Abstract
Purpose
The purpose of this study is to investigate the molecular mechanism of miR-192 in colon cancer.
Experimental Design
Human colon cancer cell lines with different p53 status were used as our model system to study the impact of miR-192 on cell proliferation, cell cycle control and mechanism of regulation.
Results
Our results show that one of the key miR-192 target genes is dihydrofolate reductase (DHFR). miR-192 impacts cellular proliferation through the p53-miRNA circuit. Western immunoblot analyses indicated that the expression of DHFR was significantly decreased by miR-192. Further investigation revealed that such suppression was due to translational arrest rather than mRNA degradation. More profound inhibition of cellular proliferation was observed by ectopic expression of miR-192 in colon cancer cell lines containing wild type p53 than cells containing mutant p53. Thus, the effect of miR-192 on cellular proliferation is mainly p53 dependent. Over-expression of miR-192 triggered both G1 and G2 arrest in HCT-116 (wt-p53) cells but not in HCT-116 (null-p53) cells. The cell cycle check point control genes p53 and p21 were highly over-expressed in cells that over-expressed miR-192. Endogenous miR-192 expression was increased in HCT-116 (wt-p53) and RKO (wt-p53) cells treated with methotrexate, which caused an induction of p53 expression. Chromatin immunoprecipitation (ChIP)-qPCR analysis revealed that the p53 protein interacted with the miR-192 promoter sequence.
Conclusion
These results indicate that miR-192 may be another miRNA candidate that is involved in the p53 tumor suppressor network with significant impact on cell cycle control and cell proliferation.
Keywords: miR-192, dihydrofolate reductase, p53, cell proliferation
INTRODUCTION
miRNAs are non-coding, single-stranded RNAs of ∼22 nucleotides, processed from larger pre-miRNAs by the RNase III enzyme, Dicer, into miRNA duplex complexes (1). One strand of this duplex can associate with the RNA-induced silencing complex (RISC), with the other strand generally degraded by cellular nucleases (1). The miRNA-RISC complex has been shown to bind to specific mRNA targets resulting in translational repression or cleavage of these mRNA sequences. miRNAs modulate protein expression by promoting RNA degradation, inhibiting mRNA translation, and also affecting transcription. Currently, there are over 450 human miRNAs that have been identified and the final number will likely be much higher (2). Although miRNA-mediated mRNA degradation occurs in mammals, in most cases, the impact of miRNA on its targets are thought to use a secondary mechanism of gene regulation via imperfect base-pairing to the 3′-untranslated regions (3′-UTRs) of the mRNA targets. This results in the repression of target gene expression post-transcriptionally, most likely at the translational level of gene expression (3-5). Such translational regulation provides the cell with a more precise, immediate and energy-efficient way of controlling expression of a given protein (6). Such regulation can induce rapid changes in protein synthesis without the need for transcriptional activation and subsequent steps in mRNA processing. Additionally, the translational control of gene expression has the advantage of being readily reversible, providing the cell with great flexibility in responding to various cytotoxic stresses (7). Clearly, it is essential to know not only the expression levels of individual mRNAs, but also to what extent mRNAs are being translated into their corresponding proteins.
Post-transcriptional control mediated by miRNAs has become an area of intense research over the last few years. The notion of miRNAs mediating gene expression at the translational level is based on the study of the first two identified miRNAs, lin-4 and let-7, in Caenorhabditis elegans (8). The lin-4 miRNA attenuates the translation, but not the mRNA level, of two target genes, lin-14 and lin-28, by imperfect base-pairing to complementary sequences in the 3′-UTR of the target mRNAs (3, 8). Although the exact function of many newly discovered miRNAs are just emerging, their ability to regulate cell proliferation and cell death has been recently demonstrated (9). Recent reports have shown that the expression of miRNAs is altered in cancer cells, and that some miRNAs can function as translational attenuators (5).
The first indication that miRNAs may function as tumor suppressors was derived from a study by Calin et al., who found that miR-15a and miR-16-1 were commonly deleted in >65% of patients with B-cell chronic lymphocytic leukemia (CLL) (10). Further, Cimmino et al. showed that miR-15a and miR-16-1 negatively regulated Bcl2, an anti-apoptotic protein that is often over-expressed in many tumor types (11). Several expression profiling studies have also reported de-regulated miRNAs in colon cancer, breast cancer and other types of solid tumors (2, 12-14).
miRNAs are shown to be critical in oncogenesis, and alterations in miRNA expression are found to be associated with several human cancers (9, 10, 14-16). miRNAs can act as either oncogenes or tumor suppressor genes based on their mRNA targets (14). We have previously identified that the expression of a number of miRNAs was altered due to the loss of p53 tumor suppressor gene (12). Subsequently, a number of groups have reported that miR-34 was directly involved in the p53 tumor suppressor network and regulated directly by p53 (17-19). Based on these results, we reasoned that there should be additional miRNAs that are involved in the p53 tumor suppressor network. By performing an in silico analysis coupled with experimental validations to search for these potential miRNAs, we report here the discovery that the miR-192 promoter contains a conserved p53 binding site and that one of the key miR-192 target genes is dihydrofolate reductase (DHFR), an important target for antifolate based anti-cancer chemotherapy such as methotrexate (MTX) (20). We also experimentally confirmed that the downstream pathway of miR-192 in cell cycle control is mediated through increased p53 expression by the induction of p21. Many studies have indicated that expression of DHFR is regulated at least in part at the translational level (21-26). Recently, Mishra et al. reported that a miR-24 binding-site polymorphism in DHFR gene led to MTX resistance (27). Translational control provides cells with acute response to growth condition changes and it is readily reversible (28). We provide evidence that miR-192 is another player in the p53-miRNA circuit and contributes to the regulation of cell cycle control and cellular proliferation.
Materials and Methods
Cell Culture and Reagents
The human colon cancer cell lines HCT-116 (wt-p53) and HCT-116 (null-p53) were gifts from Professor Bert Vogelstein (The Johns Hopkins University), and were maintained in McCoy’s 5A medium (Gibco Laboratories). The other two human colon cancer cell lines, RKO (wt-p53) and HT-29 (mut-p53) were obtained from the American Type Culture Collection. The HT-29 (mut-p53) cell line has a missense mutation in codon 273 of p53 resulting in an Arg to His substitution. RKO (wt-p53) and HT-29 (mut-p53) cells were maintained in Eagle’s Minimum Essential Medium and Iscove’s Modified Dulbecco’s Medium at ATCC respectively. All the media were supplemented with 10% dialyzed fetal bovine serum (HyClone Laboratories, Inc.). All cell lines were grown at 37°C in a humidified incubator with 5% CO2. MTX was purchased from Sigma-Aldrich.
Transfections of miRNA and siRNA Specific to Dihydrofolate Reductase
HCT-116 (wt-p53), HCT-116 (null-p53), RKO (wt-p53), and HT-29 cells (2×105) were plated in six-well plates, and transfected with 100 nM of either miR-192, miR-24 precursors or non-specific control miR (Ambion) after 24 h with Oligofectamine (Invitrogen) according to the manufacturer’s instructions. siRNA specific to DHFR (ON-TARGET plus SMARTpool L-008799-00-0010, human DHFR, NM_000791) was purchased from Dharmacon and transfected with Oligofectamine (Invitrogen) according to the manufacturer’s protocols at a final concentration of 100 nM. siRNA specific to DHFR was used as the positive control. miR-24, a recently reported miRNA that also targets DHFR (27), was also used as a positive control.
RNA Isolation
Total RNA, including miRNA, was isolated from cell lines by using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions at 24 h after transfection.
Real Time qRT-PCR Analysis of miRNA
cDNA synthesis was carried out with the High Capacity cDNA synthesis kit (Applied Biosystems) using 10 ng of total RNA as template. The miRNA sequence-specific RT-PCR primers for miR-192, miR-24 and endogenous control RNU6B were purchased from Ambion. Real-time quantitative reverse transcription-PCR (qRT-PCR) analysis was carried out using Applied Biosystems 7500 Real-Time PCR System. The PCR master mix containing TaqMan 2× Universal PCR Master Mix (No Amperase UNG), 10× TaqMan assay and RT products in 20 μl volume were processed as follows: 95°C for 10 min, and then 95°C for 15 sec, 60°C for 60 sec for up to 40 cycles (n=3). Signal was collected at the endpoint of every cycle. The gene expression ΔCT values of miRNAs from each sample were calculated by normalizing with internal control RNU6B and relative quantitation values were plotted.
Real Time qRT-PCR Analysis of mRNA Expression
cDNA was synthesized with the High Capacity cDNA synthesis kit (Applied Biosystems) using 2μg of total RNA as the template and random primers. Real-time qRT-PCR analysis was done on the experimental mRNAs. The PCR primers and probes for DHFR, and the internal control gene GAPDH were purchased from Applied Biosystems. qRT-PCR was performed on an ABI 7500HT instrument under the following conditions: 50°C, 2 min of reverse transcription; 95°C, 10 min; 95°C, 15 s; 60°C, 1 min for up to 40 cycles (n =3).
Western Immunoblot Analysis
At 48 h after transfection with miR-192, miR-24 precursors or non-specific control miRNA, the cells were scraped and lysed in RIPA buffer (Sigma). Equal amount of proteins were resolved by SDS-PAGE on 12% gels by the method of Laemmli (29), and transferred to polyvinylidene fluoride membranes (BIO-RAD Laboratories). The membranes were then blocked by 5% nonfat milk in TBS-T (Tris-buffered saline and 0.5% Tween-20) at room temperature for 1 h. The primary antibodies used for the analysis included mouse anti-DHFR mAb (1:250, BD Bioscience), mouse anti-p53 mAb (1:1000, DO-1), mouse anti-p21mAb (1:1000, F-5), and mouse anti-α-tubulin mAb (1:1000, TU-02) purchased from Santa Cruz Biotechnology. Horseradish peroxidase-conjugated antibodies against mouse or rabbit (1:1000, Santa Cruz Biotechnology) were used as the secondary antibodies. Protein bands were visualized with a chemiluminescence detection system using the Super Signal substrate.
MTX Cytotoxicity
HCT-116 (wt-p53) cells were plated in 96-well plates at 1×103 cells/well in triplicate. They were transfected with miR-192 precursor, non-specific control miRNA, or siRNA against DHFR in 100 μl of medium. Twenty-four hours later, MTX in 100 μl medium (final concentration 25 nM) was added, and incubated for 72 h. 10 μl of WST-1 (Roche Applied Science) was added to each well. After 2 h incubation, absorbance was measured at 450 and 630 nm respectively (n=3). Non-specific control miRNA alone was used as a negative control, and siRNA incubation with MTX was used as a positive control.
Cell Proliferation Analysis
HCT-116 (wt-p53), HCT-116 (null-p53), RKO (wt-p53), and HT-29 cells were plated in 96-well plates in triplicate at 1×103 cells/well after transfection with miR-192 precursor or non-specific control miRNA. Cells were cultured for 24, 48, 72, 96 h. The absorbance at 450 and 630 nm was measured after incubation with 10 μl of WST-1 for 2 h.
Cell Cycle Analysis
HCT-116 (wt-p53) and HCT-116 (null-p53) cells were transfected with miR-192 precursor and the non-specific control miRNA described as above. At 36 h after transfection, cells were harvested and resuspended at 0.5-1×105 cells/ml in modified Krishan buffer (17, 18) containing 0.1% sodium citrate and 0.3% NP-40 and kept at 4°C. Before being analyzed by flow cytometry, cells were treated with 0.02mg/ml RNase H and stained with 0.05mg/ml propidium iodide (Sigma).
MTX Treatment
HCT-116 (wt-p53), HCT-116 (null-p53), RKO (wt-p53), and HT-29 cells were seeded in six-well plates at a density of 2×105 cells per well and then incubated with or without MTX (25 nM). The cells were collected at 24 h after incubation, and total RNA and protein were extracted respectively. The subsequent real-time qRT-PCR of miRNA and western immunoblot analysis for p53 and α-tubulin were performed as described above.
Chromatin Immunoprecipitation (ChIP) and qPCR Analysis
To show that p53 protein directly interacts with the miR-192 promoter region, we performed ChIP-qPCR analysis using p21, a known cell cycle regulator transcriptionally regulated by p53, as a positive control. Mouse monoclonal antibody (DO-1) against p53 (Santa Cruz Biotechnology) was used for immunoprecipitation of the p53 binding complex. Non-related antibody α-tubulin (TU-02, Santa Cruz Biotechnology) was used as a negative control. Immunoprecipitation was performed based on the manufacturer’s protocols of Active Motif. The primer sequences for the miR-192 promoter and the p21 promoter are listed as follows:
miR-192 promoter (forward primer): 5′-AGCACCTCCCATGTCACC-3′
miR-192 promoter (reverse primer): 5′-CAAGGCAGAGCCAGAGC-3′
p21 promoter (forward primer): 5′-GCTGGTGGCTATTTTGTCCTTGGGC-3′
p21 promoter (reverse primer): 5′-CAGAATCTGACTCCCAGCACACACTC-3′
Conserved P53 Binding Mir-192 Promoter Reporter Activity Assay
Luciferase reporter assay was used to determine the transcriptional activation of conserved p53 binding promoter of miR-192. pGL3-Basic promoterless luciferase reporter plasmid (Promega) was used in this study. Double stranded DNA oligonucleotides of conserved p53 binding sequence of miR-192 was synthesized and annealed and cloned upstream of firefly luciferase in the pGL3-Basic plasmid (miR-192-pGL3). The p53 binding site oligonucleotide (underline) contains MluI at 5′-end and BglII sequence (italic) at the 3′-end (5′-ACGCGTCCATGTCACCACCAGGGGTCGCCATGCCTCCTGGCCTTGCCCAGCAGATCT-3′). Control vector and miR-192-pGL3 vector were transfected into both HCT-116 (wt-p53) cells and HCT116 (null-p53) cells. To further induce p53 expression, tranfected HCT-116 (wt-p53) cells and HCT116 (null-p53) cells were also treated with 5 μM 5-FU for 24h. The promoter activity of each construct was quantified by dual luciferase assay (Promega) 24 h after transfection. Firefly luciferase activity was normalized with Renilla luciferase internal control under each condition.
Statistical analysis
All experiments were repeated at least twice. Statistical significance was evaluated by Student’s t test (two tailed) comparison between two groups of data. Asterisks indicate significant differences of experimental groups compared with the corresponding control condition. Statistical analysis was done using GraphPad Prism software (GraphPad, Inc.). Differences were considered statistically significant at P < 0.05.
Results and Discussion
miRNA regulates the mRNA translation rate by perfect or imperfect base pairing with the 3′-UTR regions of their targets (1). It has been predicted that one miRNA can potentially regulate translation of up to hundreds of mRNAs (1). This has created a challenge for experimentally validating miRNA specific targets. Previous studies from our laboratory have discovered that nearly half of the miRNAs promoter regions contain putative p53 binding site(s) (12). To further validate the significance of some of these candidate miRNAs, we took a systematic approach by first analyzing which miRNA may target critical anticancer target genes. We also select the miRNA candidates with high predicted ranking scores of p53 binding promoter. This allows us to focus on miR-192 with DHFR as one of its predictive target, an important anticancer target. DHFR is the key enzyme responsible for intracellular folate metabolism and a target of MTX (30). The regulation of DHFR is complex and includes post-transcriptional control by an auto feedback mechanism (31).
Translational regulation of DHFR expression by miR-192
We investigated the roles of miRNAs in regulating the expression of DHFR. Based on the structural analysis of 3′-UTR of the DHFR gene and miRNA target analysis (TargetScan, PicTar, miRnaDa), we identified several miRNAs which potentially interact with the 3′-UTR region of DHFR mRNA. Bioinformatic analysis of the secondary structure of the 3′-UTR of the DHFR mRNA and miRNA binding sites led us to reduce the candidate miRNAs to a small number. This allows us to efficiently identify miRNAs that involved in regulating key targets like DHFR.
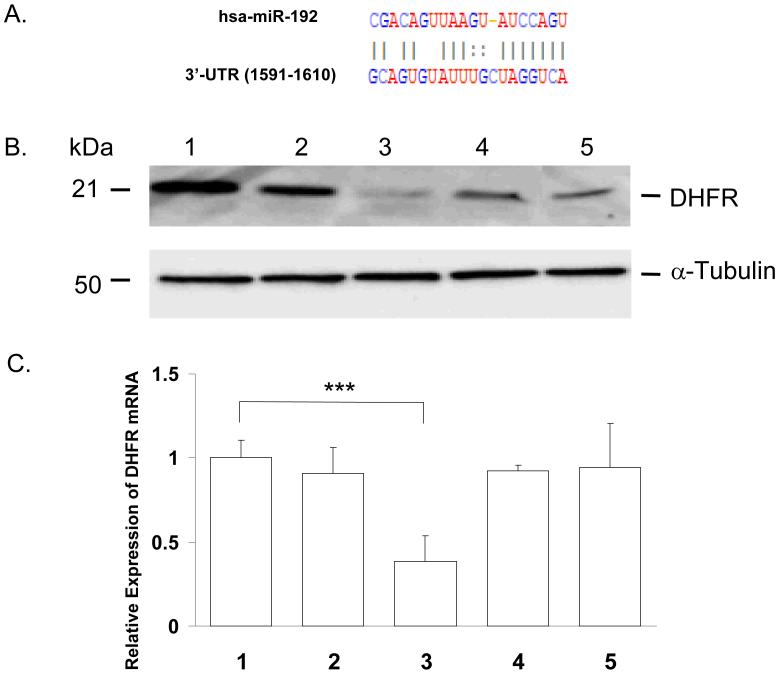
Fig. 1A shows the target sequence of 3′-UTR of the DHFR mRNA that interacts with miR-192. To experimentally confirm that the expression of DHFR was regulated by miR-192, a miR-192 precursor was transfected into HCT-116 (wt-p53) cells. A non-specific miR was used as a negative control. It has been reported that the expression of DHFR is regulated by miR-24 (27). We thus used both a DHFR siRNA and miR-24 as positive controls. Over-expression of miR-192 and miR-24 was confirmed by real time qRT-PCR analysis using U6 RNA to normalize the expression (supplemental data file 1). The expression of DHFR protein was analyzed using Western immunoblot analysis and the results are shown in Fig. 1B. Over-expression of miR-192 clearly decreased the expression of DHFR protein (Fig. 1B, lane 4). The potency of miR-192 for decreasing DHFR expression was comparable to miR-24 (Fig. 1B, lane 5). We also analyzed the expression level of DHFR mRNA using real time qRT-PCR analysis and the results (Fig. 1C) indicated that there was no reduction in DHFR mRNA expression by miR-192 (lane 4) and miR-24 (lane 5). Thus, the suppression of DHFR expression was regulated at the translational level without the degradation of DHFR mRNA. By contrast, the decreased expression of DHFR by siRNA was clearly caused by mRNA degradation (lane 3, Fig. 1C).
Figure 1.
The miR-192 binding site at 3′-UTR of DHFR mRNA (A). Western immunoblot analysis of DHFR protein expression levels in HCT-116 (wt-p53) cells transfected with miR-192 (lane 1, vehicle control; lane 2, non specific-miR control; lane 3, DHFR siRNA positive control; lane 4, miR-192; lane 5, positive control miR-24), α-tubulin was used as protein loading control (B). Expression analysis of DHFR mRNA in HCT-116 (wt-p53) cells by real time qRT-PCR analysis (lane 1, vehicle control; lane 2, non-specific miR control; lane 3, DHFR siRNA positive control; lane 4, miR-192; lane 5, positive control miR-24). GAPDH was used as internal standard for normalization (C) (*** P < 0.0001).
miR-192 sensitizes HCT-116 (wt-p53) cells to MTX
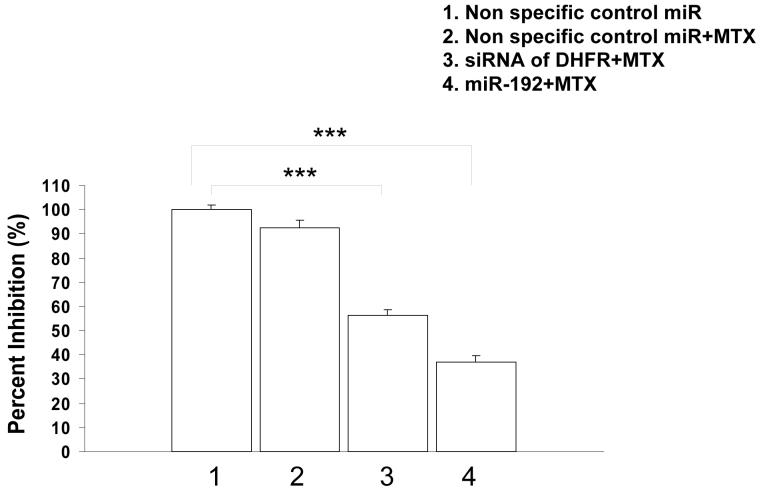
Because DHFR is a target for MTX, the increased expression of miR-192 may also contribute to the sensitivity to MTX treatment. With equal molar concentration of MTX at 25 nM (IC-10), cell proliferation was reduced by 10% in non specific control miR treated cells (Fig. 2, lane 2). However, cell proliferation was reduced by nearly 70% in cells transfected with miR-192, demonstrating a synergistic effect in combination with MTX (Fig. 2, lane 4). By contrast, cells treated with siRNA against DHFR were inhibited by 55% (Fig. 2, lane 3). The more potent effect of miR-192 plus MTX compared to siRNA targeting specifically to DHFR suggests that miR-192 may also target additional mRNA targets through imperfect base pairing. Future studies are clearly needed to systematically identify additional miR-192 regulated mRNA transcripts.
Figure 2.
Impact of miR-192 on cell proliferation with MTX treatment in HCT-116 (wt-p53) cells transfected with DHFR specific siRNA or miR-192 (lane 1, non-specific control miR; lane 2, 100 nM non-specific control miR + 25 nM MTX; lane 3, 100 nM DHFR siRNA + 25 nM MTX; lane 4, 100 nM miR-192 + 25 nM MTX) (* P < 0.006).
Effect of overexpression of miR-192 on cellular proliferation
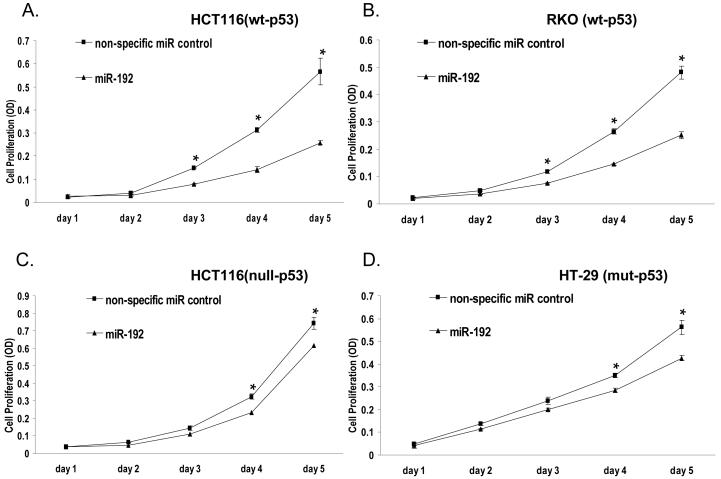
To assess the functional significance of miR-192, we evaluated the impact of miR-192 on cellular proliferation using HCT-116 (wt-p53), HCT-116 (null-p53), RKO (wt-p53) and HT-29 (mut-p53) colon cancer cell lines. A non-specific miR was used as a negative control. Our results show that the overexpression of miR-192 can suppress cellular proliferation in HCT-116 (wt-p53) cells by over 55% (n=3) (Fig. 3A) and RKO (wt-p53) cells by 48% (n=3) (Fig. 3B), with less impact on HCT-116 (null-p53) (15%, n=3) (Fig. 3C) and HT-29 cell lines (24%, n=3) (Fig. 3D). By contrast, the non-specific control miR has no effect on cellular proliferation, indicating that this effect caused by miR-192 is highly specific. These results clearly show that the effect of miR-192 on the inhibition of cellular proliferation is more potent in colon cancer cell lines containing wild type p53 than in the p53-null or mutant p53 cell lines, further indicating that miR-192’s function depends on the status of p53.
Figure 3.
Impact of miR-192 on cell proliferation using WST1 assay in HCT-116 (wt-p53) cells (A) (* P < 0.001), RKO (wt-p53) cells (B) (* P < 0.001), HCT-116 (null-p53) cells (* P < 0.02) (C), HT-29 (mut-p53) cells (D) (* P < 0.029). Each cell type was transfected with 100 nM non-specific control miR or miR-192; cell numbers were determined by the WST-1 assay.
Impact of cell cycle control by miR-192
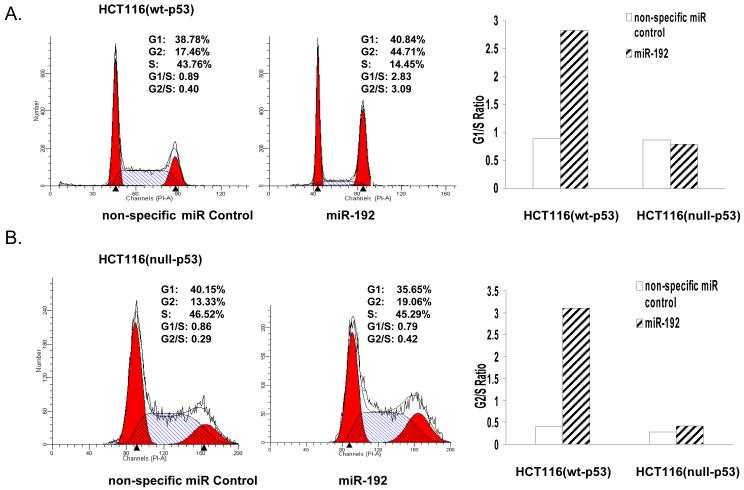
To determine whether the miR-192’s impact on cellular proliferation was related to cell cycle control, we analyzed the effect of miR-192 on cell cycle control by flow cytometry using HCT-116 (wt-p53) and HCT-116 (null-p53) cells transfected with non-specific control miR or miR-192. Our results show that miR-192 increases G1/S ratio (>2-fold) and G2/S ratio (>3-fold) in HCT-116 (wt-p53) cells (Fig. 4A). By contrast, this effect has not been observed in HCT-116 (null-p53) cells (Fig. 4B). Thus, the cell cycle analysis data is highly consistent with the cell proliferation results that the function of miR-192 is dependent on the presence of wild type p53 for cell cycle control.
Figure 4.
Cell cycle analysis by flow cytometry in HCT-116 (wt-p53) (A) or HCT-116 (null-p53) (B) transfected with 100 nM non-specific miR or miR-192. The bar graphs show the fold increase of G1/S and G2/S ratio in both HCT-116 (wt-p53) and HCT (null-p53) cells transfected with miR-192.
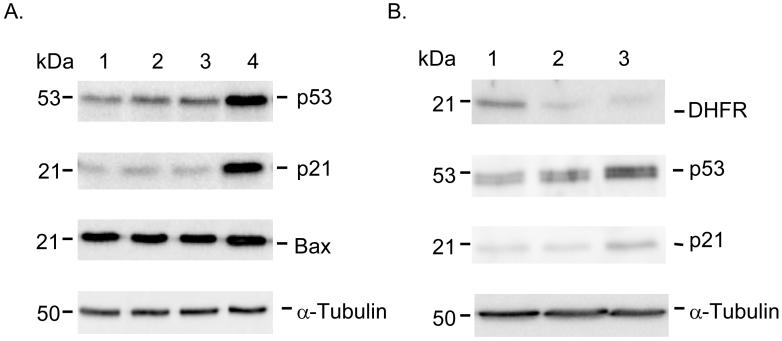
Effect of miR-192 on cell cycle control genes
To further analyze the cell cycle control genes involved in miR-192 overexpression, we analyzed a number of cell cycle control genes (p53, p21, Bax). Fig. 5 shows the results of Western immunoblot analysis in HCT-116 (wt-p53) cells (Fig. 5A) and RKO (wt-p53) cells (Fig. 5B). Ectopic expression of miR-192 increased the expression of the p53 protein (Fig. 5A, lane 4) over 10-fold and p21 10-fold. By contrast, siRNA against DHFR (Fig. 5A, lane 3) did not cause an increase in expression of p53 and p21. The expression of Bax was not altered by miR-192, indicating that miR-192 may not trigger apoptosis. Similar results were obtained for RKO (wt-p53) cells (Fig. 5B, lane 3, miR-192, lane 1, non-specific miR; lane 2, siRNA of DHFR). It has been well characterized that the induction of the p53 dependent cell cycle check point control gene p21 is the key to trigger cell cycle arrest at both the G1 and G2 phase (32, 33). Thus, our results further confirm the notion that the function of miR-192 is clearly dependent on the status of wild type p53.
Figure 5.
Western immunoblot analysis of p53, p21 and Bax expression in HCT-116 (wt-p53) cells (lane 1, vehicle control; lane 2, non specific control; lane 3, DHFR siRNA positive control; lane 4, 100 nM miR-192). α-tubulin was used as a protein loading control. B. Western immunoblot analysis of p53 and p21 expression in RKO (wt-p53) cells (lane 1, non-specific miR control; lane 2, DHFR siRNA positive control; lane 3, 100 nM miR-192). α-tubulin was used as a protein loading control.
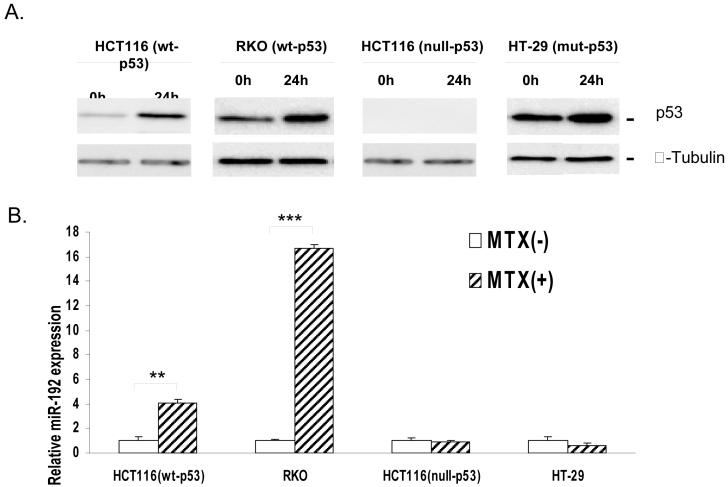
miR-192 expression was dependent on p53
With ectopic over-expression of miR-192 by transfection, both HCT-116 (wt-p53) and RKO (wt-p53) cells undergo cell cycle arrest at the G1 and G2 phase leading to decreased cellular proliferation. Bioinformatic analysis also reveals that there is a putative p53 binding site in the miR-192 promoter region. To confirm the direct regulatory relationship with p53, we performed the following experiments. First, the expression of the p53 protein was induced by treatment with MTX. The induction of p53 protein expression by MTX treatment in HCT-116 (wt-p53) and RKO (wt-p53) cells (Fig 6A) caused a significant increase of miR-192 expression (Fig 6B). By contrast, MTX treatment in HCT-116 (null-p53) and HT-29 (mut-p53) cells did not cause any change in the expression of miR-192 (Fig 6B). Furthermore, the expression level of miR-192 in HCT116 (wt-p53) cells was nearly 4-fold higher than HCT-116 (null-p53) cells (supplemental data file 4). These results suggest that the endogenous expression of miR-192 depends on the wild type p53 after genotoxic stress by MTX treatment. These findings combined with the data in Fig. 5, which shows that p53 is induced by ectopic expression of miR-192, indicate that there might be a positive feedback loop between p53 and miR-192 to ensure proper cell cycle control. miR-192 may down regulate some key cell cycle related genes to trigger p53 induction. Further studies are currently underway to define the detailed mechanism of this positive feedback loop between miR-192 and p53.
Figure 6.
(A) Effect of p53 induction by MTX treatment on the expression of endogenous miR-192 expression in HCT-116 (wt-p53) cells (** P < 0.001), RKO (wt-p53) cells (*** P < 0.0001), HCT-116 (null-p53) cells and HT-29 (mut-p53) cells. The p53 protein expression levels were analyzed by Western immunoblot analysis and α-tubulin was used as a protein loading control. (B) The expression of endogenous mature miR-192 was analyzed by real time qRT-PCR analysis using RNU6B as an internal standard for normalization (open bar, control; dashed bar, plus MTX treatment).
We have predicted in our previous study that the promoter site of miR-192 contains a well conserved p53 binding sequence (12). The binding sequence is 5′-CGCCATGCCT...GGCCTTGCCC-3′ with a 3-base gap with a ranking site score of 90 based on TFBS (34). Our results are subsequently confirmed by a recent report using a similar type of analysis which shows that the promoter of miR-192 contains a p53 binding site (35). To experimentally confirm a direct interaction between the p53 protein and the miR-192 promoter, we utilized ChIP-qPCR analysis to isolate p53 bound chromosome DNA. The isolated p53 specific binding DNA was PCR amplified using primers which span the predicted p53 binding sites of the miR-192 promoter or the positive control p21 promoter transcriptionally regulated by p53 protein. We show that the p53 protein directly interacts with the miR-192 promoter based on ChIP-qPCR analysis with a 4-fold enriched signal with p53 specific monoclonal antibody compared to the non-specific antibody control DNA (supplemental data file 2A-B). These results validate our bioinformatic prediction of the existence of a conserved p53 binding site at the promoter region of miR-192 (12). We further demonstrated directly that the conserved p53 binding site at the promoter region of miR-192 can activate luciferase expression only in HCT116 (wt-p53) cells. The activation was further enhanced by induced p53 expression in HCT (wt-p53) cells treated with 5-FU. By contrast, the induction of luciferase activity was completely absent from the HCT116 (null-p53) cells (supplemental data file 3). This suggests that miR-192, like miR-34, is another miRNA that is involved in the p53 tumor suppressor network. It is well established that p53 is one of the most frequent altered tumor suppressor gene in colorectal cancer (36). We hypothesize that the potential function of multiple miRNAs involved in p53 tumor suppressor network is to provide the p53 with greater flexibility in rapidly responding to different growth condition changes, perhaps by having unique miRNAs (e.g. miR-34, miR-192) mediate the regulation of the key mRNA targets. We also discovered recently that the expression of miR-192 was decreased in a panel of colorectal tumor specimens compared to the adjacent normal tissues (data not show). This is consistent with a recent report by Schetter et al. showed that miR-192 was one of the miRNAs with reduced expression in a large cohort of colon cancer patient samples, further supporting the potential impact and clinical relevance of miR-192 in colon cancer (37). We speculate that the decrease or loss of the suppressive function of miR-192 in colon cancer may be an important factor related to cell cycle control and chemosensitivity to anti-folate based therapy.
In conclusion, our study provides evidence that miR-192 is another candidate microRNA that is directly involved in the regulation of a key anticancer target DHFR. The expression and function of the miR-192 is largely dependent on the presence of functional wild type p53. This raises the potential of using miR-192 as a novel therapeutic option for treating cancer via an effective delivery system either alone or in combination with anti-folate compounds. Further studies are needed to identify additional miR-192 mediated targets and its relationship with other miRNAs involved in the same pathway.
Supplementary Material
Supplemental Figure 1. Real time qRT-PCR analysis of ectopic expressed miR-192 (*** P < 0.0001) (A) and miR-24 (*** P < 0.0001) (B) by transient transfection in HCT-116 (wt-p53) cells. U6 RNA was used to normalize the expression.
Supplemental Figure 2. Schematic diagram of putative miR-192 promoter with p53 binding site located between 3172 and 3150 upstream of the miR-192 precursor (A). Chromatin immunoprecipitation and quantitative PCR analysis of p53 binding activity to miR-192 promoter. The binding activity of p53 protein to p21 promoter sites was used as positive control (B) (*** P < 0.0001).
Supplemental Figure 3. Transcription activation analysis using luciferase reporter assay. Control promoterless luciferase plasmid pGL3 or miR-192-pGL3 plasmid were transfected to HCT-116 (wt-p53) cells, HCT116 (null-p53) cells and both cell lines treated with 5-FU. Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well in triplicate. miR-192 promoter activity was activated only in HCT-116 (wt-p53) cells and further enhanced by 5-FU treatment to further induce p53 expression level (*** P < 0.0001). This activation was largely missing from HCT116 (null-p53) cells.
Supplemental Figure 4. Absolute expression levels of endogenous miR-192 by real time qRT-PCR analysis. The level of miR-192 was decreased due to the loss of wild type p53 in HCT116 (null-p53) cells compared to HCT-116 (wt-p53) cells (*** P < 0.0001).
ACKNOWLEDGEMENTS
This project was supported by the Mitchell Cancer Institute Start-up Funds and by NIH CA114043 to (J. Ju) and MH075020 (J. Ju).
Footnotes
Statement of Translational Relevance.
We provide experimental evidence in this study that miR-192 is another candidate microRNA that is directly involved in the regulation of a key anticancer target Dihydrofolate Reductase (DHFR). Over expression of miR-192 suppresses the cellular proliferation and restored cell cycle control. The expression and function of the miR-192 is largely dependent on the presence of functional wild type p53. This raises the potential of using miR-192 as a novel therapeutic option for treating cancer via an effective delivery system either alone or in combination with anti-folate compounds. miR-192 may also serve as a potential biomarker for clinical prognosis.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Cummins JM, He Y, Leary RJ, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 4.Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 5.Ruvkun G. Clarifications on miRNA and cancer. Science. 2006;311:36–7. doi: 10.1126/science.311.5757.36d. [DOI] [PubMed] [Google Scholar]
- 6.Dony C, Kessel M, Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985;317:636–9. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- 7.Sheikh MS, Fornace AJ., Jr. Regulation of translation initiation following stress. Oncogene. 1999;18:6121–8. doi: 10.1038/sj.onc.1203131. [DOI] [PubMed] [Google Scholar]
- 8.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 9.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res. 2006;12:2014–24. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 16.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarasov V, Jung P, Verdoodt B, et al. Differential Regulation of microRNAs by p53 Revealed by Massively Parallel Sequencing: miR-34a is a p53 Target That Induces Apoptosis and G(1)-arrest. Cell Cycle. 2007:6. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 18.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee D, Mayer-Kuckuk P, Capiaux G, Budak-Alpdogan T, Gorlick R, Bertino JR. Novel aspects of resistance to drugs targeted to dihydrofolate reductase and thymidylate synthase. Biochim Biophys Acta. 2002;1587:164–73. doi: 10.1016/s0925-4439(02)00079-0. [DOI] [PubMed] [Google Scholar]
- 21.Chu E, Takimoto CH, Voeller D, Grem JL, Allegra CJ. Specific binding of human dihydrofolate reductase protein to dihydrofolate reductase messenger RNA in vitro. Biochemistry. 1993;32:4756–60. doi: 10.1021/bi00069a009. [DOI] [PubMed] [Google Scholar]
- 22.Tai N, Ding Y, Schmitz JC, Chu E. Identification of critical amino acid residues on human dihydrofolate reductase protein that mediate RNA recognition. Nucleic Acids Res. 2002;30:4481–8. doi: 10.1093/nar/gkf562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai N, Schmitz JC, Chen TM, Chu E. Characterization of a cis-acting regulatory element in the protein-coding region of human dihydrofolate reductase mRNA. Biochem J. 2004;378:999–1006. doi: 10.1042/BJ20031396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai N, Schmitz JC, Chen TM, O’Neill MB, Chu E. Identification of a cis-acting element of human dihydrofolate reductase mRNA. Biochem Biophys Res Commun. 2007 doi: 10.1016/j.bbrc.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 25.Tai N, Schmitz JC, Liu J, et al. Translational autoregulation of thymidylate synthase and dihydrofolate reductase. Front Biosci. 2004;9:2521–6. doi: 10.2741/1413. [DOI] [PubMed] [Google Scholar]
- 26.Ercikan E, Banerjee D, Waltham M, Schnieders B, Scotto KW, Bertino JR. Translational regulation of the synthesis of dihydrofolate reductase. Adv Exp Med Biol. 1993;338:537–40. doi: 10.1007/978-1-4615-2960-6_109. [DOI] [PubMed] [Google Scholar]
- 27.Mishra PJ, Humeniuk R, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci U S A. 2007;104:13513–8. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ju J, Pedersen-Lane J, Maley F, Chu E. Regulation of p53 expression by thymidylate synthase. Proc Natl Acad Sci U S A. 1999;96:3769–74. doi: 10.1073/pnas.96.7.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–5. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Hillcoat BL, Swett V, Bertino JR. Increase of dihydrofolate reductase activity in cultured mammalian cells after exposure to methotrexate. Proc Natl Acad Sci U S A. 1967;58:1632–7. doi: 10.1073/pnas.58.4.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ercikan-Abali EA, Banerjee D, Waltham MC, Skacel N, Scotto KW, Bertino JR. Dihydrofolate reductase protein inhibits its own translation by binding to dihydrofolate reductase mRNA sequences within the coding region. Biochemistry. 1997;36:12317–22. doi: 10.1021/bi971026e. [DOI] [PubMed] [Google Scholar]
- 32.Bunz F, Dutriaux A, Lengauer C, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 33.Waldman T, Kinzler KW, Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 1995;55:5187–90. [PubMed] [Google Scholar]
- 34.Lenhard B, Wasserman WW. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–6. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 35.Sinha AU, Kaimal V, Chen J, Jegga AG. Dissecting microregulation of a master regulatory network. BMC Genomics. 2008;9:88. doi: 10.1186/1471-2164-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogelstein B, Kinzler KW. p53 function and dysfunction. Cell. 1992;70:523–6. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 37.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Real time qRT-PCR analysis of ectopic expressed miR-192 (*** P < 0.0001) (A) and miR-24 (*** P < 0.0001) (B) by transient transfection in HCT-116 (wt-p53) cells. U6 RNA was used to normalize the expression.
Supplemental Figure 2. Schematic diagram of putative miR-192 promoter with p53 binding site located between 3172 and 3150 upstream of the miR-192 precursor (A). Chromatin immunoprecipitation and quantitative PCR analysis of p53 binding activity to miR-192 promoter. The binding activity of p53 protein to p21 promoter sites was used as positive control (B) (*** P < 0.0001).
Supplemental Figure 3. Transcription activation analysis using luciferase reporter assay. Control promoterless luciferase plasmid pGL3 or miR-192-pGL3 plasmid were transfected to HCT-116 (wt-p53) cells, HCT116 (null-p53) cells and both cell lines treated with 5-FU. Firefly luciferase activity was normalized to Renilla luciferase activity for each transfected well in triplicate. miR-192 promoter activity was activated only in HCT-116 (wt-p53) cells and further enhanced by 5-FU treatment to further induce p53 expression level (*** P < 0.0001). This activation was largely missing from HCT116 (null-p53) cells.
Supplemental Figure 4. Absolute expression levels of endogenous miR-192 by real time qRT-PCR analysis. The level of miR-192 was decreased due to the loss of wild type p53 in HCT116 (null-p53) cells compared to HCT-116 (wt-p53) cells (*** P < 0.0001).