequent loss of chromosome arm 6q. Virchows Arch 443:164-169, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Harada K, Nishizaki T, Kubota H, et al: Distinct primary central nervous system lymphoma defined by comparative genomic hybridization and laser scanning cytometry. Cancer Genet Cytogenet 125:147-150, 2001 [DOI] [PubMed] [Google Scholar]
- 16.Nakamura M, Kishi M, Sakaki T, et al: Novel tumor suppressor loci on 6q22-23 in primary central nervous system lymphomas. Cancer Res 63:737-741, 2003 [PubMed] [Google Scholar]
- 17.Rickert CH, Dockhorn-Dworniczak B, Simon R, et al: Chromosomal imbalances in primary lymphomas of the central nervous system. Am J Pathol 155:1445-1451, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber T, Weber RG, Kaulich K, et al: Characteristic chromosomal imbalances in primary central nervous system lymphomas of the diffuse large B-cell type. Brain Pathol 10:73-84, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaffe ES, Harris NL, Stein H, et al: World Health Organisation Classification of Tumours: Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France, IARC Press, 2001
- 20.Cataldo KA, Jalal SM, Law ME, et al: Detection of t(2;5) in anaplastic large cell lymphoma: Comparison of immunohistochemical studies, FISH, and RT-PCR in paraffin-embedded tissue. Am J Surg Pathol 23:1386-1392, 1999 [DOI] [PubMed] [Google Scholar]
- 21.Remstein ED, Dogan A, Einerson RR, et al: The incidence and anatomic site specificity of chromosomal translocations in primary extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue (MALT lymphoma) in North America. Am J Surg Pathol 30:1546-1553, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Remstein ED, Law M, Mollejo M, et al: The prevalence of IG translocations and 7q32 deletions in splenic marginal zone lymphoma. Leukemia 22:1268-1272, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bastard C, Deweindt C, Kerckaert JP, et al: LAZ3 rearrangements in non-Hodgkin's lymphoma: Correlation with histology, immunophenotype, karyotype, and clinical outcome in 217 patients. Blood 83:2423-2427, 1994 [PubMed] [Google Scholar]
- 24.Lo Coco F, Ye BH, Lista F, et al: Rearrangements of the BCL6 gene in diffuse large cell non-Hodgkin's lymphoma. Blood 83:1757-1759, 1994 [PubMed] [Google Scholar]
- 25.Otsuki T, Yano T, Clark HM, et al: Analysis of LAZ3 (BCL-6) status in B-cell non-Hodgkin's lymphomas: Results of rearrangement and gene expression studies and a mutational analysis of coding region sequences. Blood 85:2877-2884, 1995 [PubMed] [Google Scholar]
- 26.Barrans SL, O'Connor SJ, Evans PA, et al: Rearrangement of the BCL6 locus at 3q27 is an independent poor prognostic factor in nodal diffuse large B-cell lymphoma. Br J Haematol 117:322-332, 2002 [DOI] [PubMed] [Google Scholar]
- 27.Iqbal J, Greiner TC, Patel K, et al: Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia 21:2332-2343, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bea S, Zettl A, Wright G, et al: Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood 106:3183-3190, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cigudosa JC, Parsa NZ, Louie DC, et al: Cytogenetic analysis of 363 consecutively ascertained diffuse large B-cell lymphomas. Genes Chromosomes Cancer 25:123-133, 1999 [PubMed] [Google Scholar]
- 30.Bernicot I, Douet-Guilbert N, Le Bris MJ, et al: Characterization of IGH rearrangements in non-Hodgkin's B-cell lymphomas by fluorescence in situ hybridization. Anticancer Res 25:3179-3182, 2005 [PubMed] [Google Scholar]
- 31.Shipp MA, Ross KN, Tamayo P, et al: Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med 8:68-74, 2002 [DOI] [PubMed] [Google Scholar]
- 32.Iqbal J, Sanger WG, Horsman DE, et al: BCL2 translocation defines a unique tumor subset within the germinal center B-cell-like diffuse large B-cell lymphoma. Am J Pathol 165:159-166, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McClure RF, Remstein ED, Macon WR, et al: Adult B-cell lymphomas with Burkitt-like morphology are phenotypically and genotypically heterogeneous with aggressive clinical behavior. Am J Surg Pathol 29:1652-1660, 2005 [DOI] [PubMed] [Google Scholar]
- 34.Rubenstein JL, Fridlyand J, Shen A, et al: Gene expression and angiotropism in primary CNS lymphoma. Blood 107:3716-3723, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montesinos-Rongen M, Brunn A, Benink S, et al: Gene expression profiling suggests primary central nervous system lymphomas to be derived from a late germinal center B cell. Leukemia 22:400-405, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flavell JR, Baumforth KR, Wood VH, et al: Downregulation of the TGF-Beta target gene, PTPRK, by the epstein-barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin's lymphoma cells. Blood 111:292-301, 2008 [DOI] [PubMed] [Google Scholar]
- 37.Alonso A, Sasin J, Bottini N, et al: Protein tyrosine phosphatases in the human genome. Cell 117:699-711, 2004 [DOI] [PubMed] [Google Scholar]
- 38.Mustelin T, Vang T, Bottini N: Protein tyrosine phosphatases and the immune response. Nat Rev Immunol 5:43-57, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Tonks NK: Protein tyrosine phosphatases: From genes, to function, to disease. Nat Rev Mol Cell Biol 7:833-846, 2006 [DOI] [PubMed] [Google Scholar]
- 40.Barber M, Fail M, Shields M, et al: Validity and reliability of estimating the scandinavian stroke scale score from medical records. Cerebrovasc Dis 17:224-227, 2004 [DOI] [PubMed] [Google Scholar]
- 41.Weir NU, Counsell CE, McDowall M, et al: Reliability of the variables in a new set of models that predict outcome after stroke. J Neurol Neurosurg Psychiatry 74:447-451, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batchelor T, Loeffler JS: Primary CNS lymphoma. J Clin Oncol 24:1281-1288, 2006 [DOI] [PubMed] [Google Scholar]
- 43.Nguyen PL, Chakravarti A, Finkelstein DM, et al: Results of whole-brain radiation as salvage of methotrexate failure for immunocompetent patients with primary CNS lymphoma. J Clin Oncol 23:1507-1513, 2005 [DOI] [PubMed] [Google Scholar]
- 44.Panageas KS, Elkin EB, DeAngelis LM, et al: Trends in survival from primary central nervous system lymphoma, 1975-1999: A population-based analysis. Cancer 104:2466-2472, 2005 [DOI] [PubMed] [Google Scholar]