Abstract
Disease introduction into the New World during colonial expansion is well documented and had a major impact on indigenous populations; however, few diseases have been associated with early human migrations into North America. During the late Pleistocene epoch, Asia and North America were joined by the Beringian Steppe ecosystem which allowed animals and humans to freely cross what would become a water barrier in the Holocene. Anthrax has clearly been shown to be dispersed by human commerce and trade in animal products contaminated with Bacillus anthracis spores. Humans appear to have brought B. anthracis to this area from Asia and then moved it further south as an ice-free corridor opened in central Canada ∼13,000 ybp. In this study, we have defined the evolutionary history of Western North American (WNA) anthrax using 2,850 single nucleotide polymorphisms (SNPs) and 285 geographically diverse B. anthracis isolates. Phylogeography of the major WNA B. anthracis clone reveals ancestral populations in northern Canada with progressively derived populations to the south; the most recent ancestor of this clonal lineage is in Eurasia. Our phylogeographic patterns are consistent with B. anthracis arriving with humans via the Bering Land Bridge. This northern-origin hypothesis is highly consistent with our phylogeographic patterns and rates of SNP accumulation observed in current day B. anthracis isolates. Continent-wide dispersal of WNA B. anthracis likely required movement by later European colonizers, but the continent's first inhabitants may have seeded the initial North American populations.
Introduction
The basic premises of disease tracking have changed little since John Snow first described the London cholera epidemic of 1854. The use of molecular genotyping technologies has allowed the epidemiological linkage of geographically disparate isolates, generating hypotheses about patterns and modes of disease dispersal. As might be expected, the distribution of human pathogens that cause persistent infections such as Helicobacter pylori, the Typhi serovar of Salmonella enterica, Mycobacterium tuberculosis and Polyomavirus JC reflect both recent and ancient human migratory patterns [1]–[7]. Conversely, pathogens that cause acute infections remain only briefly within a host and are therefore less likely to follow long term host distribution patterns [3]. The dispersal of Bacillus anthracis, Yersinia pestis, and human RNA viruses often reflect short term human movement frequently associated with trading contaminated animal products or inadvertently transporting primary vectors or hosts [1], [3], [8]–[12]. Such potentially frequent and long range dispersal of pathogens can obscure more ancient phylogeographic patterns. The history of B. anthracis in North America has certainly been affected by recent trade, and livestock movement [13], however here we present evidence that the introduction of this pathogen can be traced to much more ancient human migrations. We believe this to be an example of an opportunistic human pathogen reflecting ancient human dispersal patterns.
The recent and dramatic increase in the ability for extensive genomic sampling through whole genome sequencing coupled with extensive strain collections should enhance our ability to reconstruct even ancient epidemiological events. The strictly clonal reproductive patterns and low polymorphism frequency of evolutionarily stable molecular markers in B. anthracis makes this a model organism for tracking ancient epidemiological patterns. Whole genome sequencing of multiple B. anthracis strains has led to the construction of a highly accurate phylogenetic backbone [14] based upon an expansive world-wide strain collection [13]. Whereas it would be advantageous to sequence all available isolates within a lineage, this approach is still prohibitively expensive and requires SNP detection by whole genome comparisons. Targeting characters and taxa within specific lineages can further enhance the detection of evolutionary patterns which, when combined with sample spatial data, enables precise epidemiological tracking of disease, even for pre-historical events.
B. anthracis has dispersed globally via large and sequential radiations associated with human commerce and trade of animal products contaminated with B. anthracis spores [13]. Without human involvement, infected animals typically die within 7–10 days, seeding only the surrounding soil with spores thus keeping the spread of the disease relatively contained [15]. The potential for dispersion even among migratory herds is limited since infected animals typically die quickly before extensive dispersal can occur. Historically, an animal that died of anthrax was scavenged by people for its hair, hide, bones, and even consumed as food, facilitating the dispersal of spores away from a carcass. Indeed, imported spore-contaminated animal hides account for many of the recent US human anthrax cases [16], though such modern cases infrequently result in subsequent ecological establishment or further dispersal. Therefore, with the exception of the most recent human cases, the current distribution of B. anthracis can be traced to historical human dispersion, trade, and migratory patterns.
The most dramatic dispersal and clonal expansion of B. anthracis was the A-radiation [13], which is phylogenetically rooted in the Old World. Nested within the A-radiation is the highly successful trans-Eurasian subpopulation (TEA). Its prevalence in Europe and Asia is thought to be mediated by the east-west human trade routes, such as the “Silk Road”. One sublineage of this TEA population, western North America (WNA), was introduced into North America and has become highly successful within this geographic region. The WNA sublineage is dominant today in central Canada and much of the western United States.
In North America, two distinct types of anthrax cases are seen. Many have been observed along the East Coast and are associated with trade and industrial processing of contaminated animal products, often wool in textile mills [17]–[19]. These cases contribute to the overall genetic diversity of North American B. anthracis isolates, but generally represent small case clusters that do not become ecologically founded. This lack of establishment could be due to a requirement of suitable habitat for natural disease cycling [20]. In contrast, western North American grasslands are ideal for the ecological establishment of anthrax and may have persisted for much of the Holocene epoch, possibly over 10,000 years [21].
Indeed, at least two B. anthracis clades are ecologically established in North America on a sub-continental scale [13]. The “Ames” clade (A.Br.Ames) has been associated with highly localized and sporadic outbreaks in south Texas since at least the early 1980s [22]. Only a short evolutionary time period, very few SNPs, separate Texas Ames isolates from Asian near relatives, suggesting a recent or perhaps colonial animal importation. In contrast, the WNA clade has been widely successful both in distribution and frequency across central and northerly North American regions and is clearly ecologically established in many geographic areas. WNA isolates have been recovered from near the Artic Circle in Canada to the U.S. Mexican border and even in insular Haiti, and account for 89% of non-human anthrax cases in North America. The WNA clade also exhibits greater genetic diversity than the Ames clade, and a longer evolutionary separation (106 SNPs) from its nearest Old World relatives (TEA), suggesting a more ancient introduction into North America. The ecological dominance and disease importance of the WNA clade led us to examine its evolutionary history in greater detail using whole genome sequence analysis and highly accurate phylogenetic reconstructions.
Results
SNP Analyses
Whole genome sequencing of seven diverse strains led to the discovery of 2,850 SNPs suitable for conversion to whole genome tiling microarrays. One of these seven strains was the WNA strain (A0193) [14]. These SNPs were screened among 128 diverse isolates (described by MLVA15 analyses) and identified WNA as a monophyletic group rooted in the Old World TEA group. These data also showed the WNA group to be separated by a long phylogenetic branch (106 SNPs) (Fig. 1) representing 53% of the total distance from the initiation of the A branch radiation to the sequenced WNA isolate. Ten of these 106 SNPs were developed into Real-time PCR assays and used to screen all 387 isolates in the study. These SNPs identified six sub-clades within the previously described WNA lineage.
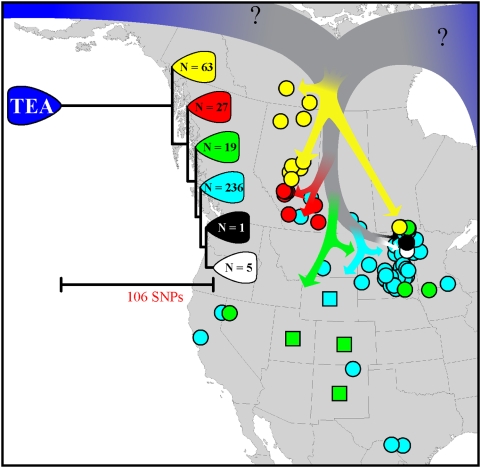
Figure 1. Phylogeography of the WNA Bacillus anthracis clone.
The evolutionary tree of the dominant North American B. anthracis lineage (WNA) and mapping of isolate locations supports southern dispersal of anthrax in North America after founding by a European or Asian ancestor (TEA). Circles indicate precise GIS coordinates, squares indicate state-level information, and colors indicate phylogenetic grouping.
Phylogenetic analyses
SNP characters are rare in the B. anthracis genome and almost never have character state reversals (∼0.1% homoplasy) [14]. Using a cladistic evolutionary model, there was one character-state inconsistency in these data (<1% homoplasy). Given such robust data, all approaches (e.g., MP, NJ) to construct phylogenetic trees generate the same evolutionary hypothesis (Fig. 1). Including any B. anthracis strain from outside this lineage allows us to accurately root this lineage using standard outgroup rooting methods.
Geographical mapping of WNA isolates
The six sub-clades identified by SNP analyses revealed a north-south phylogeographic pattern (Fig. 1), with short terminal branches indicating rapid radiation of the WNA group following the initial establishment in northern Canada. Additionally, there were correlations among the nodes identified with recovery from their respective hosts. For example, the yellow group was mostly associated with wildlife (n = 63) whereas the other groups were almost exclusively from livestock.
Discussion
SNPs are stable phylogenetic characters in B. anthracis and impart a highly accurate evolutionary hypothesis both in terms of branch lengths and branching order [14]. Our phylogenetic topologies are highly suggestive of an initial introduction of B. anthracis into the far north of North America and subsequent southerly dispersal (Fig. 1). The ancestral WNA phylogenetic nodes are northernmost, followed by progressively more southern locations for the more recently derived clades. More recent populations in southern states and a single human isolate from Haiti (not shown) are probably the result of recent commodity trading or infected livestock transport. In contrast, the northern-most and more ancient populations of WNA B. anthracis remain relatively localized, possibly due to association with bison (Bison bison) and restricted human commerce in this region. SNP-estimated evolutionary branch lengths provide additional support for a pre-Columbian introduction of WNA anthrax to North America. The accumulation of up to 106 SNPs since the split of the WNA lineage from the TEA lineage is a relatively large number for B. anthracis clades [14]. Interestingly, this indicates a long evolutionary separation followed by a genetic bottleneck and a single founding event in North America. In contrast, the Texas ecologically-established Ames lineage accumulated only ∼8 SNPs since introduction from Asia [23]. Molecular clock calibration is always problematic, but this >10-fold difference offers dramatic contrast between a recent and ancient New World introduction. Moreover, divergence times within branches of the A-radiation may be greatly affected by the number of generations in natural disease cycling. The model introduced in Van Ert et al (2007) was based upon 1 and 0.5 generations per year (approximately 3500 to 7000 ybp respectively for the major A-radiation). However, a recent overview of early outbreaks in Northern Canada provides greater insights into natural disease cycling in this region. Dragon and Elkin noted [24] that there were eight sporadic outbreaks between 1962 and 1991, and since 1993, there have been five more. Using these empirical data we can estimate 0.28 generations per year (13 outbreaks in 46 years). This empirical based estimation of this parameter significantly increases divergence times within the TEA-WNA lineage and suggests that divergence times provided in Van Ert et al. (2007) were underestimates.
Human migrations are the most likely source for the introduction and establishment of the WNA lineage of anthrax in North America through an ice-free corridor that connected Beringia to the southern areas east of the Rocky Mountains. Contiguous grassland habitat created by the partial retreat of the Laurentide and Cordilleran ice sheets would have been ideal for anthrax susceptible grazing herd animals. During the late Pleistocene epoch, Asia and North America were joined by the Beringian Steppe ecosystem [25]. This grassland refugium allowed animals and humans to freely cross what would become a water barrier (Bering Sea) in the Holocene. Humans could have transported B. anthracis to this land bridge from Asia and then moved it further south as the ice-free corridor and developing grassland opened in central Canada ∼13,000 ybp. Bison and other potential herding hosts were already widespread in North America, yet the limited range of the most ancient northern WNA B. anthracis populations suggest that anthrax was not present at this time south of the bottleneck created by the coalescing glaciers. Furthermore, as the ice-free corridor expanded, there was a simultaneous northward movement of these grazers like bison, yet the phylogenetic directionality of anthrax spread is southerly. Humans, however, did move southward through this corridor and could have brought contaminated animal products with them, which eventually enabled the ecological introduction of WNA B. anthracis in North America.
This northern-origin hypothesis is highly consistent with phylogeographic patterns and rates of SNP accumulation observed in recent B. anthracis isolates. While isolates from many different lineages have been observed in North America, their presence can be attributed to post colonial trade. Continent-wide dispersal of WNA B. anthracis may have involved later European colonizers, but some of the first inhabitants of the continent likely seeded the initial North American populations.
Materials and Methods
Strains
The 387 strains (Table 1) used in this study were obtained from various sources and linked to both livestock and wildlife outbreaks in North America. Four of these isolates were associated with human infections. Of the 387 strains, 352 cluster with the major ecologically established Western North American clade (WNA: A.Br.WNA) whereas 17 isolates were included to represent the TEA population as an outgroup. Genotypic data for each strain is provided in Table 2.
Table 1. Strains used in this study (Epidemiologic data)1.
| Sample ID 2 | Country of Origin | Host | State/Province of Infection 3 | County | Nearest City/Town | Latitude 4 | Longitude |
| A1055 | USA | soil | LA | ||||
| A1065 | USA | bovine | WY | ||||
| A2051 | USA | Bovine | CA | Santa Clara | San Jose | ||
| A2052 | USA | Bovine | CA | Santa Clara | San Jose | ||
| A0394 | USA | goat kid | TX | Terrell | |||
| A1115 | USA | Bovine | TX | Leakey | |||
| A1117 | USA | Bovine | TX | ||||
| A2012 | USA | Human | FL | Palm Bay | |||
| A0169 | Canada | bovine | |||||
| A0517 | USA | Vaccine | |||||
| A0248 | USA | Human | |||||
| A1057 | USA | bovine | OK | ||||
| A1056 | USA | Bovine | |||||
| A0078 | USA | ||||||
| A0156 | USA | sheep | |||||
| A1058 | USA | bovine | TX | ||||
| A1062 | USA | MS | |||||
| A1063 | USA | MD | Ft. Detrick | ||||
| A0032 | China | Fur | |||||
| A0033 | China | Wool | |||||
| A0149 | Turkey | human | |||||
| A0241 | Turkey | goat | |||||
| A0245 | Turkey | bovine | |||||
| A0264 | Turkey | human | |||||
| A0280 | Italy | sheep | |||||
| A0303 | Canada | bovine | |||||
| A0324 | Slovakia | ||||||
| A0343 | Hungary | ||||||
| A0362 | Norway | bovine | |||||
| A0417 | Hungary | ||||||
| A0463 | Pakistan | Sheep | |||||
| A0480 | Russia | vaccine | |||||
| A0596 | China | Marmot | Xinjiang | ||||
| A0604 | China | Soil | Xinjiang | ||||
| A0610 | China | Soil | Xinjiang | ||||
| A0669 | China | Soil | Xinjiang | ||||
| 2000031001 | Canada | ||||||
| 2000031006 | USA | NM | |||||
| 2000031036 | USA | Moose | WY | ||||
| 2000031764 | Goat | ||||||
| 2000031765 | |||||||
| 2000031766 | USA | Puritan Loom | NC | ||||
| 2000031767 | |||||||
| 2000031770 | |||||||
| 2000031774 | |||||||
| 2000032818 | |||||||
| 2000032819 | |||||||
| 2000032820 | |||||||
| 2000032821 | |||||||
| 2000032822 | |||||||
| 2000032826 | |||||||
| 2000032828 | |||||||
| 2000032829 | |||||||
| 2000032831 | |||||||
| 2000032832 | |||||||
| 2000032833 | |||||||
| 2000032834 | |||||||
| 2000032888 | |||||||
| 2000032989 | |||||||
| 2000032990 | |||||||
| 2000032993 | |||||||
| 2000032994 | |||||||
| 2000032995 | |||||||
| 2000032996 | |||||||
| 2000032997 | |||||||
| 2002013003 | |||||||
| 2002013004 | |||||||
| 2002013007 | |||||||
| 2002013008 | |||||||
| 2002013009 | |||||||
| 2002013010 | |||||||
| 2002013011 | |||||||
| 2002013012 | |||||||
| 2002013013 | |||||||
| 2002013014 | |||||||
| 2002013015 | |||||||
| 2002013016 | |||||||
| 2002013017 | |||||||
| 2002013019 | |||||||
| 2002013020 | |||||||
| 2002013021 | |||||||
| 2002013022 | |||||||
| 2002013023 | |||||||
| 2002013024 | |||||||
| 2002013025 | |||||||
| 2002013026 | |||||||
| 2002013027 | |||||||
| 2002013028 | |||||||
| 2002013029 | |||||||
| 2002013030 | |||||||
| 2002013031 | |||||||
| 2002013032 | |||||||
| 2002013033 | |||||||
| 2002013034 | |||||||
| 2002013035 | |||||||
| 2002013053 | |||||||
| 2002013054 | |||||||
| 2002013055 | |||||||
| 2002013056 | |||||||
| 2002013057 | |||||||
| 2002013058 | |||||||
| 2002013059 | |||||||
| 2002013063 | |||||||
| 2002013064 | |||||||
| 2002013065 | |||||||
| 2002013068 | |||||||
| 2002013070 | Canada | Bison | |||||
| 2002013083 | USA | Mice | WY | ||||
| 2002013086 | |||||||
| 2002013098 | |||||||
| 2002721542 | USA | Bovine | UT | ||||
| 2002734033 | USA | UT | |||||
| A0055 | USA | ||||||
| A0056 | USA | ||||||
| A0057 | USA | ||||||
| A0071 | USA | ||||||
| A0072 | USA | ||||||
| A0073 | USA | ||||||
| A0074 | USA | ||||||
| A0075 | USA | ||||||
| A0076 | USA | ||||||
| A0117 | USA | Bovine | NV | ||||
| A0130 | USA | ||||||
| A0142 | Canada | Moose | |||||
| A0143 | Canada | bovine | |||||
| A0144 | Canada | Bison | |||||
| A0157 | USA | Pig | |||||
| A0162 | Canada | Bison | |||||
| A0163 | Canada | bovine | |||||
| A0165 | Canada | Bison | |||||
| A0166 | Canada | Bison | |||||
| A0167 | Canada | bovine | |||||
| A0168 | Canada | bovine | |||||
| A0170 | Canada | bovine | |||||
| A0171 | Canada | bovine | |||||
| A0172 | Canada | bovine | |||||
| A0173 | Canada | bovine | |||||
| A0174 | Canada | bovine | |||||
| A0175 | Canada | Bison | |||||
| A0176 | Canada | Bison | |||||
| A0177 | Canada | Bison | |||||
| A0178 | Canada | Bison | |||||
| A0179 | Canada | Bison | |||||
| A0180 | Canada | bovine | |||||
| A0181 | Canada | Ravens | |||||
| A0182 | Canada | bovine | |||||
| A0192 | Canada | bovine | |||||
| A0193 | USA | bovine | |||||
| A0194 | Canada | bovine | |||||
| A0195 | Canada | bovine | |||||
| A0197 | Canada | bovine | |||||
| A0198 | Canada | bovine | |||||
| A0246 | USA | bovine | |||||
| A0268 | USA | Dog | |||||
| A0295 | Canada | Bison | |||||
| A0296 | Canada | Bison | |||||
| A0297 | Canada | Bison | |||||
| A0300 | Canada | bovine | |||||
| A0301 | Canada | bovine | |||||
| A0302 | Canada | bovine | |||||
| A0304 | Canada | Bison | |||||
| A0305 | Canada | Bison | |||||
| A0306 | Canada | Bison | |||||
| A0307 | Canada | bovine | |||||
| A0308 | Canada | bovine | |||||
| A0309 | Canada | bovine | |||||
| A0310 | Canada | bovine | |||||
| A0311 | Canada | bovine | |||||
| A0312 | Canada | Bison | |||||
| A0313 | Canada | Bison | |||||
| A0369 | Canada | Bison | |||||
| A0374 | USA | Bison | |||||
| A0377 | Haiti | man | |||||
| A0383 | USA | man | CO | ||||
| A0392 | USA | bovine | |||||
| A0393 | USA | bovine | |||||
| A0395 | USA | deer | TX | Uvalde | |||
| A0396 | USA | bovine | TX | Del Rio | |||
| A0397 | USA | deer | TX | Del Rio | |||
| A0398 | USA | deer | TX | Del Rio | |||
| A0407 | USA | bovine | |||||
| A0408 | USA | bovine | |||||
| A0409 | USA | bovine | |||||
| A0410 | USA | bovine | |||||
| A0418 | Canada | Bison | |||||
| A0444 | Canada | bovine | |||||
| A0445 | Canada | bovine | |||||
| A0486 | USA | bovine | |||||
| A0520 | USA | WT deer | |||||
| A0526 | USA | Bovine | MT | Billings | |||
| A0767 | Canada | Red Fox Scat | WBNP | Parson's Lake | 59 45′ 53.9″ N | 112 16′ 55.4″W | |
| A0768 | Canada | Red Fox Scat | WBNP | Parson's Lake | 59 45′ 53.9″ N | 112 16′ 55.4″W | |
| A0770 | Canada | Sand | WBNP | Parson's Lake | 59 45′ 14.4″N | 112 16′ 24.2″ W | |
| A0771 | Canada | Sand | WBNP | Parson's Lake | 59 45′ 14.4″N | 112 16′ 42.5″ W | |
| A0772 | Canada | Soil | WBNP | Falaise Lake | 61 28′ 52.0″ N | 116 15′ 54.0″ W | |
| A0773 | Canada | Soil | WBNP | Falaise Lake | 61 28′ 52.0″ N | 116 15′ 54.0″ W | |
| A0774 | Canada | Soil | WBNP | Falaise Lake | 61 28′ 52.0″ N | 116 15′ 54.0″ W | |
| A0775 | Canada | Soil | WBNP | Falaise Lake | 61 28′ 52.0″ N | 116 15′ 54.0″ W | |
| A0776 | Canada | Soil | WBNP | Falaise Lake | 61 28′ 52.0″ N | 116 15′ 54.0″ W | |
| A0777 | Canada | Soil | WBNP | Falaise Lake | 61 28′ 52.0″ N | 116 15′ 54.0″ W | |
| A0778 | Canada | Soil | WBNP | Falaise Lake | 61 28′ 52.0″ N | 116 15′ 54.0″ W | |
| A0779 | Canada | Soil | WBNP | Falaise Lake | 61 28′ 52.0″ N | 116 15′ 54.0″ W | |
| A0780 | Canada | Soil | WBNP | Falaise Lake | 61 28′ 52.0″ N | 116 15′ 54.0″ W | |
| A0782 | Canada | Soil | WBNP | Falaise Lake | 61 29′ 7.0″ N | 116 13′ 29.0″ W | |
| A0783 | Canada | Soil | WBNP | Falaise Lake | 61 29′ 7.0″ N | 116 13′ 29.0″ W | |
| A0784 | Canada | Soil | WBNP | Falaise Lake | 61 29′ 7.0″ N | 116 13′ 29.0″ W | |
| A0785 | Canada | Soil | WBNP | Falaise Lake | 61 29′ 7.0″ N | 116 13′ 29.0″ W | |
| A0786 | Canada | Soil | WBNP | Falaise Lake | 61 29′ 7.0″ N | 116 13′ 29.0″ W | |
| A0788 | Canada | Soil | WBNP | Falaise Lake | 61 27′ 36.0″ N | 116 18′ 23.0″ W | |
| A0791 | Canada | ||||||
| A0792 | Canada | Bovine | Rocky Mountain House | ||||
| A0793 | Canada | Bovine | Rocky Mountain House | ||||
| A0794 | Canada | Bovine | Rocky Mountain House | ||||
| A0795 | Canada | Bovine | Rocky Mountain House | ||||
| A0796 | Canada | Bovine | Rocky Mountain House | ||||
| A0797 | Canada | Horse | Rocky Mountain House | ||||
| A0798 | Canada | Bovine | Eckville | ||||
| A0799 | Canada | Bovine | Eckville | ||||
| A0800 | Canada | Bovine | Eckville | ||||
| A0801 | Canada | Bovine | Eckville | ||||
| A0802 | Canada | Bovine | Eckville | ||||
| A0803 | Canada | Bovine | Rocky Mountain House | ||||
| A0804 | Canada | Bovine | Caroline | ||||
| A0805 | Canada | Bovine | Rocky Mountain House | ||||
| A0806 | Canada | Bovine | Rocky Mountain House | ||||
| A0807 | Canada | Bovine | Alhambra | ||||
| A0808 | Canada | Bovine | Alhambra | ||||
| A0809 | Canada | Bovine | Alhambra | ||||
| A0810 | Canada | Bovine | Alhambra | ||||
| A0812 | Canada | Bovine | Rocky Mountain House | ||||
| A0913 | USA | Bovine | NV | ||||
| A0917 | USA | ||||||
| A0948 | USA | Bovine | ND | Walsh | Park River | ||
| A0949 | USA | Equine | ND | Steele | Sharon | ||
| A0950 | USA | Bison | ND | Grand Forks | Grand Forks | ||
| A0951 | USA | Equine | ND | Traill | Portland | ||
| A0952 | USA | Bovine | MN | Clay | Hawley | ||
| A0953 | USA | Bovine | ND | Stutsman | Parkhurst | ||
| A0954 | USA | Bovine | ND | Foster | Grace City | ||
| A0955 | USA | Bovine | ND | Griggs | Cooperstown | ||
| A0956 | USA | Bovine | ND | Steele | Sharon | ||
| A0957 | USA | ||||||
| A0958 | USA | Bovine | ND | Nelson | Aneta | ||
| A0959 | USA | Bovine | ND | Grand Forks | Northwood | ||
| A0960 | USA | Bovine | ND | Traill | Portland | ||
| A0961 | USA | Equine | ND | Steele | Sharon | ||
| A0962 | USA | Bovine | ND | Grand Forks | Emerado | ||
| A0963 | USA | Bovine | MN | Becker | Lake Park | ||
| A0964 | USA | Bovine | ND | Grand Forks | Grand Forks | ||
| A0965 | USA | Bovine | ND | Pembina | Walhalla | ||
| A0966 | USA | Bovine | ND | Grand Forks | Emerado | ||
| A0967 | USA | Bovine | ND | Pembina | Hensel | ||
| A0968 | USA | Bovine | MN | Roseau | Greenbush | ||
| A0969 | USA | Bovine | ND | Grand Forks | Larimore | ||
| A0970 | USA | Bovine | ND | Adams | Reeder | ||
| A0971 | USA | Bovine | ND | Nelson | Aneta | ||
| A0972 | USA | Bovine | ND | Nelson | Pekin | ||
| A0973 | USA | Bovine | ND | Eddy | Hamar | ||
| A0974 | USA | Bovine | ND | Nelson | Tolna | ||
| A0975 | USA | Equine | ND | Nelson | Petersburg | ||
| A0976 | USA | ||||||
| A0977 | USA | Bovine | ND | Grand Forks | Larimore | ||
| A0978 | USA | Bovine | MN | Roseau | Badger | ||
| A0979 | USA | Bovine | MN | Mower | Lyle | ||
| A0980 | USA | Bovine | MN | Pennington | Thief River Falls | ||
| A0981 | USA | Bovine | ND | Grand Forks | Larimore | ||
| A0982 | USA | Bovine | ND | Pembina | Hensel | ||
| A0983 | USA | ||||||
| A0984 | USA | ||||||
| A0985 | USA | ||||||
| A0988 | Canada | Bison | WBNP | ||||
| A0989 | Canada | ||||||
| A0990 | Canada | ||||||
| A0991 | Canada | Bear | |||||
| A0993 | Canada | Bison | |||||
| A0994 | Canada | Bison | |||||
| A0995 | Canada | Wolf | |||||
| A0996 | Canada | Bovine | |||||
| A0997 | Canada | Bovine | |||||
| A0998 | Canada | Moose | |||||
| A0999 | Canada | Bovine | |||||
| A1000 | Canada | Bovine | |||||
| A1001 | Canada | Bovine | Manitoba | Vita | |||
| A1002 | Canada | Bovine | Manitoba | Vita | |||
| A1003 | Canada | Bovine | Manitoba | Vita | |||
| A1004 | Canada | Bovine | Manitoba | Vita | |||
| A1005 | Canada | Bovine | Manitoba | Vita | |||
| A1006 | Canada | Bovine | Manitoba | Vita | |||
| A1007 | Canada | Bovine | Manitoba | Vita | |||
| A1008 | Canada | Bovine | Manitoba | Vita | |||
| A1009 | Canada | Bovine | Manitoba | Vita | |||
| A1010 | Canada | Bovine | Manitoba | Vita | |||
| A1011 | Canada | Bovine | Manitoba | Vita | |||
| A1012 | Canada | Bovine | Manitoba | Vita | |||
| A1013 | Canada | Bovine | Manitoba | Vita | |||
| A1014 | Canada | Bovine | Manitoba | Vita | |||
| A1015 | Canada | Bovine | Manitoba | Vita | |||
| A1016 | Canada | Bovine | Manitoba | Vita | |||
| A1017 | Canada | Bovine | Manitoba | Vita | |||
| A1018 | Canada | Bovine | Manitoba | Vita | |||
| A1019 | Canada | Manitoba | Vita | ||||
| A1020 | Canada | Manitoba | Vita | ||||
| A1021 | Canada | Manitoba | Vita | ||||
| A1022 | Canada | Bison | Manitoba | Vita | |||
| A1023 | Canada | Bovine | Manitoba | Vita | |||
| A1024 | Canada | Bovine | Manitoba | Vita | |||
| A1025 | Canada | Bovine | Manitoba | Vita | |||
| A1026 | Canada | Bovine | Manitoba | Vita | |||
| A1027 | Canada | Bovine | Manitoba | Vita | |||
| A1028 | Canada | Bovine | Manitoba | Vita | |||
| A1029 | USA | Bovine | NV | Vita | |||
| A1030 | USA | Bovine | NV | Washoe | Reno | ||
| A1031 | USA | Bovine | NV | Washoe | Reno | ||
| A1040 | USA | bovine | SD | ||||
| A1041 | USA | bovine | SD | ||||
| A1042 | USA | bovine | SD | ||||
| A1047 | USA | bovine | IA | ||||
| A1118 | USA | Bison | MN | Greenbush | |||
| A1119 | USA | Bovine | MN | Greenbush | |||
| A1120 | USA | Bovine | MN | Greenbush | |||
| A1121 | USA | Bovine | MN | Greenbush | |||
| A1122 | USA | Bovine | MN | Greenbush | |||
| A1123 | USA | Bovine | MN | Greenbush | |||
| A1124 | USA | Bovine | MN | Greenbush | |||
| A1125 | USA | Bovine | MN | Greenbush | |||
| A1126 | USA | Bovine | MN | Greenbush | |||
| A1127 | USA | Bovine | MN | Greenbush | |||
| A1128 | USA | Bovine | MN | Greenbush | |||
| A1129 | USA | Bovine | MN | Greenbush | |||
| A1130 | USA | Bovine | MN | Greenbush | |||
| A1131 | USA | Bovine | MN | Greenbush | |||
| A1132 | USA | Bovine | MN | Greenbush | |||
| A1133 | USA | Bovine | MN | Greenbush | |||
| A1134 | USA | Bovine | MN | Greenbush | |||
| A1135 | Canada | Manitoba | Sprague | ||||
| A1137 | USA | SD | |||||
| A1138 | USA | SD | |||||
| A1139 | USA | SD | |||||
| A2014 | Mexico | Bovine | Acuna | ||||
| A2015 | Mexico | Bovine | Acuna | ||||
| A3455 | USA | Bovine | SD | 44.83663 | 100.33589 | ||
| A3456 | USA | Bovine | SD | 45.66154 | 98.41541 | ||
| A3457 | USA | Bovine | SD | 46.90731 | 98.10566 | ||
| A3458 | USA | Bovine | SD | 44.77967 | 100.42004 | ||
| A3459 | USA | Bovine | SD | 44.83672 | 100.33584 | ||
| A3460 | USA | Bovine | SD | 44.70675 | 100.19478 | ||
| A3461 | USA | Bovine | SD | North | |||
| A3462 | USA | Bovine | SD | North | |||
| A3463 | USA | Bovine | SD | 44.98544 | 100.36005 | ||
| A3464 | USA | Bovine | SD | 44.69881 | 100.46175 | ||
| A3465 | USA | Bovine | SD | 44.98094 | 100.30310 | ||
| A3466 | USA | Bovine | SD | 44.68186 | 98.05983 | ||
| A3467 | USA | Bovine | SD | Central | |||
| A3468 | USA | Bovine | SD | 44.38436 | 99.61700 | ||
| A3469 | USA | Bovine | SD | 44.41030 | 99.80588 | ||
| A3470 | USA | Bovine | SD | 44.72853 | 100.45644 | ||
| A3471 | USA | Bovine | SD | 44.91887 | 99.79668 | ||
| A3472 | USA | Bovine | SD | 44.72853 | 100.45644 | ||
| A3473 | USA | Bovine | SD | Central | |||
| A3474 | USA | Bovine | SD | 44.67489 | 98.05903 | ||
| A3475 | USA | Bovine | SD | 44.90751 | 100.48686 | ||
| A3476 | USA | Bovine | SD | North | |||
| A3477 | USA | Bovine | SD | Central | |||
| A3478 | USA | Bovine | SD | 44.95312 | 100.49400 | ||
| A3479 | USA | Bovine | SD | 45.70536 | 98.39379 | ||
| A3480 | USA | Bovine | SD | 45.25863 | 100.23270 | ||
| A3481 | USA | Bovine | SD | 44.58392 | 99.87045 | ||
| A3482 | USA | Bovine | SD | 45.92200 | 98.05015 | ||
| A3483 | USA | Bovine | SD | North | |||
| A3484 | USA | Bovine | SD | 44.94745 | 100.51275 | ||
| A3485 | USA | Bovine | SD | 45.03621 | 100.30700 | ||
| A3486 | USA | Bovine | SD | 45.16650 | 100.11835 | ||
| A3487 | USA | Bovine | SD | 44.79834 | 100.55977 | ||
| A3488 | USA | Bovine | SD | 44.95903 | 99.90616 | ||
| A3489 | USA | Bovine | SD | 45.13020 | 100.33318 | ||
| A3490 | USA | Bovine | SD | 44.84012 | 98.87670 | ||
| A3491 | USA | Bovine | SD | 45.52951 | 97.72276 | ||
| A3492 | USA | Bovine | SD | 45.35902 | 97.49963 | ||
| A3493 | USA | Bovine | SD | 44.53633 | 103.51009 | ||
| A3494 | USA | Bovine | SD | 44.91887 | 99.79668 | ||
| A3495 | USA | Bovine | SD | 44.10121 | 99.86757 | ||
| A3496 | USA | Bovine | SD | Central | |||
| A3497 | USA | Bovine | SD | 45.51513 | 100.48982 | ||
| A3498 | USA | Bovine | SD | 43.78920 | 99.26104 | ||
| A3499 | USA | Bovine | SD | 44.17555 | 99.50194 | ||
| A3500 | USA | Bovine | SD | 44.10859 | 99.45870 | ||
| A3501 | USA | Bovine | SD | 45.06766 | 100.39238 |
Missing or Unknown data has been intentionally left blank.
Samples having an “A” preceeding a four digit code are samples for which we possess live culture material. Samples having a 10-digit code were received as DNA from part of a historic collection maintained at the Centers for Disease Control and Prevention.
Two letter state codes are used when referencing states within the United States, otherwise the state is written out. WBNP is Wood Buffalo National Park.
Centroids were used for locations where state of origin information was available but Global Positioning System data was unavailable.
Table 2. Strains used in this study (Genotyping data)1.
| Sample ID2 | canSNP group3 | A.Br.WNA4 | wna237471 | wna1141774 | wna2994131 | wna3368524 | wna3631093 | wna3682247 | wna3732539 | wna3774186 | wna4461234 | wna4718500 |
| A1055 | C.Br.A1055 | A | T | C | C | A | G | G | T | T | G | G |
| A1065 | C.Br.A1055 | A | T | C | C | A | G | G | T | T | G | G |
| A2051 | B.Br.001/002 | A | T | C | C | A | G | G | T | T | G | G |
| A2052 | B.Br.001/002 | A | T | C | C | A | G | G | T | T | G | G |
| A0394 | A.Br.001/002 | A | T | C | C | A | G | G | T | T | G | G |
| A1115 | A.Br.001/002 | A | T | C | C | A | G | G | T | T | G | G |
| A1117 | A.Br.001/002 | A | T | C | C | A | G | G | T | T | G | G |
| A2012 | A.Br.Ames | A | T | C | C | A | G | G | T | T | G | G |
| A0169 | A.Br.001/002 | A | T | C | C | A | G | G | T | T | G | G |
| A0517 | A.Br.001/002 | A | T | C | C | A | G | G | T | T | G | G |
| A0248 | A.Br.Aust94 | A | T | C | C | A | G | G | T | T | G | G |
| A1057 | A.Br.Aust94 | A | T | C | C | A | G | G | T | T | G | G |
| A1056 | A.Br.003/004 | A | T | C | C | A | G | G | T | T | G | G |
| A0078 | A.Br.Vollum | A | T | C | C | A | G | G | T | T | G | G |
| A0156 | A.Br.Vollum | A | T | C | C | A | G | G | T | T | G | G |
| A1058 | A.Br.Vollum | A | T | C | C | A | G | G | T | T | G | G |
| A1062 | A.Br.Vollum | A | T | C | C | A | G | G | T | T | G | G |
| A1063 | A.Br.Vollum | A | T | C | C | A | G | G | T | T | G | G |
| A0032 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0033 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0149 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0241 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0245 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0264 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0280 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0324 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0343 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0362 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0417 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0463 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0480 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0596 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0604 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0610 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| A0669 | A.Br.008/009 | A | T | C | C | A | G | G | T | T | G | G |
| 2000031001 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| 2000031006 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| 2000031036 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000031764 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000031765 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000031766 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000031767 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000031770 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000031774 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032818 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032819 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032820 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | ? |
| 2000032821 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032822 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032826 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032828 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032829 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032831 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032832 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032833 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| 2000032834 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032888 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032989 | A.Br.WNA | G | T | C | C | A | G | G | T | T | G | G |
| 2000032990 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032993 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032994 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032995 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032996 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2000032997 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013003 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013004 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013007 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013008 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013009 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013010 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013011 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013012 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013013 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013014 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013015 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013016 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013017 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013019 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013020 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013021 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013022 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013023 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013024 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013025 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013026 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013027 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013028 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013029 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013030 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013031 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013032 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013033 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013034 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013035 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013053 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013054 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013055 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013056 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013057 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013058 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013059 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013063 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013064 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013065 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013068 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013070 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| 2002013083 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013086 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002013098 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| 2002721542 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| 2002734033 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A0055 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0056 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0057 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0071 | A.Br.WNA | G | G | T | G | C | A | A | C | C | A | A |
| A0072 | A.Br.WNA | G | G | T | G | C | A | A | T | C | A | A |
| A0073 | A.Br.WNA | G | G | T | G | C | A | A | C | C | A | A |
| A0074 | A.Br.WNA | G | G | T | G | C | A | A | C | C | A | A |
| A0075 | A.Br.WNA | G | G | T | G | C | A | A | C | C | A | A |
| A0076 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0117 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A0130 | A.Br.WNA | G | G | T | G | C | A | A | C | C | A | A |
| A0142 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0143 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0144 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0157 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0162 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0163 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0165 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0166 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0167 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0168 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0170 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0171 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0172 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0173 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0174 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0175 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0176 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0177 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0178 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0179 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0180 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0181 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0182 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0192 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0193 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A0194 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0195 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0197 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0198 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0246 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A0268 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0295 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0296 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0297 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0300 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0301 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0302 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0303 | A.Br.WNA | A5 | G | T | C | C | G | G | T | T | G | G |
| A0304 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0305 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0306 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0307 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0308 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0309 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0310 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0311 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0312 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0313 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0369 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0374 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A0377 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0383 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A0392 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0393 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0395 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0396 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0397 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0398 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0407 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0408 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0409 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0410 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0418 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0444 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0445 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0486 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0520 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0526 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0767 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0768 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0770 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0771 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0772 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0773 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0774 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0775 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0776 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0777 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0778 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0779 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0780 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0782 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0783 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0784 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0785 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0786 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0788 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0791 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0792 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0793 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0794 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0795 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0796 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0797 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0798 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0799 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0800 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0801 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0802 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0803 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0804 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0805 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0806 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0807 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0808 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0809 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0810 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0812 | A.Br.WNA | G | G | T | C | C | A | G | T | T | G | A |
| A0913 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A0917 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A0948 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0949 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0950 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0951 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0952 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0953 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0954 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0955 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0956 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0957 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0958 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0959 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0960 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0961 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0962 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0963 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0964 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0965 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0966 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0967 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0968 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0969 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0970 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0971 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0972 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0973 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0974 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0975 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0976 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0977 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0978 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0979 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0980 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0981 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0982 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0983 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0984 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0985 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0988 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0989 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0990 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0991 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0993 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0994 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0995 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0996 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0997 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A0998 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A0999 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1000 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1001 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1002 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1003 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1004 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1005 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1006 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1007 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1008 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1009 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1010 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1011 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1012 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1013 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1014 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1015 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1016 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1017 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1018 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1019 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1020 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1021 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1022 | A.Br.WNA | G | G | T | C | C | G | G | T | T | G | G |
| A1023 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1024 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1025 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1026 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1027 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1028 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A1029 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A1030 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1031 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1040 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A1041 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A1042 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A1047 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1118 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1119 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1120 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1121 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1122 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1123 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1124 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1125 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1126 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1127 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1128 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1129 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1130 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1131 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1132 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1133 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1134 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1135 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A1137 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A1138 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A1139 | A.Br.WNA | G | G | T | C | C | A | G | T | C | G | A |
| A2014 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A2015 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3455 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3456 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3457 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3458 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3459 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3460 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3461 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3462 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3463 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3464 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3465 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3466 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3467 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3468 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3469 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3470 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3471 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3472 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3473 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3474 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3475 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3476 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3477 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3478 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3479 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3480 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3481 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3482 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3483 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3484 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3485 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3486 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3487 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3488 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3489 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3490 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3491 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3492 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3493 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3494 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3495 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3496 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3497 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3498 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3499 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3500 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
| A3501 | A.Br.WNA | G | G | T | G | C | A | G | T | C | G | A |
Genotyping data are presented as the SNP state at the particular locus. Each locus is presented with reference to the genome position in the Ancestral Ames strain (GenBank ID: AE017334).
Sample IDs with “A” numbers indicate isolates for which we have live culture. Sample IDs bearing a 10-digit number are from an historical strain collection maintained by the Center for Disease Control and Prevention.
Canonical SNP groups as defined in Van Ert et al. (2007).
The A.Br.WNA canonical SNP was also definied in Van Ert et al. (2007).
Rules of parsimony place this isolate (A0303) in the A.Br.WNA group. This single data point is homoplastic.
DNA Extraction
DNA for Affymetrix whole genome tiling microarrays was extracted on 21 geographically dispersed and genetically diverse (MLVA15) strains within the A.Br.009 clade by a modification of the chloroform∶isoamyl alcohol method described in Keim et al. (1997). DNA for sub-clade specific TaqMan® MGB dual-probe real-time PCR SNP assays was extracted by a simplified heat lysis protocol described in Keim et al. (2000).
SNP Discovery
Seven diverse B. anthracis strains, including one from the WNA clade, which had previously been characterized by Multi-Locus Variable Number of Tandem Repeats Analyses (MLVA), were further characterized by shotgun cloning and whole genome sequencing by the Sanger method. These efforts lead to the discovery of ∼3,000 SNPs within the B. anthracis genome.
Construction of a whole genome tiling microarray
A custom Affymetrix genotyping microarray was constructed using 2850 SNPs. We genotyped 128 diverse strains using this format; relevant to this study, 10 strains were from the trans-Eurasian population (TEA: A.Br.008/009) and 21 were from the Western North American clade (WNA: A.Br.WNA). Of the 2,850 SNPs, 78 separated TEA from WNA, while 28 split the WNA clade into 6 genotypes (Fig. 1).
Real-Time PCR analyses
We selected 10 SNPs for conversion into Taqman® MGB dual-probe real-time PCR SNP assays (Table 3). Assays were designed using Primer Express (Applied Biosystems, Foster City, CA). This is a highly sensitive technology that is fast and cost-effective when analyzing hundreds of samples. Table 4 describes the associated phylogenetic groups for each SNP: Branch separating TEA from WNA (4 SNPs) and WNA subtypes (6 SNPs). These rapid PCR assays were used to genotype a total of 352 isolates from the WNA clade (Table 2). Most of the WNA members had been previously identified using the canonical SNP assay for A.Br.009 [13].
Table 3. Primers and Probes used to detect WNA clade specific SNPs.
| Locus Name 1 (GenBank ID: AE017334) | Primers 5′→3′ 2 | Probes 5′→3′ 3 |
| wna2994131 | F-GCACGGTCTTTCTAAATTCATTGTT | VIC-AAAGAACATAGGAGTTTAC |
| R-TGCGATTGGAGTTGCAAATAAT | FAM-AAAGAACATACGAGTTTAC | |
| wna3631093 | F-CAGAACCTACAGAACCATCATTAAAGAA | VIC-ATTGCTTACACTTCAGA |
| R-GTAAACCCATTACCACCACACTATGT | FAM-ATTGCTTACACTTCGGAC | |
| wna3682247 | F-ACATGTTCACTTCACACATTTTCTCA | VIC-ACTCTTGAACAAACCA |
| R-GCAATTGCAACAAGTCATCCA | FAM-ACTCTTGAACAAGCCAA | |
| wna3732539 | F-CTAAAAGCTCCAAATGCAATAGCA | VIC-CTGTTCCTGATAACAA |
| R-TGGTGGATCAAATGCAGTTAACTT | FAM-CTGTTCCTGATAATAA | |
| wna3774186 | F-CTTTGGTTTTCCTTTGGTATAATCTCTT | VIC-ATGGCACCTTTACATCT |
| R-TGACGTTGAAGGATGGAATATTTTTA | FAM-CAGATGGCATCTTT | |
| wna4461234 | F-TTTTGATGGAGAGATTTTGCTTTCT | VIC-TACGTTCTACAAATGGTACGT |
| R-GCGAAATCGAGCAAGGATTC | FAM-TACGTTCTACAGATGGT | |
| wna4718500 | F-GCATCACCATTTAGATCATAAACCA | VIC-AGCCATGTGTATAGAA |
| R-TGTCGTCGTACAGAAAGAAACGTT | FAM-AGCCATGTGTATGGAA | |
| wna237471 | F-TCGATGGTGCGAGCTTTTATATT | VIC-AATGAGCTCGGCACCAT |
| R-TGGTCATTGGTGGTATTTGCA | FAM-AATGAGCTCTGCACCA | |
| wna1141774 | F-CGGCTTTTTTTCATTACGCATTA | VIC-CATTTACCGTATTGTTTTG |
| R-AAAGAAAACAGAACATGCATTGATG | FAM-ATTTACCGCATTGTTTTG | |
| wna3368524 | F-CACGCTTATCGCCATCGAT | VIC-TCTACTGGCATTTCA |
| R-TGACGGAAGTGTAACGGAAGGT | FAM-TCTACTGGAATTTC |
The locus name correlates to the position on the whole genome sequence of the Ancestral Ames strain (AE017334).
F refers to the Forward primer whereas R refers to the Reverse primer.
Allele states are designated in red. VIC is the fluor conjugated to the 5′ end of the probe for one allele whereas FAM is the fluor conjugated to the 5′end of the alternate allele.
Table 4. SNPs used in this study to define clades in WNA.
| Branch Name 1 | Locus Name | Base Change (Ancestral→Derived) |
| A.Br.0092 | canSNP2589947* | A→G |
| A.Br.0182 | wna0237471 | T→G |
| A.Br.0182 | wna1141774 | C→T |
| A.Br.0182 | wna3368524 | A→C |
| A.Br.0193 | wna3631093 | A→G |
| A.Br.0193 | wna4718500 | G→A |
| A.Br.0204 | wna3774186 | T→C |
| A.Br.0215 | wna2994131 | C→G |
| A.Br.0226 | wna3682247 | A→G |
| A.Br.0226 | wna4461234 | G→A |
| A.Br.0237 | wna3732539 | T→C |
SNPs are arranged in this table from basal to derived nodes.
The A.Br.009 canonical SNP was previously reported5 and was used to identify most of the WNA clade samples used in this study.
These four SNPs are located on the Fig. 1 basal node separating TEA from the “yellow” clade.
These two SNPs are located on the Fig. 1 node separating the “yellow” from “red” clades.
This SNP is located on the Fig. 1 node separating the “red” from “green” clades.
This SNP is located on the Fig. 1 node separating the “green” from “blue” clades.
These two SNPs are located on the Fig. 1 node separating the “blue” from “black” clades.
This SNP is located on the Fig. 1 node separating the “black” and the terminal clades.
Spatial Analysis
Spatial data were then linked to genotypic analysis for each isolate and plotted with a Geographic Information System (ArcView 3.3). Some isolates were retrieved from archival collections and were not associated with geographic information, hence only 285 isolates were spatially mapped.
Phylogenetic analysis
Phylogenetic analysis using a cladistic approach was accomplished with PAUP 4.0 [26].
Acknowledgments
We are grateful to Paul Jackson and Martin Hugh-Jones for strain acquisition efforts.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by NIH NIGMS (1R01GM060795), DHS (NBCH2070001) and NIH NIAID (N01-AI15447). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 2.Devi SM, Ahmed I, Khan AA, Rahman SA, Alvi A, et al. Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island. BMC Genomics. 2006;7:191. doi: 10.1186/1471-2164-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes EC. The phylogeography of human viruses. Mol Ecol. 2004;13:745–756. doi: 10.1046/j.1365-294x.2003.02051.x. [DOI] [PubMed] [Google Scholar]
- 4.Linz B, Balloux F, Moodley Y, Manica A, Liu H, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mokrousov I, Ly HM, Otten T, Lan NN, Vyshnevskyi B, et al. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Res. 2005;15:1357–1364. doi: 10.1101/gr.3840605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavesi A. Detecting Traces of Prehistoric Human Migrations by Geographic Synthetic Maps of Polyomavirus JC. J Mol Evol. 2004;58:304–313. doi: 10.1007/s00239-003-2552-0. [DOI] [PubMed] [Google Scholar]
- 7.Roumagnac P, Weill F-X, Dolecek C, Baker S, Brisse S, et al. Evolutionary History of Salmonella Typhi. Science. 2006;314:1301–1304. doi: 10.1126/science.1134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eskey CR, Haas VH. Plague in the western part of the United States. Public Health Bulletin. 1940;254:1–83. [Google Scholar]
- 9.Green DM, Jamieson WM. Anthrax and bone-meal fertiliser. Lancet. 1958;2:153–154. doi: 10.1016/s0140-6736(58)92237-2. [DOI] [PubMed] [Google Scholar]
- 10.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10:100–103. doi: 10.1016/s0966-842x(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 11.Klemm DM, Klemm WR. A history of anthrax. J Am Vet Med Assoc. 1959;135:458–462. [PubMed] [Google Scholar]
- 12.Link VB. A history of plague in the United States of America. Public Health Monographs. 1955;26:1–120. [PubMed] [Google Scholar]
- 13.Van Ert MN, Easterday WR, Huynh LY, Okinaka RT, Hugh-Jones ME, et al. Global genetic population structure of Bacillus anthracis. PLoS ONE. 2007;2:e461. doi: 10.1371/journal.pone.0000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson T, Busch JD, Ravel J, Read TD, Rhoton SD, et al. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc Natl Acad Sci U S A. 2004;101:13536–13541. doi: 10.1073/pnas.0403844101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragon DC, Bader DE, Mitchell J, Woollen N. Natural dissemination of Bacillus anthracis spores in northern Canada. Appl Environ Microbiol. 2005;71:1610–1615. doi: 10.1128/AEM.71.3.1610-1615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MMWR. Inhalation anthrax associated with dried animal hides–Pennsylvania and New York City, 2006. MMWR Morb Mortal Wkly Rep. 2006;55:280–282. [PubMed] [Google Scholar]
- 17.Brachman PS. Anthrax. Ann N Y Acad Sci. 1970;174:577–582. doi: 10.1111/j.1749-6632.1970.tb45583.x. [DOI] [PubMed] [Google Scholar]
- 18.Hanson RP. The earliest account of anthrax in man and animals in North America. J Am Vet Med Assoc. 1959;135:463–465. [PubMed] [Google Scholar]
- 19.Plotkin SA, Brachman PS, Utell M, Bumford FH, Atchison MM. An epidemic of inhalation anthrax, the first in the twentieth century: I. Clinical features. 1960. Am J Med. 2002;112:4–12; discussion 12–13. doi: 10.1016/S0002-9343(01)01058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackburn JK, McNyset KM, Curtis A, Hugh-Jones ME. Modeling the geographic distribution of Bacillus anthracis, the causative agent of anthrax disease, for the contiguous united states using predictive ecologic niche modeling. Am J Trop Med Hyg. 2007;77:1103–1110. [PubMed] [Google Scholar]
- 21.Guthrie RD. Frozen Fauna of the Mammoth Steppe: The Story of Blue Babe. Chicago: The University of Chicago Press; 1990. [Google Scholar]
- 22.Kenefic L, Pearson T, Chung W, Max T, Van Ert M, et al. Recent Texas Isolates are Near-Relatives to the Bacillus anthracis Ames strain. Emerg Infect Dis. 2008 doi: 10.3201/eid1409.080076. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Ert MN, Easterday WR, Simonson TS, U'Ren JM, Pearson T, et al. Strain-specific single-nucleotide polymorphism assays for the Bacillus anthracis Ames strain. J Clin Microbiol. 2007;45:47–53. doi: 10.1128/JCM.01233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dragon DC, Elkin BT. An Overview of Early Anthrax Outbreaks in Northern Canada: Field Reports of the Health of Animals Branch, Agriculture Canada, 1962–71. Artic. 2001;54:32–40. [Google Scholar]
- 25.Shapiro B, Drummond AJ, Rambaut A, Wilson MC, Matheus PE, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. [DOI] [PubMed] [Google Scholar]
- 26.Swofford DL. Phylogenetic Analysis Using Parsimony (PAUP 40) Sunderland, , Massachusetts: Sinauer Associates; 2003. [Google Scholar]