Abstract
Although the enzymes enabling Hypocrea jecorina (anamorph Trichoderma reesei) to degrade the insoluble substrate cellulose have been investigated in some detail, little is still known about the mechanism by which cellulose signals its presence to the fungus. In order to investigate the possible role of a G-protein/cyclic AMP signaling pathway, the gene encoding GNA3, which belongs to the adenylate cyclase-activating class III of G-alpha subunits, was cloned. gna3 is clustered in tandem with the mitogen-activated protein kinase gene tmk3 and the glycogen phosphorylase gene gph1. The gna3 transcript is upregulated in the presence of light and is almost absent in the dark. A strain bearing a constitutively activated version of GNA3 (gna3QL) exhibits strongly increased cellulase transcription in the presence of the inducer cellulose and in the presence of light, whereas a gna3 antisense strain showed delayed cellulase transcription under this condition. However, the gna3QL mutant strain was unable to form cellulases in the absence of cellulose. The necessity of light for stimulation of cellulase transcription by GNA3 could not be overcome in a mutant which expressed gna3 under control of the constitutive gpd1 promoter also in darkness. We conclude that the previously reported stimulation of cellulase gene transcription by light, but not the direct transmission of the cellulose signal, involves the function and activation of GNA3. The upregulation of gna3 by light is influenced by the light modulator ENVOY, but GNA3 itself has no effect on transcription of the light regulator genes blr1, blr2, and env1. Our data for the first time imply an involvement of a G-alpha subunit in a light-dependent signaling event in fungi.
Hypocrea jecorina (anamorph Trichoderma reesei) is a saprophytic fungus that grows on wood trunks. To this end it forms a very potent cellulolytic enzyme system, which has also found widespread industrial application (11, 21, 22). The regulation of expression of these cellulases has been subject to extensive research (2, 60), and at least three transcriptional activators (XYR1 [70], ACE2 [3], and the HAP2/3/5 complex [80]) as well as the two repressors CRE1 (33) and ACE1 (56) have been found to be involved.
In contrast, the mechanism by which cellulose signals its presence to the cell and induces cellulase gene expression has remained enigmatic. Cellulase formation is absolutely dependent on the presence of an inducer (69), which suggests the involvement of a signaling cascade. In addition, this induction is modulated by several environmental conditions, of which the stimulation by light has recently been investigated in some molecular detail (59). The biochemical nature of this cellulose signaling cascade is not yet known, however. Sestak and Farkas (66) demonstrated that the rate of cellulase induction in H. jecorina by the soluble β-linked disaccharide sophorose can be doubled by increasing the intracellular concentration of cyclic AMP (cAMP). However, at concentrations exceeding an optimum, cellulase synthesis is repressed by cAMP (62). In the chestnut blight fungus Cryphonectria parasitica, Wang and Nuss (75) showed that cellobiohydrolase I gene expression requires the function of the G-alpha protein CPG-1. Together these two findings would give rise to the hypothesis that cellulase induction could involve signaling by a G-protein/cAMP pathway.
Fungal heterotrimeric G proteins, consisting of G-alpha, G-beta, and G-gamma subunits, have been shown to play a major role in signaling to various processes such as regulation of growth, reproduction, morphogenesis, virulence, secondary metabolite production, and pathogenic development (6, 16, 17, 50, 53, 81). After binding of a specific ligand to the cognate receptor, GDP bound to the G-alpha subunit is replaced by GTP, leading to activation of the G protein, which then can interact with its effectors. Thereafter the intrinsic GTPase activity of the G-alpha subunit catalyzes hydrolysis of GTP to GDP and Pi, thus inactivating the G-alpha protein and preventing continuous transmission of the signal (6, 79). Based on phylogenetic analysis and functional characteristics, the G-alpha proteins can be classified in three major subgroups: subgroup I proteins, which inhibit adenylate cyclase, a function also reported for the mammalian Gαi proteins; subgroup II proteins, which have no homology with mammalian G proteins; and subgroup III proteins, which in most fungi where tested activate adenylate cyclase and are thus functionally related to mammalian Gαs proteins (6, 77). However, exceptions to this classification have also been reported (35). The genome of H. jecorina encodes a set of G-alpha subunits comparable to that in Neurospora crassa (7, 39), i.e., three G-alpha subunits which can be assigned to the groups described above (58).
The objective of this study was therefore to clone a subgroup III G-alpha subunit-encoding gene from H. jecorina and to determine its role in the induction and/or regulation of cellulase gene expression. As a member of subgroup III, such a G protein should cause elevated intracellular cAMP levels upon activation due to its positive effect on adenylate cyclase. We will show that, in contrast to our initial hypothesis, the cellulose signal is not transmitted via this G-alpha subunit (GNA3). However, we will provide evidence that GNA3 has a significant impact on the regulation of cellulase gene expression by positively modulating its stimulation by light.
MATERIALS AND METHODS
Microbial strains and culture conditions.
H. jecorina (T. reesei) wild-type strains QM9414 (ATCC 26921) and TU-6 (ATCC MYA-256) (Δpyr4; uridine auxotroph) (25); the env1PAS− mutant strain, which lacks the PAS domain of ENVOY but still transcribes a truncated transcript (59); gna3AS (gpd1p::gna3::gpd1t; an antisense strain) (this study); and gna3S (gpd1p::gna3::gpd1t; a strain expressing gna3 under control of the constitutive gpd1 promoter) (this study) were used throughout this study and maintained on malt extract agar. The uridine auxotrophic mutant strain H. jecorina TU-6 (ATCC MYA-256; Δpyr4) (25) was maintained on malt extract agar supplemented with 10 mM uridine (Sigma-Aldrich Co.). The recombinant gna3QL1 and -2 mutant strains (Δpyr4; gna3Q206L+::pyr4+) (this study) were maintained on selective minimal medium [1 g/liter MgSO4·7H2O, 10 g/liter 1% KH2PO4, 6 g/liter (NH4)2SO4, 3 g/liter trisodium citrate·2H2O, 10 g/liter glucose, 20 ml/liter 50× trace elements solution (0.25 g/liter FeSO4·7H2O, 0.07 g/liter ZnSO4·2H2O, 0.1 g/liter CoCl2·6H2O, 0.085 g/liter MnSO4·H2O), 2% (wt/vol) agar] (all chemicals were from Merck, Darmstadt, Germany). Strains were grown in 1-liter Erlenmeyer flasks at 28°C on a rotary shaker (200 rpm) in 200 ml of minimal medium as described by Mandels and Andreotti (47), which was supplemented with 0.1% (wt/vol) peptone to induce germination and 1% (wt/vol) of microcrystalline cellulose (no. 14204; Serva, Heidelberg, Germany) or glycerol (Merck, Darmstadt, Germany) as a carbon source. For cultivation of the uridine auxotrophic strain TU-6, the medium was supplemented with 10 mM uridine. When TU-6 was used as a control for other strains, they were also supplemented with 10 mM uridine to ensure equal conditions. Approximately 108 conidia/liter (final concentration) were used as the inoculum.
Strains were grown either in the presence of constant illumination (25 μmol photons m−2 s−1; 1,800 lx) or in constant darkness. In the latter case, cultures were harvested under a red safety light. This has previously been shown to have no effect on the fungus (59). Growth in race tubes (49, 55) was performed at 28°C in constant light or constant darkness. Escherichia coli JM109 (78) was used for the vector propagation and DNA manipulation.
DNA and RNA manipulations.
Fungal mycelia were harvested by filtration, frozen, and ground in liquid nitrogen. For cultivation in constant darkness, cultures were harvested under a red safety light. Genomic DNA was isolated as described previously (61). For Northern blotting, total RNA was isolated by the guanidinium-phenol procedure (14, 61). Standard methods (57) were used for electrophoresis, blotting, and hybridization of nucleic acids. The probes used for hybridization were as follows: for gna3, the 3.2-kb EcoRI/XbaI fragment (all restriction enzymes were from Fermentas, Vilnius, Lithuania unless stated otherwise) introduced into pBgna3Q206L (see below); a 1.2-kb cbh1 fragment generated by PCR amplification with primers cbh1SF and cbh1SR; and a 1.0-kb cbh2 fragment generated by PCR amplification with primers cbh2SF and cbh2SR. Primers for amplification of PCR fragments to be used for analysis of transcription of blr1, blr2, and env1 were trblr1F and trblr1R, trblr2F and trblr2R, and env1neu1F and env1neu1R, respectively. For 18S rRNA hybridization, primers 18SRF and 18SRR were used for amplification of the probe. Sequences of the oligonucleotides used are given in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| cbh1SF | 5′-TCGGCCTGCACTCTCCAATCC-3′ |
| cbh1SR | 5′-TGGAGTCCAGCCACAGCATG-3′ |
| cbh2SF | 5′-ATTCTCACCACGCTGGTCAC-3′ |
| cbh2SR | 5′-CGGCGTAGTTGATGCACTC-3′ |
| trblr1F | 5′-TGTGCCTTTGTCGTTTGTG-3′ |
| trblr1R | 5′-GACCGATATGACGTGGACC-3′ |
| trblr2F | 5′-GCATGAGGAAGAAGGACG-3′ |
| trblr2R | 5′-GGAACTGTACCGCAGTCAG-3′ |
| env1neu1F | 5′-ATGCCGGCGTTGACATTAACCC-3′ |
| env1neu1R | 5′-ACGCATCTATTGGATATCTCCC-3′ |
| 18SRF | 5′-GGTGGAGTGATTTGTCTG-3′ |
| 18SRR | 5′-CTTACTAGGGATTCCTCG-3′ |
| gna3aa5R | 5′-CCACTTCTTCCGCTCACTCCGTAGCC-3′ |
| gna3aa5F | 5′-GTTTGGCCCGGATTGAAG-3′ |
| gna3aa3R | 5′-ATATAGCTCCACGGCCAATTC-3′ |
| gna3aa3F | 5′-GGCTACGGAGTGAGCGGAAGAAGTG-3′ |
| gna3aa5NF | 5′-ATGAATTCCACGGCCAATTCTTTG-3′ |
| gna3aa3NR | 5′-AATCTAGATTGAAGCGATCCCAGGATC-3′ |
| gna3cDNA1F | 5′-AACGTCGTCACTCTCGTC-3′ |
| gna3cDNA1R | 5′-ATGGGAAAGAACGTGATG-3′ |
| pgpdF | 5′-GAGAGCTACCTTACATCAA-3′ |
| GNA3S1F | 5′-ACGGAGTCAAAGAGCAATAAG-3′ |
| GNA3S1R | 5′-ATGAGATGGACGTGCTGC-3′ |
Mutated bases are in bold, and artificially introduced restriction sites are underlined.
Densitometric scanning of autoradiograms was done using the Bio-Rad GS-800 calibrated densitometer for three different exposures and two independent experiments. Measurements were normalized to 18S rRNA signals. As a control, the wild-type strain was included in every cultivation and subsequent Northern analysis to allow for representative evaluation of the data obtained.
Cloning of gna3 and construction of an H. jecorina gna3QL strain.
Using sequence information for the corresponding gene from Trichoderma atroviride (81), a tga3 orthologue was identified in the genome sequence database of H. jecorina (http://genome.jgi-psf.org/Trire2/Trire2.home.html), and its sequence was used to design primers for amplification and cloning.
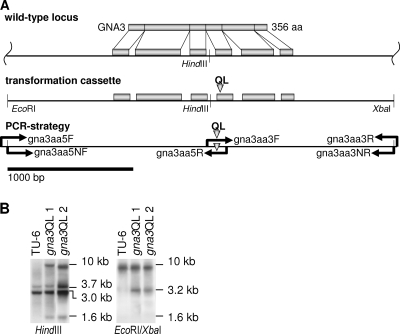
For construction of a modified copy for expression of a constitutively activated version of GNA3, an overlap extension mutagenesis approach was used. This procedure resulted in the single amino acid modification Q206L. A corresponding mutation has been reported to impair the intrinsic GTPase activity of this protein in mammals (5). Similar mutations were introduced into other fungi in order to assess the function of G-alpha subunits (48, 52, 65). Four oligonucleotides (Fig. 1A; Table 1) were designed based on the genomic sequence to generate the desired mutation in three steps. In a first PCR, primers gna3aa5F and gna3aa5R, which contains the mutation to be introduced into the wild-type template DNA, were used to amplify the 1,919-bp 5′ region. In a second PCR, primers gna3aa3F, which contains the mutation, and gna3aa3R were used to amplify the 1,386-bp 3′ region. Afterwards, these fragments were mixed and used as the template in a third PCR to obtain a 3,270-bp fragment bearing the intended mutation as well as approximately 1 kb of 3′ and 5′ flanking regions by using the two nested primers gna3aa5NF and gna3aa3NR, which in addition contains an artificial EcoRI and an XbaI restriction site to facilitate cloning. The PCR product was cloned into EcoRI-XbaI sites of pBluescript SK+ (Stratagene, La Jolla, CA), thereby generating pBgna3Q206L. The gna3 coding region was completely sequenced to ensure that only the desired mutations had been introduced. After linearization with SacI, which cuts within the backbone of pBluescript, 10 μg of this fragment was used to transform protoplasts of H. jecorina TU-6 in cotransformation with 2 μg of a 2.7-kb SalI pyr4 fragment excised from vector pFG1 (25), conferring uridine prototrophy. Transformants were selected on minimal medium as described above. Stable transformants were obtained by at least two rounds of single-spore isolation or three rounds of transfer to selection medium lacking uridine in the case of nonsporulating mutants. Integration into the H. jecorina genome and copy number were analyzed by Southern blotting using EcoRI and XbaI, which had been introduced to facilitate cloning, thereby showing the presence of the cassette by the presence of an additional 3.2-kb band (Fig. 1B). The presence of the 8,843-bp wild-type EcoRI/XbaI band in both wild-type and mutant strains confirmed that the cassette integrated ectopically. Southern blotting using HindIII, which has only one restriction site within the cassette and one within the vector backbone, revealed that one copy had integrated ectopically due to the additional 1.6-kb band resulting from integration of the cassette and only one additional band resulting from the integration locus, with this band being slightly different for gna3QL1 and gna3QL2 (Fig. 1B). Two strains bearing a single ectopically integrated copy of the constitutively activated gna3 gene were chosen for further analysis. In order to confirm the actual expression of the additional copy, cDNA fragments of gna3QL1 grown for 72 h on microcrystalline cellulose as described above were amplified and cloned into pGEM-T Easy (Promega, Madison, WI). Sequencing of the resulting plasmids confirmed an approximately equal representation of wild-type and mutated alleles.
FIG. 1.
Gene structure of gna3, transformation with the gna3Q206L allele, and PCR strategy. (A) The gene structure of gna3 is shown along with its exons and introns, the predicted GNA3 protein, and construction of a transformation cassette to introduce the single amino acid exchange at position 206 as indicated by a triangle. The EcoRI and XbaI restriction sites have been introduced using nested primers to facilitate cloning. (B) Southern blot analysis of TU-6 and two Q206L mutants. Genomic DNA was digested with HindIII or EcoRI/XbaI and probed with an ∼3.2-kb EcoRI/XbaI fragment excised from pBgna3Q206L. The additional HindIII band at 1.6 kb results from one restriction site within gna3 and a second site within the vector backbone, and the bands around 10 kb result from ectopic integration of the cassette and indicate the presence of only one copy in either of the two strains used for further experiments. The second Southern blot resulting from digestion with EcoRI/XbaI confirms ectopic integration of the mutant allele, since the 8,843-bp wild-type fragment is present in all three strains. Only the mutant strains contain the 3.2-kb fragment resulting from the artificially introduced restriction sites.
Construction of a gna3 antisense mutant strain and a mutant strain constitutively expressing gna3.
A fragment spanning the cDNA of gna3 was amplified using primers gna3cDNA1F and gna3cDNA1R (Table 1) and cloned into pGEM-T Easy, resulting in plasmids pgna3CS (sense) and pgna3CAS (antisense), depending on the orientation. The plasmid pLH1hph (27) comprises the hph open reading frame under the control of the constitutive gpd1 promoter and gpd1 terminator. The hph open reading frame was excised by digestion with NsiI, Klenow filling in to obtain a blunt end, and digestion with XbaI. The gna3 fragment was isolated from pgna3CS or pgna3CAS by digestion with EcoRI, Klenow filling in to create a blunt end, and digestion with SpeI, which creates overhangs corresponding to those of XbaI. The respective fragments were ligated with the digested vector to result in plasmids pg3Sgpd (constitutive expression) and pg3ASgpd (antisense). These plasmids were digested using BamHI and SmaI and ligated to a pki1p::hph::cbh2t fragment excised from pBSXH (59) by digestion with XhoI and XmnI, Klenow filling in to create a blunt end, and digestion with BamHI. The resulting plasmids, pg3SgpdH and pg3ASgpdH, were linearized by digestion with NarI and used for transformation as described above. The resulting strains gna3S (gpd1p::gna3::gpd1t; sense) and gna3AS (gpd1p::gna3::gpd1t; antisense) were tested for integration of the construct by PCR using primers GNA3cDNA1F or GNA3cDNA1R, respectively, in combination with pgpdF, which binds within the gpd1 promoter region to specifically amplify the newly introduced cassette. This analysis revealed one gna3S strain and one gna3AS strain. Reverse transcription-PCR (RT-PCR) using primers GNA3S1F and GNA3S1R was applied to verify constitutive (gna3 sense strain gna3S) or knocked-down (gna3 antisense strain gna3AS) transcription of gna3. The amplified fragment spans the third and fourth introns (correct fragment, 250 bp) and thus prevents misinterpretation of background bands due to residual DNA (fragment size, 410 bp) as positives. Amplification of the transcript of translation elongation factor 1 alpha (tef1) with primers TEF1F and TEF1R was used as a control.
cDNA preparation and RT-PCR.
Total RNA was treated with DNase I (Fermentas, Vilnius, Lithuania) to remove contaminating chromosomal DNA. cDNA was prepared from total RNA using the Clontech Creator SMART cDNA library construction kit. GoTaq polymerase (Promega, Madison, WI) was used for PCR amplification, with 25 cycles for tef1 and 35 cycles for gna3.
Measurement of intracellular cAMP levels.
Mycelia were grown on malt extract (3%, wt/vol) agar plates covered with cellophane for 72 h. Inoculation of the plates was done using a 3-mm-diameter agar slice with sporulated mycelium of the respective strain, which was placed in the middle of the malt extract plate. To extract cAMP, mycelia were ground to a fine powder under liquid nitrogen, weighed, and suspended in 10 ml 0.1 N HCl per g mycelium. Samples were centrifuged at 600 × g at 22°C for 10 min, and the cAMP concentration was measured with the Direct cAMP enzyme immunoassay kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. cAMP levels were related to the protein content of the sample and were expressed as means ± standard deviations from three independent experiments.
Nucleotide sequence accession number.
The gna3 sequence was deposited at NCBI (GenBank accession number DQ993172).
RESULTS
Characterization of H. jecorina gna3.
The gna3 gene consists of a predicted 1,423-bp open reading frame interrupted by five introns, which encodes a protein of 356 amino acids (Fig. 1A). The amino acid sequence of GNA3 (GenBank accession number ABJ55985) shares 97% identity with Tga3 (GenBank accession number AAM69919) of Hypocrea atroviridis (Trichoderma atroviride) and has high identity to other fungal G proteins (91% to Gibberella zeae [Fusarium graminearum] GP-3 alpha [EAA76506.1], 88% to Magnaporthe grisea MagA [AAB65425.1], 86% to N. crassa GNA-3 [AAG21364], and 77% to Aspergillus nidulans GanB [AAF12813]). Neighbor-joining analysis of the amino acid sequences of H. jecorina GNA3 and of these proteins, together with other fungal G-alpha proteins, provided evidence that GNA3 is an orthologue of these genes (58).
Analysis of the gna3 cDNA by RT-PCR confirmed the model provided in the Trichoderma reesei genome database v2.0 (http://genome.jgi-psf.org/Trire2/Trire2.home.html) with five introns. Following the guidelines defined for the H. jecorina genome annotation, which recommends the use of N. crassa gene names when the orthologue has already been characterized, we named this G-alpha protein GNA3.
Promoter analysis of 1 kb upstream of the predicted gna3 translational start codon revealed a CCAAT box at −120, eight CRE1-binding sites (5′ SYGGRG 3′) (among them one double site as described previously [15]), and the sequence 5′ CTGTGCTGTGCTGTGCTGTGCTGTGC 3′ at −774, comprising five overlapping EUM1-binding motifs (5′ CTGTGC 3′) and a single EUM1-binding site at −924. The EUM1-binding motif has recently been described to occur in genes regulated by light, especially in Neurospora crassa vvd as well as in H. jecorina env1 (59). Further, the gna3 promoter also comprises two motifs (at −808 and −851 relative to the ATG) which can be recognized by SRY family HMG box DNA-binding proteins (5′ CAAAG 3′) (4).
Characterization of the H. jecorina gna3 genomic locus.
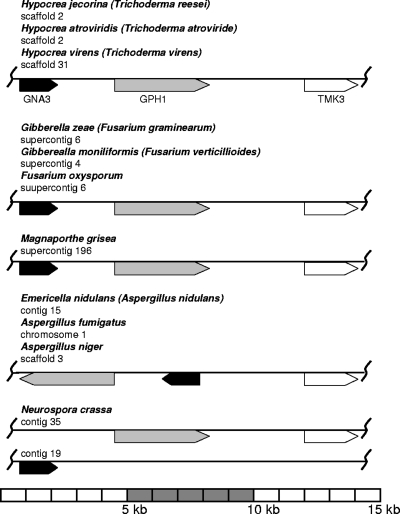
Analysis of the gna3 sequence in the Trichoderma reesei genome database v2.0 revealed that gna3 is present as a single copy, located on scaffold 2, and is flanked downstream by gph1, a gene encoding a glycosyl hydrolase family 35 protein, which is the only member of this family in H. jecorina and encodes an orthologue of the Saccharomyces cerevisiae glycogen phosphorylase Gph1p (NP_015486; 61% amino acid identity) required for the mobilization of glycogen (Fig. 2). Downstream of gph1 is located the tmk3 gene (encoding mitogen-activated protein [MAP] kinase), which encodes the orthologue of S. cerevisiae HOG1 involved in osmosensing (Fig. 2). Interestingly, the activity of Gph1p is regulated by cAMP-mediated phosphorylation (40), and its expression is regulated by the HOG-MAP kinase pathway (71). These findings suggest that these three genes may constitute a functional gene cluster. For simplicity we name the proposed cluster MGG (MAP kinase, glycogen phosphorylase, G protein). BLAST searches against numerous fungal genomes (http://www.broad.mit.edu/annotation/fungi/; http://genome.jgi-psf.org/) revealed that this clustering of gna3, gph1, and tmk3 is syntenic in Hypocrea atroviridis (Trichoderma atroviride), Hypocrea virens (Trichoderma virens), Gibberella zeae (Fusarium graminearum), Gibberella moniliformis (Fusarium verticillioides), Fusarium oxysporum, and Magnaporthe grisea, thus further substantiating this hypothesis. In Emericella nidulans (Aspergillus nidulans), Aspergillus niger, and Aspergillus fumigatus, the order of the genes in this cluster is different (Fig. 2), while in N. crassa only Hog1p and the Gph1p orthologue are located nearby, whereas the G-alpha protein GNA-3 is located on a different contig. In Saccharomyces cerevisiae, Rhizopus oryzae, Phanerochaete chrysosporium, Laccaria bicolor, and Cryptococcus neoformans, orthologous genes are present but unlinked. In Coprinopsis cinerea, a MAP kinase is located next to the Gph1p orthologue, but it shares highest similarity with Fus7p. Ustilago maydis seems to lack an orthologue of Gph1p but has a hypothetical protein with low similarity to a glycosyltransferase family 35 protein in the vicinity of the TMK3 orthologue. In Phycomyces blakesleeanus also, no Gph1p orthologue was detected and the loci of the Hog1p and GNA3 orthologues are unlinked, but interestingly, a MAP kinase kinase orthologue is located next to the GNA3 orthologue (for genomic coordinates and characterized orthologues of the respective genes, see Table S1 in the supplemental material). Phylogenetic analysis of the orthologues of GNA3, GPH1, and TMK3 did not reveal striking peculiarities and showed evolutionary relationships as expected for these organisms (data not shown).
FIG. 2.
Composition of the MGG cluster in ascomycetes. Contig or scaffold numbers are represented as specified in the respective genome databases (http://www.broad.mit.edu/annotation/fungi/; http://genome.jgi-psf.org/). Genes encoding orthologues of GNA3 are given in black, those encoding orthologues of GPH1 are in gray, and those encoding orthologues of TMK3 are in white. The scheme is drawn to scale. For exact genomic coordinates and known orthologues of the respective genes, see Table S1 in the supplemental material.
gna3 transcription is stimulated by light.
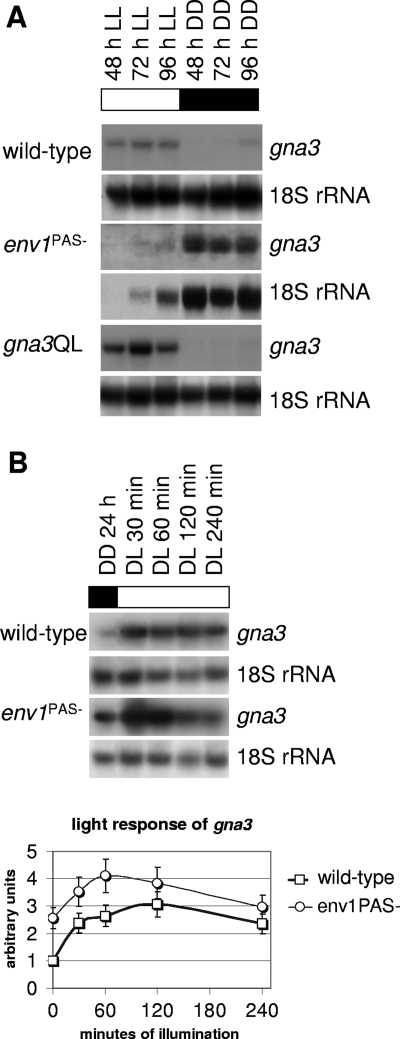
As a prerequisite to studying a possible function of gna3 in cellulase induction, we first investigated whether gna3 would be expressed under the conditions relevant for this study. In order to be able to relate these data to subsequent analyses of cellulase gene transcription, cellulose was used as a carbon source. Since light has been shown to enhance cellulase gene transcription, all experiments were done both in the presence of light and in darkness (59). Figure 3A shows that expression of gna3 was influenced by the presence of light; whereas the transcript was below the detection limit in darkness, it accumulated in the presence of light. In order to test whether the light modulator protein ENV1, which was previously shown by us to be responsible for the upregulation of expression of several genes by light in H. jecorina (62), was involved in this light regulation of gna3 transcription, we compared gna3 transcript accumulation in the parent strain QM 9414 and the env1PAS− mutant strain (59). Figure 3A shows that the env1PAS− strain accumulates the gna3 transcript also in the dark, whereas the transcript is hardly detectable in light. The clear difference from the wild-type strain in darkness indicates that gna3 transcription is indeed influenced by ENV1. However, due to the decreased light tolerance of the env1PAS− strain and the consequent slow growth observed in light (59), we cannot rule out that despite the clear difference in darkness in the env1PAS− strain, regulation of gna3 in light on cellulose might be comparable to that for the wild-type. In order to analyze a potential short-term light response of the gna3 transcript, the cellulase-noninducing carbon source glycerol and shorter periods of illumination instead of steady-state conditions as applied for cellulose were used (Fig. 3B). These conditions also allowed for comparison with the results of a study by Schuster et al., which reported different darkness- and light-related functions of ENVOY (62). Stimulation of gna3 gene expression on glycerol occurred within 30 min of illumination and reached a peak after 60 min (Fig. 3B). Thereafter transcript levels decreased until they were close to the initial level after 4 h. An essentially similar pattern was observed in the env1PAS− strain, but transcript levels are clearly increased. We therefore conclude that GNA3 belongs to the category of light-regulated genes which are repressed by env1 in darkness, as shown by Schuster et al. (62).
FIG. 3.
gna3 transcription is enhanced by light. (A) Mycelia of the wild-type strain TU-6 and of the env1PAS− and gna3QL mutant strains were cultivated in Mandels-Andreotti medium with 1% (wt/vol) microcrystalline cellulose as the sole carbon source in constant darkness (DD) or constant light (LL). Twenty micrograms of total RNA was loaded per lane, and an α-32P-radiolabeled PCR fragment spanning the cDNA of gna3 was used as a probe. 18S rRNA was used as control. (B) The short-term light response of gna3 was analyzed in the wild-type strain QM9414 and the env1PAS− mutant strain upon growth in Mandels-Andreotti medium with 1% (wt/vol) glycerol as a carbon source in constant darkness (DD) and after 30, 60, 120, and 240 min of illumination (DL). Twenty micrograms of total RNA was loaded per lane, and an α-32P-radiolabeled PCR fragment spanning the cDNA of gna3 was used as a probe. Results were quantified, and the amount of light-induced transcription of gna3 in the specified strain was normalized to the 18S rRNA control hybridization. Graphs show transcription levels above background.
Generation of an H. jecorina mutant carrying a constitutively activated gna3 allele.
In order to learn whether GNA3 is involved in transmitting the cellulose signal to cellulase gene transcription, we first produced a mutant strain which carried a gna3 mutant allele whose product is locked in the active GTP-bound state, resulting in a constitutively activated protein and permanent signal transmission. To this end, Q206 was replaced by L. Analogous mutations in mammalian Gαs and Gαi subunits have been shown to lower the GTPase activity, and thus these mutants cannot return to the inactive state. In fungi such mutations have been used to analyze the function of G-alpha subunits (48, 52, 65). Segers and Nuss (65) showed that ectopic integration of the mutated allele results in the same phenotype as integration in a deletion mutant of the respective G-alpha subunit. Thus, the activated G-alpha allele is considered dominant over the wild-type allele. Cotransformation of the uridine auxotrophic H. jecorina strain TU-6 with a respectively altered copy of gna3 and the pyr4 marker cassette (25) conferring uridine prototrophy resulted in two different transformants which contained a single additional copy of the constitutively activated allele (Fig. 1A), which yielded consistent results in all further experiments. Data from only one of them are given in this paper.
Phenotype of H. jecorina mutants bearing a constitutively activated gna3 allele.
The transcription pattern of gna3 was not altered in the H. jecorina mutant gna3QL, as its transcript still accumulated in the presence of light but remained below the detection limit in darkness (Fig. 3A). Densitometry of the gna3 transcript in the parent strain and the gna3QL mutant showed that it is about threefold more abundant in the latter. Since both alleles are present in single copies, this disproportional accumulation may indicate feedback regulation by active GNA3. Sequencing of several random cDNA clones of gna3QL further confirmed that the wild-type and mutant alleles are transcribed in approximately equal proportions in this strain.
The gna3QL mutant showed considerably decreased sporulation (by approximately 60%) in comparison to the wild-type strain TU-6 on full medium (malt extract) and no sporulation at all on minimal medium with glucose as the sole carbon source. Vegetative growth, on the other hand, was not affected by the Q206L mutation, as shown by similar hyphal extension rates of the parent and the mutant strain on plates or in race tubes, in darkness and in light.
A constitutively activated GNA3 protein should permanently activate adenylate cyclase and hence increase the intracellular concentration of its product, cAMP. In order to verify that the Q206L mutation indeed results in this expected adenylate cyclase activation, we measured the intracellular cAMP concentration in the mutant and in the parent strain. In support of the hypothesis, mycelia of the wild-type strain TU-6 grown on malt extract plates in constant light showed intracellular cAMP levels of 18.2 ± 4.1 pmol/mg protein, whereas mycelia of the gna3QL strain grown under the same conditions contained 103.9 ± 24.2 pmol cAMP/mg protein.
Constitutive activation of GNA3 does not lead to inducer-independent cellulase gene expression.
The availability of a mutant strain with a constitutively activated gna3 allele now allowed us to test the main hypothesis of this paper, i.e., that GNA3 may be involved in transmitting the signal from cellulose to cellulase gene expression. If this hypothesis was correct, this mutant should form cellulases also in the absence of their inducer. We therefore cultivated the gna3QL mutant on the two noninducing carbon sources glucose and glycerol in the presence of light and in darkness. However, no cellulase mRNA (cbh1 or cbh2) was detected, and the presence or absence of light had no influence on this result (data not shown). We note that carbon catabolite repression would not have interfered with this experiment, because it acts only on the basal expression level of cbh1 and not at all on cbh2 (82). Thus, we conclude that constitutive activation of GNA3 does not result in inducer-independent cellulase gene expression.
GNA3 modulates the stimulation of cellulase gene expression by light.
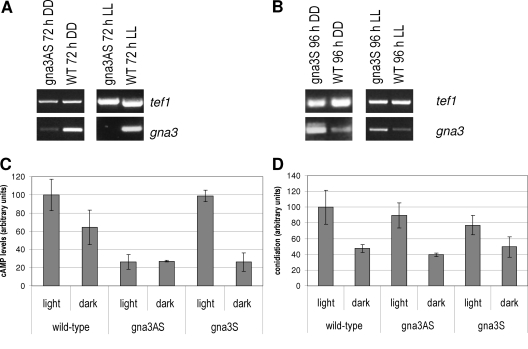
Although the above results caused us to reject the hypothesis of direct cellulose signaling by GNA3, we considered it still possible that GNA3 would modulate cellulase formation in response to another environmental cue in the presence of an inducer. We therefore investigated this by reverse genetics, using the gna3QL strain described above and a gna3 antisense strain, gna3AS (under the control of the H. jecorina gpd1 promoter, which allows strong constitutive expression), in order to knock down expression of GNA3. RT-PCR showed that the transcript of gna3 was indeed decreased or even below the detection limit in the antisense strain (Fig. 4A). As test conditions we chose 72 h of cultivation in constant light with cellulose as a carbon source, since these conditions promote gna3 transcription in the wild type. No gna3 transcript was detected in the gna3AS strain. In constant darkness the gna3 transcript was detected in the gna3AS strain, but with significantly decreased abundance. In the sense strain we detected elevated levels of the gna3 transcript compared to those in the wild type (Fig. 4B). However, no effect on hyphal elongation was detected in this strain. As expected, cAMP levels in the gna3AS strain are clearly reduced relative to those in the wild-type strain. Interestingly, overexpression of GNA3 does not increase cAMP accumulation above wild-type levels, thus emphasizing the importance of GNA3 activation for this process (Fig. 4C). In both the gna3S and gna3AS strains, conidiation was not significantly altered compared to that in the wild type (Fig. 4D). Thus, the effect of constitutive activation of GNA3 on conidiation is unlikely to be solely due the altered cAMP accumulation in this strain. If cAMP were responsible for the effect of mutations of gna3 on condidation, a contrary effect for constitutive activation and knockdown (in the antisense strain) would be expected, which is also not the case. While there is some correlation between conidiation and cAMP levels in dependence on light and darkness, the fact that conidiation in the gna3QL strain is decreased despite strongly elevated cAMP levels in this strain also contradicts a strict interdependence of cAMP accumulation and conidiation.
FIG. 4.
RT-PCR and cAMP levels of gna3 antisense and overexpressing strains. (A) RT-PCR of gna3 in the gna3AS antisense strain after 72 h of growth in Mandels-Andreotti medium with cellulose as a carbon source in constant light (LL) and constant darkness (DD). tef1 (encoding translation elongation factor 1 alpha) was used as a control. The antisense fragment of gna3 is under the control of the constitutive gpd1 promoter in the gna3AS strain. (B) RT-PCR of gna3 in the gna3S overexpressing strain under conditions where transcription of gna3 was below the detection limit in Northern blots, i.e., after 96 h of growth in the same medium as mentioned above in constant darkness (DD) and additionally after 96 h in constant light (LL). gna3 is under the control of the constitutive gpd1 promoter in the gna3S strain. (C) cAMP levels in the gna3AS and gna3S strains after 72 h of growth in constant light or constant darkness on Mandels-Andreotti medium with cellulose as a carbon source relative to the wild-type strain (21.72 pmol/mg protein). (D) Conidiation of the gna3AS and gna3S strains relative to the wild type in light and darkness.
Despite several attempts, we did not succeed in generating a gna3 knockout mutant. Although deletion of orthologues of gna3 in other fungi caused more or less severe phenotypes (13, 37, 81), this gene has not been shown to be essential.
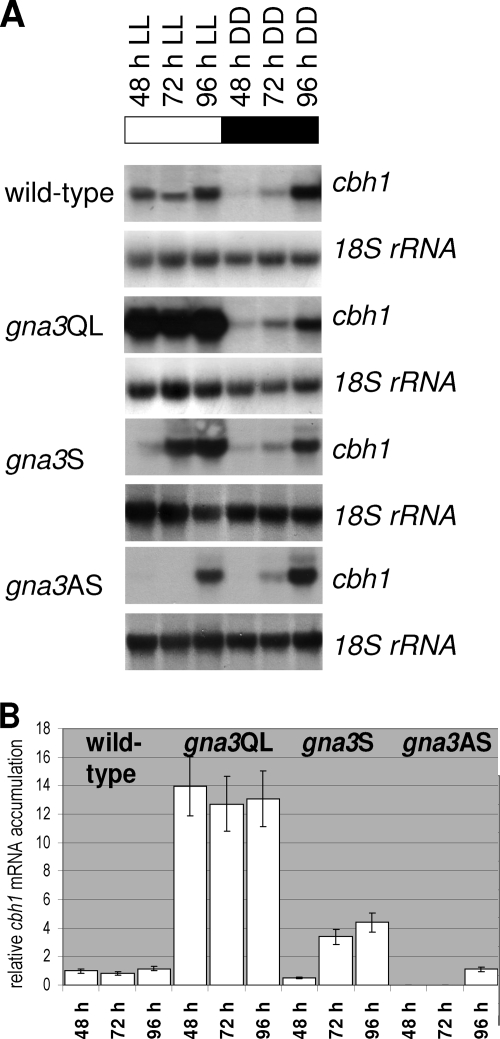
Cultivation of these two transgenic H. jecorina strains on cellulose in the dark and in the presence of light resulted in pronounced differences in the expression of the major cellulase gene cbh1 depending on the presence or absence of light (Fig. 5A). In darkness, the patterns of cbh1 gene expression were similar in the wild-type strain and in both mutants, demonstrating that gna3 has no effect on cellulase formation under this condition. In the presence of light, however, cbh1 gene expression showed a direct correlation with the putative in vivo activity of GNA3: while the gna3 antisense strain showed a strongly delayed accumulation of the cbh1 transcript, the Q206L mutant strain exhibited a significantly (10- to 12-fold) increased cbh1 transcript abundance (Fig. 5B). Hence, the function of GNA3 directly correlates with cellulase gene expression under conditions of illumination.
FIG. 5.
Northern analysis of cellulase gene transcription and gna3. (A) Mycelia of the wild-type (TU-6), gna3QL (constitutively activated GNA3, expression under the control of the original gna3 promoter), gna3AS (antisense strain, fragment under the control of the constitutive gpd1 promoter), and gna3S (overexpressing strain, gna3 under the control of the gpd1 promoter) strains were grown in Mandels-Andreotti medium supplemented with 1% microcrystalline cellulose as a carbon source in constant light (1,800 lx, 25 μmol photons m−2 s−1) (LL) or constant darkness (DD) for 48, 72, and 96 h. For Northern blotting, 20 μg of total RNA was loaded per lane and α-32P-radiolabeled PCR fragments of cbh1 and 18S rRNA (control) were used as probes. (B) Quantitative analysis of transcript abundance in constant light on cellulose in the wild-type, gna3QL, gna3AS, and gna3S strains. Values were normalized to wild type and to the corresponding 18S rRNA using the Bio-Rad GS-800 calibrated densitometer.
Stimulation of cbh1 gene expression by GNA3 requires the presence of light.
The finding that gna3 itself requires illumination for expression at high levels (see above) raised the question of whether the light-dependent GNA3 stimulation of cbh1 expression was due to the fact that gna3 is transcribed at very low levels in darkness and thus is unable to stimulate cellulase formation under these conditions or, alternatively, whether GNA3 may modulate the response of cellulase gene expression to the signal to be transmitted by its related receptor only in light and in the presence of its inducer. In order to test this, we constructed a mutant strain which expresses gna3 under control of the constitutive gpd1 promoter. This strain did not show altered growth compared to the wild type. RT-PCR showed that gna3 is indeed expressed in this strain in darkness (Fig. 4B). However, analysis of cAMP levels in this strain revealed a lower level than in the wild type (Fig. 4C), thus suggesting that increased abundance of GNA3 does not necessarily result in increased cAMP levels but seems to influence cAMP levels in a more complex way. Cellulase gene expression in darkness remained unaffected in this strain (Fig. 5A). Under illumination, elevated transcript levels compared to those in the parent strain were detected (up to fourfold), which nevertheless did not reach the level seen in the gna3QL mutant (Fig. 5B). These data therefore indicate that GNA3 per se does not influence cellulase gene transcription but modulates the stimulation of cbh1 gene transcription by light.
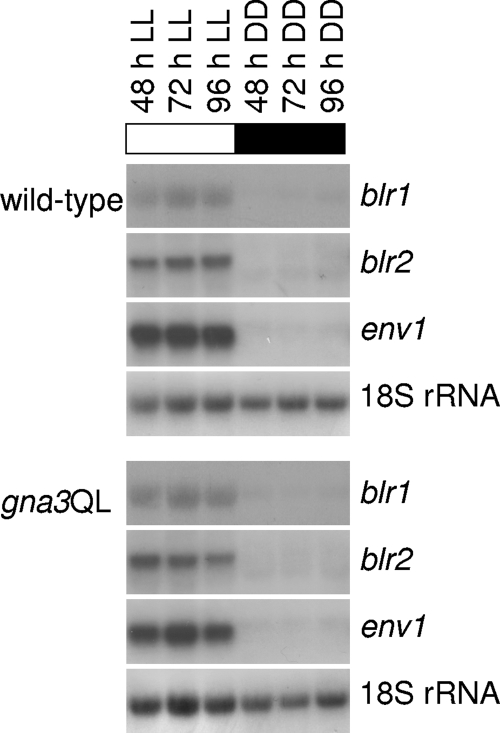
In order to test whether this effect of gna3 is due to a stimulation of transcription of the light receptor genes blr1, blr2, and env1 and thus controls the whole light signaling process, their transcripts were also monitored in the parent strain and the gna3QL mutant strain (Fig. 6). However, these data showed that these three genes display the same expression pattern in the gna3QL strain and the parent strain. Taking this together with the altered transcription pattern of gna3 in the env1PAS− mutant (see above), we therefore conclude that GNA3 does not influence transcription of the light receptor genes but rather acts downstream of the light perception machinery.
FIG. 6.
GNA3 does not influence transcription of the light receptors blr1, blr2, and env1. Mycelia of the wild-type (TU-6) and gna3QL strains were grown in Mandels-Andreotti medium supplemented with 1% (wt/vol) microcrystalline cellulose as a carbon source in constant light (1,800 lx, 25 μmol photons m−2 s−1) (LL) for 48, 72, and 96 h or in constant darkness (DD). For Northern blotting, 20 μg of total RNA was loaded per lane and α-32P-radiolabeled PCR fragments of blr1, blr2, env1, and 18S rRNA were used as probes.
DISCUSSION
The main objective of this study was to investigate whether a G-protein/cAMP signaling pathway was involved in the transduction of a putative signal derived from the presence of cellulose in the substrate of H. jecorina to the nucleus and induced cellulase gene expression, as suggested by the data reported by Sestak and Farkas (66) and Wang and Nuss (75). GNA3 was selected as the candidate G protein because it belongs to subgroup III of G-protein alpha subunits, members of which have previously been shown to be involved in the control of cAMP concentrations in fungi (6). The amino acid sequence of H. jecorina GNA3 and the physiology of the mutants bearing the constitutively activated gna3 allele of GNA3 (i.e., reduced conidiation and an elevated intracellular cAMP content) are all in agreement with previous studies of other fungi, including other Trichoderma spp. (6, 37, 50, 81), and support the conclusion that GNA3 is a functional homologue of the subgroup III G-alpha proteins.
The results obtained with strains bearing mutated alleles of gna3 allowed us to reject the hypothesis of direct cellulose signaling via GNA3, as a constitutively activated gna3 allele still did not initiate formation of cellulases in the absence of an inducer. This allows two alternative conclusions: either induction of cellulase formation does not at all involve a G-protein-coupled receptor for signal recruitment or another G protein is involved in this process. While this question must yet remain unanswered, our results nevertheless showed that GNA3 significantly stimulates cellulase gene expression under induced conditions and, moreover, that this effect is observed only in the presence of light and not in its absence. Since this obligatory dependence on the presence of light could not be overcome by overexpression of gna3 in the dark, we conclude that the prime stimulatory signal is either light or a light-specific signal and that GNA3 is a modulator of the subsequent response.
Effects of light on the physiology and metabolism of fungi have been observed widely. Orthologues of the N. crassa photoreceptor WHITE COLLAR-1 (WC-1) and the transcription factor WHITE COLLAR-2 (WC-2) (10, 18, 31, 32, 41, 54, 68, 72) are believed to act most upstream in this process. In addition, another blue light photoreceptor of N. crassa, VIVID, is regulated by the WC-1-WC-2 complex (28, 64). The H. atroviridis orthologues of WC-1 and WC-2, BLR1 and BLR2, are involved in both photoconidiation and mycelial growth in this organism (12). The downstream components transferring the light signal to the metabolic targets have not been identified, however. Our study makes it likely that GNA3 represents one of the downstream components in this cascade. First, the gna3 transcript is much more abundant in the presence of light and almost absent in darkness, implying that its function is required mainly under illuminated conditions. Further, expression of gna3 in H. jecorina is altered in an env1PAS− mutant strain, whereas its constitutive activation does not affect expression of env1 (and of neither blr1 nor blr2). This is consistent with the interpretation that gna3 acts downstream of ENV1 in the light signaling cascade and is thus involved in modulating the light response of cellulase gene expression. Moreover, our results on the light responsiveness of the gna3 transcript suggest that ENVOY may stabilize the gna3 transcript or, consistent with the known role of N. crassa VIVID in adaptation (63, 67), increase its rate of transcription in the light. The surprising lack of a gna3 transcript under steady-state conditions in the env1PAS− strain on cellulose in light could be caused by a continued decrease of transcript levels after 4 h of illumination, resulting in a transcript abundance below the detection limit. Interestingly, the delayed cellulase gene transcription in the gna3 antisense mutant gna3AS resembles the situation in the env1PAS− mutant strain (59), thus supporting this hypothesis. In this regulatory process the influence of ENVOY on gna3 transcription is likely to involve further downstream factors and maybe also a feedback mechanism. One possible target would be the H. jecorina equivalent of the white collar complex described for N. crassa. Hence, ENVOY could act via activation of this transcription factor complex, causing alteration of transcription of gna3. On the other hand, while transcription of blr1 and blr2 is not altered by constitutive activation of GNA3, it cannot be excluded that GNA3 interacts with a complex of these proteins, which in turn may act on transcription of the cellulase genes. Alternatively, although so far shown only for higher eukaryotes (30, 76), light-dependent activation of a G-alpha protein without the involvement of an equivalent of the white collar complex also appears to be possible.
An involvement of G proteins in responses to blue light has earlier been indirectly shown for N. crassa (34), Phycomyces blakesleeanus (73), and Coprinus congregatus (38). Despite these hints as to a connection of G-alpha subunits to the light response pathway, regulation of G-alpha genes by light in other fungal species has not been reported so far. However, considering the fact that both BLR1 and BLR2 represent transcription factors, we cannot rule out that, as mentioned above, GNA3 acts not on transcription but on activity of these photoreceptors and thereby causes light-dependent cellulase regulation. It will be interesting to investigate whether GNA3 is also involved in other light-dependent processes in H. jecorina.
The light-dependent modulating effect of GNA3 on cellulase gene expression is reminiscent of the modulating effect of artificial increases in the intracellular concentration of cAMP on cellulase formation in H. jecorina (66). This treatment, while enhancing the rate of cellulase induction twofold, also did not overcome inducer dependency. Unfortunately, that study was not done under controlled conditions of illumination, and a strict comparison with the present data is therefore not possible. However, we have recently shown (20) that an artificial elevation of intracellular cAMP levels in the dark in another Trichoderma sp., H. atroviridis, mimicked the stimulatory effect of light. Illumination of T. viride also leads to a transient rise in cAMP levels and subsequent protein kinase A-dependent phosphorylation of several intracellular proteins in T. viride (24). These data suggest that at least some metabolic effects caused by illumination are due to an increase in intracellular cAMP concentrations. We therefore consider it likely that the stimulation of cellulase gene expression by light in H. jecorina occurs at least partially by GNA3-dependent adenylate cyclase activation. Nevertheless, the fact that cAMP levels in the overexpressing stain hardly reach wild-type levels while the upregulation of gna3 causes increased cellulase levels in light indicates that additional downstream effectors must be targeted by GNA3.
Orthologues of GNA3 have also been reported to play a role in sporulation in fungi (13, 37, 43, 81), although no consistent relationship to the impact of this G-alpha subunit on cAMP levels could be established. Phenotypes of mutant strains could not be mimicked or rescued consistently. For H. jecorina, sporulation in the mutants studied also cannot be correlated directly to the different cAMP levels. Consequently, while sporulation in H. jecorina is influenced by GNA3, this influence is not (solely) due to the effect of GNA3 on cAMP levels.
Interestingly, expression of the constitutively activated gna3 allele led to disproportionally enhanced gna3 transcript levels, which suggests the operation of feedback regulation. Similarly, the enhancement of cbh1 gene transcription in the gna3QL mutant compared to the gna3S mutant (with gna3 under the control of the constitutive gpd1 promoter) over that in the parent strain shows a disproportional effect of the constitutively activated allele. This behavior could be explained by assuming that the activity of wild-type GNA3 is subject to negative regulation by a factor which is inactive on the constitutively activated GNA3. A possible candidate for this task would be the regulator of G-protein signaling (RGS) proteins, which enhance the intrinsic GTPase activity of G-alpha subunits (29). Because of the abolished GTPase activity in the constitutively activated GNA3 as expressed in the gna3QL mutant, this function would no longer be effective and thus could be responsible for the observed differences. In fact, the respective protein of Magnaporthe grisea, Rgs1, was shown to negatively regulate all three G-alpha subunits, including MagA, the orthologue of GNA3 (42). In this fungus, constitutive activation of MagA, caused by the introduction of an allele similar to that used in this work [magA(Q208L)], resulted in a phenotype similar to that caused by deletion of Rgs1. Regulation of activity of G-alpha subunits by RGS proteins has also been reported for Aspergillus nidulans (26) and Aspergillus fumigatus (46). Since an orthologue of Rgs1 is present in the genome of H. jecorina (RGS1; http://genome.jgi-psf.org/Trire2/Trire2.home.html), we consider the operation of this regulation in this fungus to be very likely and to be a plausible interpretation of our data. Also, such a mechanism would at least in part explain why the cAMP levels in the overexpressing strain do not exceed those in the wild type.
Investigation of gna3's neighboring genes in the genome revealed that it is located in tandem with gph1, an orthologue of S. cerevisiae GPH1 encoding a glycogen phosphorylase whose activity is regulated by cAMP-mediated phosphorylation, and tmk3, encoding the H. jecorina orthologue of the S. cerevisiae HOG1 MAP kinase. The 5′ nontranslated sequence of tmk3 contains a single copy of the EUM1 motif, is positively regulated by light as is gna3, and is influenced by ENVOY (62). On the other hand, within the promoter of gph1 no EUM1 sequence was detected, and this gene is downregulated by light. However, for this gene also a regulatory impact of ENVOY was found (62). Although these data indicate a connection of both genes to light-dependent signaling events, further studies will be necessary to understand the regulatory interdependence and the downstream targets of the genes within this cluster. The three genes within the proposed MGG cluster are syntenic in several sordariomycetes (Fusarium spp., Magnaporthe grisea, and Hypocrea spp., but not N. crassa), which are the phylogenetically the closest fungi to H. jecorina, but are still clustered in Aspergillus spp. (eurotiomycetes), albeit in a different order. The cluster could not be detected in S. cerevisiae, Phycomyces blakesleeanus, Rhizopus oryzae, and several basidiomycetes. We conclude that the MGG cluster is specific to ascomycetes, although hints as to a nonincidental proximity of G-alpha subunits, Gph1p orthologues, and MAP kinases have also been detected in basidiomycetes and zygomycetes.
The clustering of these three genes within the proposed MGG cluster may be functionally relevant, because yeast GPH1 is regulated by the HOG-MAP kinase pathway (71), and consequently this clustering may be indicative of a joint function in glycogen mobilization. Clustering of functionally related genes is a common feature in many eukaryotic regulatory pathways, including the nitrate assimilation cluster in Aspergillus (1, 36) and the gene clusters regulating secondary metabolite biosynthesis (8, 9, 23). The genes of such clusters are often coregulated, not least because of their common susceptibility to changes in chromatin rearrangement (74). In yeast, G-protein-mediated signal transduction and the HOG1 MAP kinase pathway are linked (51), and it is also known that G-protein-coupled receptors can affect both G-protein-mediated pathways and MAP kinase cascades (44, 45). Hints about a physiological role of MGG come from the finding that the glycogen content of Trichoderma is degraded upon illumination (19). Glycogen degradation requires the action of the GPH1 glycogen phosphorylase l and causes formation of glucose-6-phosphate, which is involved in regulating cellulase synthesis in H. jecorina (66).
To summarize, our study, while not supporting the hypothesis that direct cellulose signaling in H. jecorina involves GNA3, has revealed a new role of GNA3 in stimulation of a light-responsive process in this fungus. This striking dependence of the effect of GNA3 of light renders the hypothesis of light gating of cellulose signaling/regulation involving the regulatory function of ENVOY and/or GNA3 worth considering. It will be interesting to learn whether the other G proteins of this fungus are also involved in light-dependent phenomena and whether any of the other G proteins is involved in cellulase gene induction. Given the numerous reports on effects of light on other fungi, it is likely that the functions of GNA3 orthologues in these fungi also involve an influence of light.
Supplementary Material
Acknowledgments
This work was supported by grants from the Austrian Science Foundation (FWF-P17325 to C.P.K. and FWF-20004 to M.S.). M.S. is the recipient of an APART fellowship of the Austria Academy of Sciences at the Institute of Chemical Engineering, TU Vienna. The T. reesei genome sequencing project was funded by the U.S. Department of Energy.
Footnotes
Published ahead of print on 9 January 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Amaar, Y. G., and M. M. Moore. 1998. Mapping of the nitrate-assimilation gene cluster (crnA-niiA-niaD) and characterization of the nitrite reductase gene (niiA) in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Genet. 33206-215. [DOI] [PubMed] [Google Scholar]
- 2.Aro, N., T. Pakula, and M. Penttila. 2005. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol. Rev. 29719-739. [DOI] [PubMed] [Google Scholar]
- 3.Aro, N., A. Saloheimo, M. Ilmen, and M. Penttila. 2001. ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J. Biol. Chem. 27624309-24314. [DOI] [PubMed] [Google Scholar]
- 4.Baxevanis, A. D., and D. Landsman. 1995. The HMG-1 box protein family: classification and functional relationships. Nucleic Acids Res. 231604-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman, D. M., T. M. Wilkie, and A. G. Gilman. 1996. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell 86445-452. [DOI] [PubMed] [Google Scholar]
- 6.Bolker, M. 1998. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 25143-156. [DOI] [PubMed] [Google Scholar]
- 7.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. Plamann, M. Goodrich-Tanrikulu, U. Schulte, G. Mannhaupt, F. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Sachs, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, J. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 681-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brakhage, A. A. 1997. Molecular regulation of penicillin biosynthesis in Aspergillus (Emericella) nidulans. FEMS Microbiol. Lett. 1481-10. [DOI] [PubMed] [Google Scholar]
- 9.Brown, D. W., R. B. Dyer, S. P. McCormick, D. F. Kendra, and R. D. Plattner. 2004. Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet. Biol. 41454-462. [DOI] [PubMed] [Google Scholar]
- 10.Brunner, M., and T. Schafmeier. 2006. Transcriptional and post-transcriptional regulation of the circadian clock of cyanobacteria and Neurospora. Genes Dev. 201061-1074. [DOI] [PubMed] [Google Scholar]
- 11.Buchert, J., T. Oksanen, J. Pere, M. Siika-Aho, A. Suurnäkki, and L. Viikari. 1998. Applications of Trichoderma reesei enzymes in the pulp and paper industry, p. 343-363. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium, vol. 2. Taylor & Francis, London, United Kingdom. [Google Scholar]
- 12.Casas-Flores, S., M. Rios-Momberg, M. Bibbins, P. Ponce-Noyola, and A. Herrera-Estrella. 2004. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology 1503561-3569. [DOI] [PubMed] [Google Scholar]
- 13.Chang, M. H., K. S. Chae, D. M. Han, and K. Y. Jahng. 2004. The GanB Galpha-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans. Genetics 1671305-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 15.Cubero, B., and C. Scazzocchio. 1994. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 13407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Souza, C. A., J. A. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 213179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Souza, C. A., and J. Heitman. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25349-364. [DOI] [PubMed] [Google Scholar]
- 18.Dunlap, J. C., and J. J. Loros. 2004. The Neurospora circadian system. J. Biol. Rhythms 19414-424. [DOI] [PubMed] [Google Scholar]
- 19.Farkas, V., M. Gresik, N. Kolarova, Z. Sulova, and S. Sestak. 1990. Biochemical and physiological changes during photo-induced conidiation and derepression of cellulase synthesis in Trichoderma, p. 139-155. In C. P. Kubicek, D. E. Eveleigh, W. Esterbauer, W. Steiner, and E. M. Kubicek-Pranz (ed.), Trichoderma reesei cellulase: biochemistry, genetics, physiology, and application. Graham House, Cambridge, United Kingdom.
- 20.Friedl, M. A., C. P. Kubicek, and I. S. Druzhinina. 2008. Carbon source dependence and photostimulation of conidiation in Hypocrea atroviridis. Appl. Environ. Microbiol. 74245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galante, Y., A. De Conti, and R. Monteverdi. 1998. Application of Trichoderma enzymes in the food and feed industries, p. 327-342. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium. Taylor & Francis, London, United Kingdom.
- 22.Galante, Y., A. De Conti, and R. Monteverdi. 1998. Application of Trichoderma enzymes in the textile industry, p. 311-325. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium. Taylor & Francis, London, United Kingdom.
- 23.Gardiner, D. M., and B. J. Howlett. 2005. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol. Lett. 248241-248. [DOI] [PubMed] [Google Scholar]
- 24.Gresik, M., N. Kolarova, and V. Farkas. 1989. Light-stimulated phosphorylation of proteins in cell-free extracts from Trichoderma viride. FEBS Lett. 248185-187. [DOI] [PubMed] [Google Scholar]
- 25.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaff. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 1871-76. [DOI] [PubMed] [Google Scholar]
- 26.Han, K. H., J. A. Seo, and J. H. Yu. 2004. Regulators of G-protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Galpha) signalling. Mol. Microbiol. 53529-540. [DOI] [PubMed] [Google Scholar]
- 27.Hartl, L., C. P. Kubicek, and B. Seiboth. 2007. Induction of the gal pathway and cellulase genes involves no transcriptional inducer function of the galactokinase in Hypocrea jecorina. J. Biol. Chem. 28218654-18659. [DOI] [PubMed] [Google Scholar]
- 28.Heintzen, C., J. J. Loros, and J. C. Dunlap. 2001. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104453-464. [DOI] [PubMed] [Google Scholar]
- 29.Hollinger, S., and J. R. Hepler. 2002. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 54527-559. [DOI] [PubMed] [Google Scholar]
- 30.Hu, G., Z. Zhang, and T. G. Wensel. 2003. Activation of RGS9-1GTPase acceleration by its membrane anchor, R9AP. J. Biol. Chem. 27814550-14554. [DOI] [PubMed] [Google Scholar]
- 31.Idnurm, A., and J. Heitman. 2005. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 3e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Idnurm, A., and J. Heitman. 2005. Photosensing fungi: phytochrome in the spotlight. Curr. Biol. 15R829-R832. [DOI] [PubMed] [Google Scholar]
- 33.Ilmen, M., M. L. Onnela, S. Klemsdal, S. Keranen, and M. Penttila. 1996. Functional analysis of the cellobiohydrolase I promoter of the filamentous fungus Trichoderma reesei. Mol. Gen. Genet. 253303-314. [DOI] [PubMed] [Google Scholar]
- 34.Ivey, F. D., P. N. Hodge, G. E. Turner, and K. A. Borkovich. 1996. The G alpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 71283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivey, F. D., Q. Yang, and K. A. Borkovich. 1999. Positive regulation of adenylyl cyclase activity by a galphai homolog in Neurospora crassa. Fungal Genet. Biol. 2648-61. [DOI] [PubMed] [Google Scholar]
- 36.Johnstone, I. L., P. C. McCabe, P. Greaves, S. J. Gurr, G. E. Cole, M. A. Brow, S. E. Unkles, A. J. Clutterbuck, J. R. Kinghorn, and M. A. Innis. 1990. Isolation and characterisation of the crnA-niiA-niaD gene cluster for nitrate assimilation in Aspergillus nidulans. Gene 90181-192. [DOI] [PubMed] [Google Scholar]
- 37.Kays, A. M., P. S. Rowley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the Galpha protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 207693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak, K. R., and I. K. Ross. 1991. Signal transduction in Coprinus congregatus: evidence for the involvement of G proteins in blue light photomorphogenesis. Biochem. Biophys. Res. Commun. 1791225-1231. [DOI] [PubMed] [Google Scholar]
- 39.Li, L., S. J. Wright, S. Krystofova, G. Park, and K. A. Borkovich. 2007. Heterotrimeric G protein signaling in filamentous fungi. Annu. Rev. Microbiol. 61423-452. [DOI] [PubMed] [Google Scholar]
- 40.Lin, K., P. K. Hwang, and R. J. Fletterick. 1995. Mechanism of regulation in yeast glycogen phosphorylase. J. Biol. Chem. 27026833-26839. [DOI] [PubMed] [Google Scholar]
- 41.Linden, H., and G. Macino. 1997. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 1698-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, H., A. Suresh, F. S. Willard, D. P. Siderovski, S. Lu, and N. I. Naqvi. 2007. Rgs1 regulates multiple Galpha subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J. 26690-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, S., and R. A. Dean. 1997. G protein alpha subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 101075-1086. [DOI] [PubMed] [Google Scholar]
- 44.Luttrell, L. M. 2002. Activation and targeting of mitogen-activated protein kinases by G-protein-coupled receptors. Can. J. Physiol. Pharmacol. 80375-382. [DOI] [PubMed] [Google Scholar]
- 45.Luttrell, L. M. 2005. Composition and function of G protein-coupled receptor signalsomes controlling mitogen-activated protein kinase activity. J. Mol. Neurosci. 26253-264. [DOI] [PubMed] [Google Scholar]
- 46.Mah, J. H., and J. H. Yu. 2006. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot. Cell 51585-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mandels, M., and R. Andreotti. 1978. Problems and challenges in the cellulose to cellulase fermentation. Proc. Biochem. 136-13. [Google Scholar]
- 48.Masters, S. B., R. T. Miller, M. H. Chi, F. H. Chang, B. Beiderman, N. G. Lopez, and H. R. Bourne. 1989. Mutations in the GTP-binding site of GS alpha alter stimulation of adenylyl cyclase. J. Biol. Chem. 26415467-15474. [PubMed] [Google Scholar]
- 49.Merrow, M., T. Roenneberg, G. Macino, and L. Franchi. 2001. A fungus among us: the Neurospora crassa circadian system. Semin. Cell Dev. Biol. 12279-285. [DOI] [PubMed] [Google Scholar]
- 50.Mukherjee, P. K., J. Latha, R. Hadar, and B. A. Horwitz. 2004. Role of two G-protein alpha subunits, TgaA and TgaB, in the antagonism of plant pathogens by Trichoderma virens. Appl. Environ. Microbiol. 70542-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramezani-Rad, M. 2003. The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr. Genet. 43161-170. [DOI] [PubMed] [Google Scholar]
- 52.Regenfelder, E., T. Spellig, A. Hartmann, S. Lauenstein, M. Bolker, and R. Kahmann. 1997. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 161934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reithner, B., K. Brunner, R. Schuhmacher, I. Peissl, V. Seidl, R. Krska, and S. Zeilinger. 2005. The G protein alpha subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet. Biol. 42749-760. [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez-Romero, J., and L. M. Corrochano. 2006. Regulation by blue light and heat shock of gene transcription in the fungus Phycomyces: proteins required for photoinduction and mechanism for adaptation to light. Mol. Microbiol. 611049-1059. [DOI] [PubMed] [Google Scholar]
- 55.Ryan, F. J., G. W. Beadle, and E. L. Tatum. 1943. The tube method of measuring the growth rate of Neurospora. Am. J. Bot. 30784-799. [Google Scholar]
- 56.Saloheimo, A., N. Aro, M. Ilmen, and M. Penttila. 2000. Isolation of the ace1 gene encoding a Cys(2)-His(2) transcription factor involved in regulation of activity of the cellulase promoter cbh1 of Trichoderma reesei. J. Biol. Chem. 2755817-5825. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 58.Schmoll, M. 2008. The information highways of a biotechnological workhorse—signal transduction in Hypocrea jecorina. BMC Genomics 9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmoll, M., L. Franchi, and C. P. Kubicek. 2005. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (Anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot. Cell 41998-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmoll, M., and C. P. Kubicek. 2003. Regulation of Trichoderma cellulase formation: lessons in molecular biology from an industrial fungus. A review. Acta Microbiol. Immunol. Hung. 50125-145. [DOI] [PubMed] [Google Scholar]
- 61.Schmoll, M., S. Zeilinger, R. L. Mach, and C. P. Kubicek. 2004. Cloning of genes expressed early during cellulase induction in Hypocrea jecorina by a rapid subtraction hybridization approach. Fungal Genet. Biol. 41877-887. [DOI] [PubMed] [Google Scholar]
- 62.Schuster, A., C. P. Kubicek, M. A. Friedl, I. S. Druzhinina, and M. Schmoll. 2007. Impact of light on Hypocrea jecorina and the multiple cellular roles of ENVOY in this process. BMC Genomics 8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwerdtfeger, C., and H. Linden. 2001. Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol. Microbiol. 391080-1087. [DOI] [PubMed] [Google Scholar]
- 64.Schwerdtfeger, C., and H. Linden. 2003. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 224846-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Segers, G. C., and D. L. Nuss. 2003. Constitutively activated Galpha negatively regulates virulence, reproduction and hydrophobin gene expression in the chestnut blight fungus Cryphonectria parasitica. Fungal Genet. Biol. 38198-208. [DOI] [PubMed] [Google Scholar]
- 66.Sestak, S., and V. Farkas. 1993. Metabolic regulation of endoglucanase synthesis in Trichoderma reesei: participation of cyclic AMP and glucose-6-phosphate. Can. J. Microbiol. 39342-347. [DOI] [PubMed] [Google Scholar]
- 67.Shrode, L. B., Z. A. Lewis, L. D. White, D. Bell-Pedersen, and D. J. Ebbole. 2001. vvd is required for light adaptation of conidiation-specific genes of Neurospora crassa, but not circadian conidiation. Fungal Genet. Biol. 32169-181. [DOI] [PubMed] [Google Scholar]
- 68.Silva, F., S. Torres-Martinez, and V. Garre. 2006. Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Mol. Microbiol. 611023-1037. [DOI] [PubMed] [Google Scholar]
- 69.Sternberg, D., and G. R. Mandels. 1979. Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J. Bacteriol. 139761-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stricker, A. R., K. Grosstessner-Hain, E. Wurleitner, and R. L. Mach. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 52128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sunnarborg, S. W., S. P. Miller, I. Unnikrishnan, and D. C. LaPorte. 2001. Expression of the yeast glycogen phosphorylase gene is regulated by stress-response elements and by the HOG MAP kinase pathway. Yeast 181505-1514. [DOI] [PubMed] [Google Scholar]
- 72.Talora, C., L. Franchi, H. Linden, P. Ballario, and G. Macino. 1999. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 184961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsolakis, G., N. K. Moschonas, P. Galland, and K. Kotzabasis. 2004. Involvement of G proteins in the mycelial photoresponses of Phycomyces. Photochem Photobiol. 79360-370. [DOI] [PubMed] [Google Scholar]
- 74.van Driel, R., P. F. Fransz, and P. J. Verschure. 2003. The eukaryotic genome: a system regulated at different hierarchical levels. J. Cell Sci. 1164067-4075. [DOI] [PubMed] [Google Scholar]
- 75.Wang, P., and D. L. Nuss. 1995. Induction of a Cryphonectria parasitica cellobiohydrolase I gene is suppressed by hypovirus infection and regulated by a GTP-binding-protein-linked signaling pathway involved in fungal pathogenesis. Proc. Natl. Acad. Sci. USA 9211529-11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, T., and C. Montell. 2007. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 454821-847. [DOI] [PubMed] [Google Scholar]
- 77.Wilkie, T. M., and S. Yokoyama. 1994. Evolution of the G protein alpha subunit multigene family. Soc. Gen. Physiol. Ser. 49249-270. [PubMed] [Google Scholar]
- 78.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 79.Yu, J. H., J. H. Mah, and J. A. Seo. 2006. Growth and developmental control in the model and pathogenic aspergilli. Eukaryot. Cell 51577-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zeilinger, S., A. Ebner, T. Marosits, R. Mach, and C. P. Kubicek. 2001. The Hypocrea jecorina HAP 2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the cbh2 (cellobiohydrolase II-gene) activating element. Mol. Genet. Genomics 26656-63. [DOI] [PubMed] [Google Scholar]
- 81.Zeilinger, S., B. Reithner, V. Scala, I. Peissl, M. Lorito, and R. L. Mach. 2005. Signal transduction by Tga3, a novel G protein alpha subunit of Trichoderma atroviride. Appl. Environ. Microbiol. 711591-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeilinger, S., M. Schmoll, M. Pail, R. L. Mach, and C. P. Kubicek. 2003. Nucleosome transactions on the Hypocrea jecorina (Trichoderma reesei) cellulase promoter cbh2 associated with cellulase induction. Mol. Genet. Genomics 27046-55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.