Abstract
Iron-inducible transcription of a malic enzyme gene (also reputed to be ap65-1) in Trichomonas vaginalis was previously shown to involve a Myb1 repressor and a Myb2 activator, each of which may preferentially select two closely spaced promoter sites, MRE-1/MRE-2r, which comprises overlapping promoter elements, and MRE-2f. In the present study, an iron-inducible ∼32-kDa Myb3 nuclear protein was demonstrated to bind only the MRE-1 element. Changes in the iron supply, which produced antagonistic effects on the levels of Myb2 and Myb3 expression, also resulted in temporal and alternate entries of Myb2 and Myb3 into the ap65-1 promoter. Repression or activation of basal and iron-inducible ap65-1 transcription was detected in transfected cells when Myb3 was, respectively, substantially knocked down or overexpressed. In the latter case, increased Myb3 promoter entry was detected with concomitant decrease in Myb2 promoter entry under specific conditions, while Myb3 promoter entry was inhibited under all test conditions in cells overexpressing Myb2. In contrast, concomitant promoter entries by Myb2 and Myb3 diminished in cells overexpressing Myb1, except that Myb3 promoter entry was slightly affected under prolonged iron depletion. Together, these results suggest that Myb2 and Myb3 may coactivate basal and iron-inducible ap65-1 transcription against Myb1 through conditional and competitive promoter entries.
Trichomonas vaginalis is one of the most prevalent sexually transmitted human pathogens. It poses an imminent threat to public health because the protozoan infection is a risk factor in transmission of the human immunodeficiency virus (30). The parasite inhabits only the human urogenital tract where it exists as a trophozoite without an alternate life stage to escape challenges from immune surveillance and rapid changes in the host environment. The iron supply, which may periodically vary to a great extent in the human vagina, is the principal determinant in modulating expression of multiple virulence phenotypes, such as cytoadherence, phenotypic variation, and resistance to complement lysis, for T. vaginalis (1-3, 10). The roles of iron in regulating transcription, phosphorylation, and trafficking of some of the virulence factors have been well documented (1, 10, 32), providing a sound basis for studying the molecular mechanisms leading to iron-activated virulence expression.
In contrast to its simple life cycle, the parasite is reputed to have an exceedingly large genome, carrying nearly 60,000 monocistronic genes (7). Only a few of them contain introns (7, 33), suggesting that regulation of transcription initiation probably plays a primary role in the parasite's ability to adapt to and survive hostile host environments. In this regard, T. vaginalis may use a conserved initiator-like DNA sequence as the sole core promoter element to initiate transcription of most of its protein-coding genes (7, 17). This initiator element binds IBP39, a novel initiator binding protein, which also interacts with the C-terminal domain of RNA polymerase II (18, 28). In contrast to the assortments of multiple core promoter elements and their complicated interactions with numerous transcription factors in eukaryotic model systems (12, 21, 29), oversimplified DNA-protein and protein-protein interactions centered on the core initiator element imply that the parasite may have evolved unique transcription machinery, which may rely heavily on gene-specific transcription apparatus to control the transcription efficiencies of myriad protein-coding genes in a rapidly changing environment.
In T. vaginalis, several novel promoter distal elements have been identified in two protein-coding genes, α-scs and ap65-1 (16, 32), implying that gene-specific transcription in the parasite may also exhibit unusual features. For example, two discrete Myb protein-recognition sites, MRE-1/MRE-2r and MRE-2f, which are interspersed among several closely spaced DNA elements in the iron-responsive promoter region (−132 to −37 nucleotides from the transcription initiation site) of a malic enzyme gene (also reputed to be ap65-1), which encodes a 65-kDa hydrogenosomal malic enzyme that might have an ambiguous role in the cytoadherence of the parasite (4, 13), were demonstrated to play crucial roles in temporal and iron-inducible ap65-1 transcription (24-26, 32). MRE-1/MRE-2r and MRE-2f share similar but oppositely oriented DNA sequences, each of which is also the target site for several distinct Myb-like proteins (24-26). Two of these DNA-binding proteins, Myb1 and Myb2, which were identified by Southwestern library screening using a probe with concatenated MRE-2f sequences (25), were found to pose antagonistic actions on basal and iron-inducible transcription of the ap65-1 gene through dual recognition and differential promoter selection toward the MRE-1/MRE-2r and MRE-2f sites (24, 25).
The Myb family of transcription factors in vertebrates comprises three members, c-Myb, A-Myb, and B-Myb, with three conserved and repetitive DNA-binding domains reputed to be R1R2R3. They display similar DNA-biding specificities but regulate transcription of different subsets of genes in distinct cell types (9, 15, 22, 27). In contrast, the Myb family in T. vaginalis, like that in Arabidopsis thaliana (31), is composed of over 100 members, with the consensus DNA-binding domains divided mainly into the R1R2R3, R2R3, and single-repeat subfamilies (H. W. Liu and J. H. Tai, unpublished data). It is intriguing why and how this simple protozoan that does not normally undergo differentiation uses so many distinct Myb proteins for transcription.
In the present study, a myb3 gene was identified by Southwestern library screening using a probe with concatenated MRE-1/MRE-2r sequences. Myb3 was demonstrated to bind DNA in a context confined primarily to the MRE-1 moiety of the MRE-1/MRE-2r overlap. Myb3 was found to activate basal and iron-inducible ap65-1 transcription while also exhibiting temporal and differential promoter entry in a manner different from that of Myb2. A conditional competition for promoter entry between Myb2 and overexpressed Myb3, or vice versa, was observed, whereas concurrent entries of Myb2 and Myb3 diminished under most situations when Myb1 was overexpressed, suggesting that Myb2 and Myb3 may coordinate to activate ap65-1 transcription in competition with Myb1 for promoter entry.
MATERIALS AND METHODS
Cultures.
T. vaginalis T1 cells were maintained as previously described (32). Iron repletion or depletion was achieved with the respective addition of 250 μM of ferrous ammonium sulfate or 50 μM of 2,2′-dipyridyl, an iron-chelator, in normal growth medium. A Myb2-overexpressing cell line was obtained as described in a previous study (24).
DNA transfection and selection of stable transfectants.
Plasmids were electroporated into T. vaginalis for paromomycin selection of stable transfectants, and cloned cell lines were established as previously described (26).
Oligonucleotides.
Sequences of the oligonucleotides used in the present study are listed in Table 1 unless they were reported previously elsewhere (24, 25).
TABLE 1.
Sequences of oligonucleotides used in this study
| Purpose | Oligonucleotide | Oligonucleotide sequence (5′ to 3′) |
|---|---|---|
| RT-PCR | ||
| myb3-f | TCATAATTTTTTCGTATATGGGCAAAAAC | |
| myb3-r | TTGTCAAGCGCAATGGGAGTGATC | |
| Plasmid constructiona | ||
| ha-myb3-hind3-5′ | AAAGCTTTACCCATACGATGTTCCAGATTACGCTCTTATGGGCAAAAACTGGACCGC | |
| ha-myb3-nde1-3′ | ACATATGGCCATAGGTACCATAGATGCG | |
| αs-myb3-5′-nde1 | ACATATGCATAATTTTTTCGTATATGGGC | |
| αs-myb3-3′-hind3 | AAAGCTTGGAAAAATACAGAAAAGAGTG | |
| ap65-1-3′utr-sac2-5′ | ACCCCGCGGCAAGTACCTCATCGACAACG | |
| ap65-1-3′utr-nsi1-3′ | ACCAATGCATACTAGTGATTTAAATTAAGAAAGC | |
| myb3-sal1-5′ | AGTCGACATGGGCAAAAACTGGACCGCT | |
| myb3-not1-3′ | AGCGGCCGCTTAAAATTTAGCAAAAGCAAC | |
| ap65-2.2-sac2-5′ | AACCGCGGCGAATCTATATCTTATGTACAGTTTGG | |
| ap65-2.2-3ha-bgl2-3′ | AAGATCTAAGAGCGTAATCTGGAACATCGTATGGGTAAAGAGCGTAATCTGGAACATCGTATGGGTAAAGAGCGTAATCTGGAACATCGTATGGGTAGACTGAAGATGTGAGCAT |
The sequence of the restriction enzyme site as indicated in the name is underlined.
Screening of DNA-binding proteins.
A concatenated 32P-labeled double-stranded DNA probe, mre-1/2r, which is derived from the sequence spanning the MRE-1/MRE-2r region (see Fig. 3 for the iron-responsive [IR] sequence), was used for Southwestern library screening as described previously (25, 34).
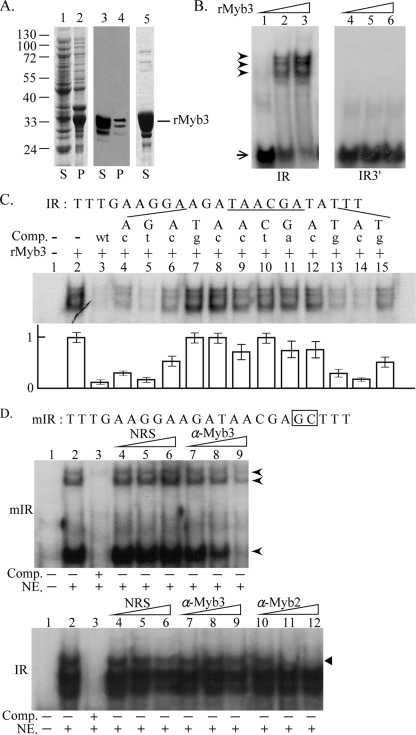
FIG. 3.
Myb3 DNA-binding specificity. (A) Soluble (lanes 1 and 3) and insoluble (lanes 2 and 4) fractions of lysates from rMyb3-expressing E. coli and purified rMyb3 (lane 5) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a 12% gel and stained with Coomassie blue (lanes 1, 2, and 5) or Western blotting using an anti-His6 antibody (lanes 3 and 4). (B) One nanogram (lanes 1 and 4), 5 ng (lanes 2 and 5), or 25 ng (lanes 3 and 6) of rMyb3 was incubated with γ32P-IR (lanes 1 to 3), which spans MRE-1/MRE-2r, or γ32P-IR3′ (lanes 4 to 6), which spans MRE-2f. Free and bound probes are indicated by an arrow and arrowheads, respectively. (C and D) Five nanograms of rMyb3 (C, lanes 2 to 15) or 10 μg of nuclear extract (NE; D, lanes 2 to 9, top, and lanes 2 to 12, bottom) was incubated with γ32P-IR (C, lanes 1 to 15; D, bottom panel, lanes 1 to 12) or γ32P-mIR (D, top panel, lanes 1 to 9). A 250× molar excess of the IR competitor (Comp.) or a series of mutant competitors (C, lanes 4 to 15, and D, lane 3), each with a single point mutation as indicated by a lowercase letter at the top of each lane, was included in the reaction mixture. The sequence of the IR probe is listed with the MRE-1 site underlined. The average signal intensities of the complexes in each reaction mixture from three experiments are depicted below. (D) Normal rabbit serum (NRS) (lanes 4 to 6), an anti-Myb3 (α-Myb3) (lanes 7 to 9), or an anti-Myb2 (α-Myb2) antibody (bottom, lanes 10 to 12) with increasing amounts (1, 2, and 4 μl for NRS and α-Myb3, 1/32, 1/16, and 1/8 μl for α-Myb2) was included in the reaction mixture. The γ32P-mIR-protein complexes in the top panel are indicated by arrowheads. The complex of γ32P-IR-Myb2 in the bottom panel is indicated by a closed triangle. The sequence of the mIR probe is listed, with the mutated residues boxed. The reaction mixtures were separated on 10% polyacrylamide gels by electrophoresis.
Cloning of the genomic myb3 gene.
The sequence flanking 5′ of the myb3 gene was amplified from a T. vaginalis T1 genomic DNA library (25) by PCR analysis using the primer pair T3 and myb3-3′-2. The amplified DNA was then cloned into pGEM_T Easy to produce a recombinant plasmid, pTAmyb3, as described by the supplier (Promega).
Construction of T. vaginalis expression plasmids.
For myb3 expression knockdown, an antisense DNA fragment spanning the myb3 coding region was amplified from genomic DNA using the primer pair αs-myb3-5′sac2 and αs-myb3-3′hind3 and was cloned into pGEM_T Easy to produce pTAαs-myb3. A DNA fragment spanning the 3′ untranslated region of the ap65-1 gene was amplified from genomic DNA using the primer pair ap65-1-3′utr-sac2 and ap65-1-3′utr-nsi1. The DNA was then cloned into pGEM_T Easy to generate pTA-AP65-1-1-3′utr. The HinDIII/SacII insert from pTAαs-myb3 and the SacII/NsiI insert from pTA-AP65-1-1-3′utr were ligated with the HinDIII/NsiI predigested pAP65-2.1-ha-myb2/TUBneo (24) to generate pAP65-2.1-αs-myb3/TUBneo (see Fig. 4A).
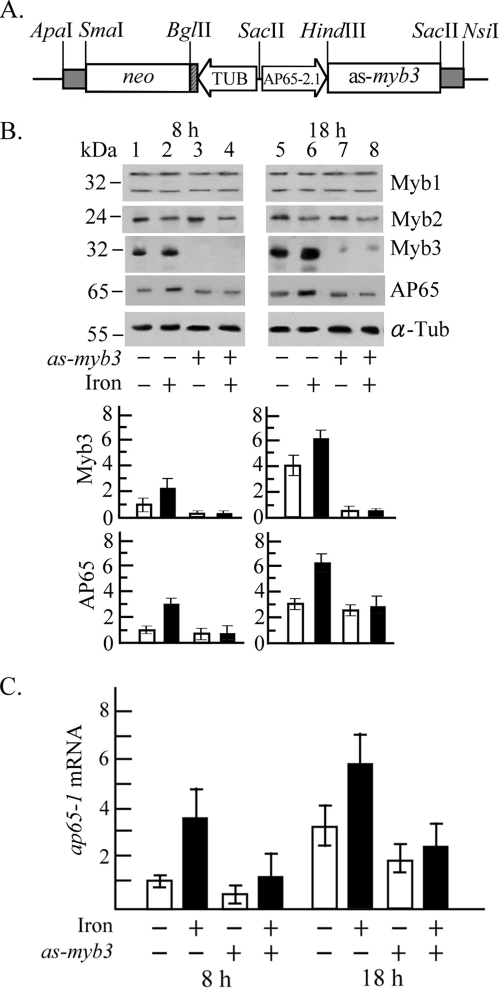
FIG. 4.
Myb3 knockdown in T. vaginalis. (A) In pAP65-2.1as-myb3, the ap65-2 proximal promoter (AP65-2.1) drives an antisense myb3 gene, and the β-tubulin (TUB) proximal promoter drives the neo gene, a selective marker. (B and C) T. vaginalis T1 (lanes 1, 2, 5, and 6) and transfected cells (lanes 3, 4, 7, and 8) were cultured under iron-depleted (open bars; lanes 1, 3, 5, and 7) and iron-replete (closed bars; lanes 2, 4, 6, and 8) conditions for 8 h (lanes 1 to 4) and 18 h (lanes 5 to 8). (B) Total cell lysates were analyzed by Western blotting using the antibodies against Myb1, Myb2, Myb3, AP65, and α-tubulin (α-Tub). Relative levels of Myb3 or AP 65 versus α-Tub are depicted in the bottom panel. (C) Relative levels of ap65-1 versus β-tubulin mRNA in 10 μg RNA from myb3 knockdown (as-myb3) or nontransfected T. vaginalis with iron depletion (open bars) or iron repletion (closed bars) for 8 or 18 h were analyzed by RT-qPCR. The results depicted in bar graphs are the averages ± the standard error from three separate experiments.
For expression of a hemagglutinin (HA)-tagged Myb3, a DNA fragment spanning the myb3 coding region was amplified from genomic DNA by PCR using the primer pair ha-myb3-hind3-5′ and myb3-sac2-3′ and was cloned into pGEM_T Easy to produce pTA-ha-Myb3. The HinDIII/SacII fragment from pTA-ha-Myb3 and the SacII/NsiI fragment from pTA-AP65-1-3′utr were cloned into HinDIII/NsiI-restricted pAPm(MRE-1) (24), which harbors a mutated ap65-1 promoter with disruption of the Myb3-binding MRE-1 site to generate pAPm(MRE-1)-ha-myb3 (see Fig. 5A).
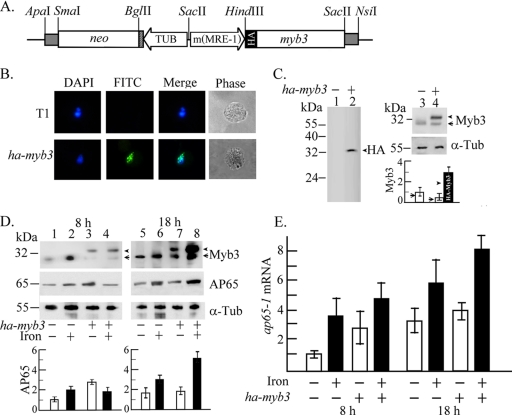
FIG. 5.
Overexpression of HA-Myb3 in T. vaginalis. (A) In pAPm(MRE-1)-ha-myb3, a mutated ap65-1 promoter, m(MRE-1) (22), drives expression of an HA-tagged myb3 gene, and the β-tubulin (TUB) promoter drives the neo gene, a selective marker. (B) HA-Myb3 in T. vaginalis T1 (top row) and transfected cells (bottom row) was detected by IFA using a mouse anti-HA-antibody (100× dilution). The nucleus was stained with DAPI (4′,6-diamidino-2-phenylindole). (C) Total lysates of T1 (lanes 1 and 3) and transfected cells (lanes 2 and 4) of 106 (lanes 1 and 2), 2 × 105 (lanes 3 and 4, top), and 4 × 104 (lanes 3 and 4, middle) were loaded, respectively, for the detection of the HA-Myb3 (arrowhead or closed bar), endogenous Myb3 (arrow or open bars), and α-tubulin (α-Tub) by Western blotting using respective antibodies as indicated. (D) Total lysates from T1 (lanes 1, 2, 5, and 6) and transfected cells (lanes 3, 4, 7, and 8) with iron depletion (open bars; lanes 1, 3, 5, and 7) and iron repletion (closed bars; lanes 2, 4, 6, and 8) for 8 h (lanes 1 to 4) and 18 h (lanes 5 to 8) were examined by Western blotting. (E) Relative levels of ap65-1 versus β-tubulin mRNA in 10 μg RNA from myb3 overexpression (ha-myb3) or nontransfected T. vaginalis with iron depletion (open bars) or iron repletion (closed bars) for 8 or 18 h were analyzed by RT-qPCR. The results depicted in bar graphs are the average ± the standard error from three separate experiments.
For stable HA-Myb1 overexpression in T. vaginalis, a shorter version (−160/+22) of the ap65-2.1 promoter was amplified from pTA-AP65-2.1 (24) using the primer pair ap65-2.2-sac2-5′ and ap65-2.2-3ha-bgl2-3′ and was cloned into pGEM_T Easy to generate pTA-AP65-2.2. The SacII/BglII fragment from pTA-AP65-2.2 and the BglII/NsiI fragment from pFLPha-myb1 (25) were ligated with a SacII/NsiI-restricted pAP65-1luc+/TUBneo (26) vector backbone to produce pAP65-2.2.ha-myb1/TUBneo (see Fig. 7A).
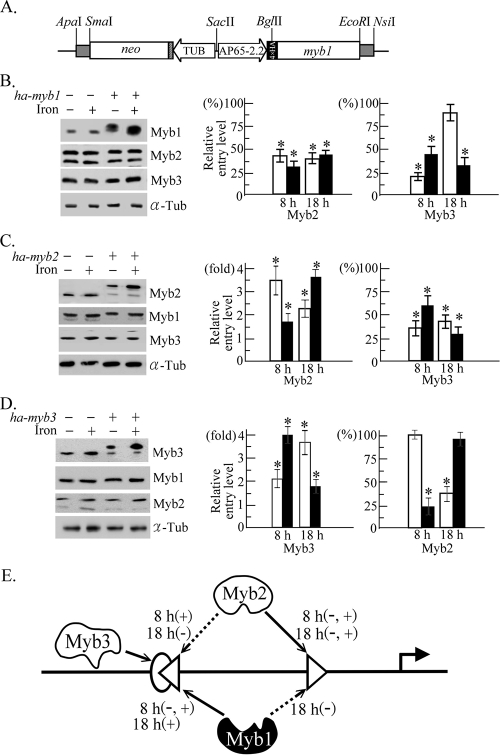
FIG. 7.
Competitive promoter entries by multiple Myb proteins in T. vaginalis. (A) In pAP65-2.2ha-myb1, the ap65-2.2 promoter (AP65-2.2) drives a myb1 gene with four copies of the HA tag, and the β-tubulin (TUB) promoter drives the neo gene, a selective marker. (B to D) Total lysates (left panel) or cells in a 50-ml culture (middle and right) from T. vaginalis T1 and transfected cells overexpressing HA-Myb1 (ha-myb1) (B), HA-Myb2 (ha-myb2) (C), and HA-Myb3 (ha-myb3) (D) under iron-depleted (open symbols) and iron-replete (closed symbols) conditions for 8 and 18 h were evaluated by Western blotting using the anti-Myb1, anti-Myb2, anti-Myb3, and anti-α-tubulin (α-Tub) antibodies (left) and by ChIP (middle and right). Relative levels of DNA pulled down by the anti-Myb2 and anti-Myb3 antibodies versus input DNA were determined by qPCR. The original levels of Myb2 and Myb3 promoter entry in wild-type cells is considered 1 or 100% to illustrate the relative Myb promoter entry levels in transfected cells. The results are the average ± the standard error from three separate experiments. The data in each set of experiments were analyzed by analysis of variance. A significant difference (P < 0.05) in promoter entry under the same situation in transfected versus wild-type cells is indicated by an asterisk. (E) Competitive promoter entries by Myb1, Myb2, and Myb3 into the MRE-1 (oval), MRE-2r (left-pointing triangle), and MRE-2f (right-pointing triangle) sites in region I of the ap65-1 promoter are depicted. The arrow with a solid line indicates a primary entry site for Myb1, Myb2, and Myb3, and the arrow with a broken line indicates the secondary entry site for a defined Myb protein under iron repletion (+) or iron depletion (−) for 8 or 18 h as indicated. The transcription activator and repressor are depicted by open and closed symbols, respectively.
Northern hybridization.
Cellular RNA was extracted from T. vaginalis by TRIzol (Invitrogen), and mRNA was further purified by oligo(dT) cellulose column chromatography. Probe labeling and Northern hybridization were performed as previously described (25). The [α-32P]dCTP-labeled myb3 DNA probe was synthesized from a pTA-myb3 template.
Reverse transcriptase qPCR (RT-qPCR).
Expression of the ap65-1, myb1, myb2, myb3, and β-tubulin genes was examined by reverse transcription of total RNA followed by PCR as previously described (24, 25), except that the relative level of a cDNA species versus β-tubulin cDNA was determined by real-time quantitative PCR (qPCR) using a SYBR Green I Master kit and LightCycler 480 as described by the supplier (Roche). myb3 cDNA was amplified using the primer pair myb3-f and myb3-r (Table 1), annealed at 55°C. Primer pairs and annealing temperatures used for the amplifications of other cDNA species were reported previously elsewhere (24, 25).
Expression of rMyb3.
To produce His-tagged recombinant Myb3 (rMyb3), the myb3 coding region was amplified from genomic DNA by PCR using the primer pair myb3-sal1-5′ and myb3-not1-3′ and was cloned into pGEM_T Easy to produce pTAmyb3. The SalI/NotI fragment from pTAmyb3 was cloned into pET28b (Novagen) predigested with SalI and NotI to produce pET28/Myb3. Escherichia coli strain BL21-CodonPlus(DE3)-RIL (Stratagene) was transformed with pET28b/Myb3 for the production and purification of rMyb3 as described by the supplier (Novagen).
Antibody production.
Purified soluble rMyb3 was used for rabbit immunization using a standard protocol (11). The antiserum was purified by protein A affinity chromatography as described by the supplier (Sigma).
Western blotting.
The cytoplasmic and nuclear fractions of T. vaginalis lysate were prepared using a cellular fractionation kit, NE-PER, as described by the supplier (Pierce). The Western blot assay was performed as previously described (25). The reaction conditions for the primary antibodies from commercial sources, including the mouse monoclonal anti-α-tubulin antibody (5,000×; DM1A, Sigma), rabbit monoclonal anti-acetyl-histone H3(Lys9) (3,000×; Upstate), rat monoclonal anti-HA antibody (2,000×; 3F10, Roche), and His6 monoclonal antibody (10,000×; Clontech), were as described by the supplier. The Myb1, Myb2, Myb3, and AP65 proteins were detected by rabbit anti-Myb1 (1,000×), anti-Myb2 (2,000×) and anti-Myb3 (2,000×) and mouse monoclonal anti-malic enzyme antibody 15D7 (10,000×) (6), respectively. The enhanced chemiluminescence system was used for signal detection as instructed by the supplier (Pierce), and relative signal intensities were analyzed by MetaMorph Offline, version 6.2r6 (Universal Imaging).
IFA.
Subcellular localization of HA-Myb3 or the neomycin (NEO)-selective marker was performed by an immunofluorescence assay (IFA) using the mouse anti-HA monoclonal antibody (100×; HA-7, Sigma) or rabbit anti-NPT II antibody (800×; Upstate), respectively, as previously described (25).
Electrophoretic mobility shift assay (EMSA).
Probe labeling and EMSA were performed as previously described (24, 32). The signal intensity of the 32P isotope was measured using a Typhoon 9410 variable mode imager (Pharmacia).
ChIP.
A chromatin immunoprecipitation (ChIP) assay was performed using the anti-HA, anti-Myb2, and anti-Myb3 antibodies to pull down promoter DNA as previously described (19, 24, 25), except that relative levels of antibody-pulled-down DNA versus input DNA were determined by qPCR as described above. Primer pairs and annealing temperatures used for the amplifications of different regions of the ap65-1 promoter were reported previously elsewhere (25).
RESULTS
Identification of an MRE-1/MRE-2r-binding protein.
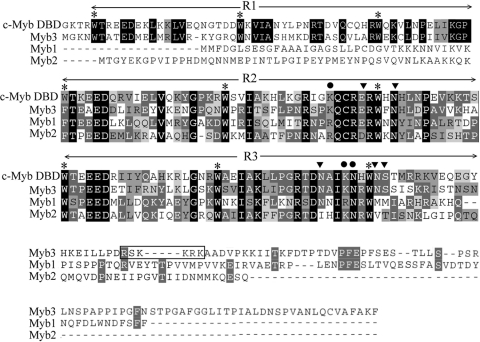
The MRE-1/MRE-2r overlap in the ap65-1 promoter was previously demonstrated to be composed of multiple DNA elements crucial for iron-inducible ap65-1 transcription (24-26). To identify the transcription factors targeting this site, a T. vaginalis cDNA expression library was screened using mre-1/2r, a 32P-labeled DNA probe, which contains multiple copies of a concatenated MRE-1/MRE-2r sequence (IR sequence). Screening of nearly 2 × 106 plaques resulted in three partial cDNA clones, λc45, λc57, and λc73, that overlap within the gene referred to as myb3. The 5′ and 3′ sequences of λc73 cDNA were extended by the PCR amplifications of respective DNA fragments from a genomic DNA library. The sequence analysis revealed the full-length myb3 gene, which carries an open reading frame encoding 250 amino acid residues (Fig. 1), with a size estimated to be 28 kDa and a pI value of 9.66. The protein sequence comprises N-terminal-conserved R1R2R3 DNA-binding domains of the Myb protein family and a unique C-terminal sequence, which contains a small motif rich in basic amino acid residues (168RSKKRK173) resembling the classical nuclear localization signals (5, 14). The R1R2R3 domains in Myb3 reveal ∼50% sequence identity to those in the human c-Myb, with complete conservation of all base-contacting amino acid residues (23), but its R2R3 domains reveal ∼42% identity to those of T. vaginalis Myb1 or Myb2, with less conservation in the base-contacting amino acid residues. Otherwise, Myb3 shows little sequence homology to Myb1 and Myb2 outside of the R2R3 domains.
FIG. 1.
Myb3 sequence conservation. The protein sequences of Myb3 (EU094478), Myb1 (AY948338), and Myb2 (AY948337) in the T. vaginalis T1 isolate were aligned with the R1R2R3 DNA-binding domains (DBD) of the human c-Myb (CAF04477). Conserved amino acids are highlighted. A putative nuclear localization signal in Myb3 sequence is boxed. The conserved tryptophan (W) residues interspersed in R1R2R3 are indicated with asterisks. The reputed strong (circles) and weak (inverted triangles) base-contacting amino acids in c-Myb (23) are indicated.
Temporal and iron-inducible myb3 expression.
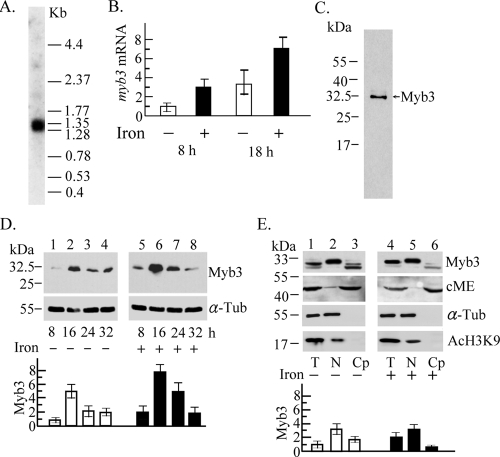
The myb3 gene was expressed as a 1.3-kb mRNA species in T. vaginalis as revealed by Northern hybridization (Fig. 2A). The expression level of myb3 mRNA in cells as examined by RT-qPCR was approximately threefold higher in samples that had iron depletion for 18 h than in samples that had iron depletion for 8 h and was approximately three- or twofold higher under iron-replete conditions than under iron-depleted conditions for 8 or 18 h, respectively (Fig. 2B). A major ∼32-kDa band was detected in cell lysate from T. vaginalis by Western blotting using the anti-Myb3 antibody (Fig. 2C). The Myb3 signal intensity increased from an initial low level at 8 h to a fivefold-higher level at 16 h and gradually declined to the original low level toward the stationary phase. In this time period, an approximately twofold-higher intensity in iron-replete than in iron-depleted samples was detected, except at the final stage of cell growth (Fig. 2D). On a duplicate blot, the intensity of α-tubulin showed little variation. None of these protein bands was detected using the preimmune serum (data not shown). The subcellular distribution of Myb3 in samples from 16-h cultures was studied by Western blotting. Myb3 was detected as doublet bands upon prolonged electrophoresis. The faster-migrating band was detected mainly in the cytoplasmic fractions, more in iron-depleted than in iron-replete samples, and the slower-migrating one was detected mainly in the nuclear fractions (Fig. 2E). The purity of the cellular fractions was validated by the presence of a 50-kDa cytosolic malic enzyme (8) only in the cytoplasmic fractions and an 18-kDa acetyl histone H3 and a 55-kDa α-tubulin only in the nuclear fractions.
FIG. 2.
Expression of the myb3 gene in T. vaginalis. (A) Ten micrograms of mRNA purified from T. vaginalis was examined by Northern hybridization using an [α-32P]dCTP-labeled DNA probe derived from pTAha-myb3. A low-molecular-weight RNA ladder was used as the size marker (Invitrogen). (B) Relative levels of myb3 versus β-tubulin mRNA in 10 μg of cellular RNA from T. vaginalis with iron depletion (open bars) and iron repletion (closed bars) for 8 and 18 h were assayed by RT-qPCR. (C to E) Lysate from T. vaginalis in normal growth medium (C) or medium with iron depletion (−) or repletion (+) (D and E) was examined by Western blotting. Myb3 (C) or relative levels of Myb3 versus α-tubulin (α-Tub) (D and E) in samples from total lysates (T), nuclear fractions (N), or cytoplasmic fractions (Cp) of T. vaginalis exposed to iron depletion (D, lanes 1 to 4, and E, lanes 1 to 3) and iron repletion (D, lanes 5 to 8, and E, lanes 4 to 6) for different time periods (D) and for 16 h (E) were examined by Western blotting (top) using the antibodies against Myb3, cytosolic malic enzyme (cME), acetyl-histone H3 (AcH3K9), and α-Tub. The levels of Myb3 in the cytoplasmic fractions were relative to the levels of cME. The results depicted in bar graphs are the averages ± the standard error from three separate experiments.
Myb3 DNA-binding specificity.
Using EMSA, as little as 5 ng of purified rMyb3 (Fig. 3A) was sufficient to form multiple complexes with a 32P-IR probe, which contains the MRE-1/MRE-2r sequence, but not for 32P-IR3′ with the MRE-2f sequence (Fig. 3B). The DNA-binding specificity was tested in competition assays using a 250× molar excess of IR or a series of mutated sequences as previously described (24) (Fig. 3C). IR completely competed for the complexes, but they were removed only partially by some of the mutant competitors. The signal intensities of the complexes in individual reaction mixtures were measured, revealing that rMyb3 interacts with the core DNA sequence, TAACGA, which resembles the target site of the reputed MRE-1-binding proteins (26). Sequences flanking the core may also contribute to optimizing interactions between Myb3 and MRE-1.
The DNA-binding activity of Myb3 in nuclear lysate was examined using a 32P-mIR probe, which has mutations in IR to disrupt the binding of MRE-2r-binding proteins (32). A major DNA-protein complex and two minor, more slowly migrating ones were detected in the binding reaction mixtures including 10 μg of nuclear lysate and 32P-mIR (Fig. 3D, top). A 250× molar excess of mIR fully competed for the complexes, and they were proportionally removed with an increasing amount of the anti-Myb3 antibody but not of normal rabbit serum. In contrast, the complexes formed in the reactions including 10 μg of nuclear lysate and 32P-IR were not affected by the anti-Myb3 antibody (Fig. 3D, bottom), but the more slowly migrating one was removed by the anti-Myb2 antibody, suggesting that Myb2 and other MRE-2r-binding proteins, which interact with 32P-IR (24), may outcompete Myb3 in binding to the MRE-1/MRE-2r overlap.
Myb3 knockdown.
To knock down Myb3 expression, T. vaginalis was transfected with pAP65-2.1-αs-myb3 (Fig. 4A), with greater than 90% of transfected cells expressing the NEO-selective marker as detected by IFA using an anti-NPTII antibody (data not shown). Protein expression in T. vaginalis was then assayed by Western blotting (Fig. 4B). The level of Myb3 as detected by the anti-Myb3 antibody was much lower in samples from transfected than nontransfected cells, with a nearly complete knockout or 90% knockdown observed in samples from 8 or 18 h, respectively, while expression of Myb1, Myb2, or α-tubulin was not affected. Basal expression levels of AP65, as defined by the expression level of the ap65-1 gene under iron depletion for 8 h (25), were similar in samples from transfected cells and nontransfected cells. Threefold increases in growth-related expression, as defined by relative expressions levels under iron depletion for 18 versus 8 h, remained similar for transfected and nontransfected cells. Iron-inducible expression, as defined by relative levels of AP65 under iron repletion versus iron depletion for 8 and 18 h that were three- and twofold, respectively, in nontransfected cells, was abolished in transfected cells.
Since the ap65 gene family is composed of multiple members with homologous coding regions but divergent promoters (25), transcription of the ap65-1 gene in the knockdown parasite was examined by RT-qPCR (Fig. 4C). Basal expression of ap65-1 mRNA in transfected cells was approximately twofold lower than that in nontransfected cells. Growth-related expression that was threefold in nontransfected cells changed slightly in transfected cells. Iron-inducible ap65-1 transcription at 8 h that was 3.5-fold in nontransfected cells was slightly repressed to an approximately twofold level in transfected cells. The repression was more severe by 18 h from an approximately twofold increase in nontransfected cells to only a ∼20% increase in transfected cells. The levels of myb1 and myb2 mRNA remained similar in the knockdown parasite and nontransfected cells (data not shown). No signal was detected if RT was omitted from RT-qPCR (data not shown). Together, these observations suggest that Myb3 is a transcription activator, which plays a significant role in basal and iron-inducible transcription of the ap65-1 gene at 8 and 18 h, respectively.
Myb3 overexpression.
T. vaginalis was transfected with a plasmid, pAPm(MRE-1)-ha-myb3, to overexpress HA-Myb3 (Fig. 5A), with greater than 80% of transfected cells expressing detectable NEO as examined by IFA using the anti-NPTII antibody (data not shown). Due to a very low expression level, a weak HA-Myb3 signal was detected only in the nuclei of a small fraction of transfected cells by IFA using a mouse anti-HA monoclonal antibody (Fig. 5B).
A ∼33-kDa band was detected by Western blotting in samples only from transfected cells, using a rat anti-HA antibody when protein samples were overloaded (Fig. 5C, left). This band, along with a ∼32-kDa band, was detected by the anti-Myb3 antibody, which detected only endogenous Myb3 in samples from nontransfected cells (Fig. 5C, right), suggesting that the addition of the HA tag changes the mobility of overexpressed Myb3 in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The signal intensity of HA-Myb3 as detected by the anti-Myb3 antibody in samples from transfected cells was threefold higher than that of endogenous Myb3 in samples from nontransfected cells as normalized by the expression levels of α-tubulin in these samples (Fig. 5C, right). Expression of endogenous Myb3 in transfected cells was substantially diminished compared to that in nontransfected cells (Fig. 5D; see 7D).
Expression of HA-Myb3 in transfected cells was iron-inducible and growth-related as was endogenous Myb3 in nontransfected cells (Fig. 5D). Basal expression of AP65 was threefold higher in transfected cells than in nontransfected cells. Growth-related expression that was detected in nontransfected cells at a twofold level was slightly repressed in transfected cells. Iron-inducible expression that was higher in transfected cells than in nontransfected cells at 18 h was repressed in transfected cells at 8 h.
The effect of HA-Myb3 overexpression on ap65-1 transcription was examined by RT-qPCR (Fig. 5E). The basal ap65-1 mRNA level was 2.5-fold higher in samples from transfected cells than from nontransfected cells. The increased basal expression has a direct impact on measuring the level of growth-related transcription, which was reduced from a threefold level in nontransfected cells to a 1.4-fold level in transfected cells. Similarly, iron-inducible transcription at 8 h was reduced from a 3.5-fold level in nontransfected cells to a 1.8-fold level in transfected cells. In contrast, the level of inducible transcription was slightly higher in transfected cells than in nontransfected cells at 18 h. No signal was detected if RT was omitted from RT-qPCR (data not shown).
Differential promoter entries by Myb3 and Myb2.
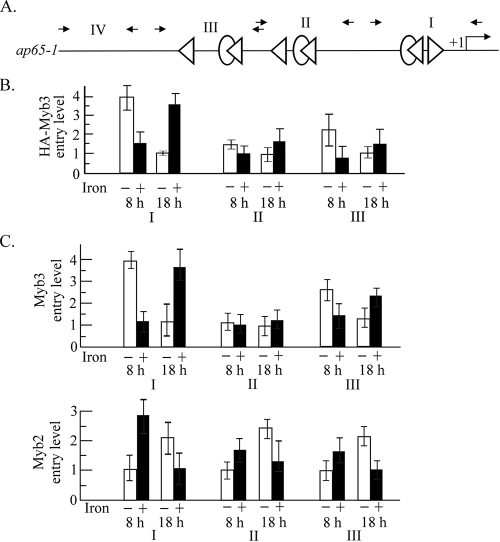
The entire ap65-1 promoter contains three clusters of two closely spaced MRE-1/MRE-2r- and MRE-2f-like sequences (Fig. 6A). The promoter selection by HA-Myb3 in transfected cells toward these regions was examined by ChIP using the anti-HA antibody (Fig. 6B). HA-Myb3 was associated with region I to similarly low levels in samples under iron repletion for 8 h and in those under iron depletion for 18 h. A fourfold-higher entry level was observed in samples under iron depletion for 8 h than in those under iron depletion for 18 h, and a ∼2.3-fold-lower entry level was detected in samples under iron repletion for 8 h than in those under iron repletion for 18 h. HA-Myb3 selected region III with a twofold-higher level in samples under iron depletion than in those under iron repletion for 8 h, but this difference was not detected in samples treated for 18 h. Its selection of region II was less affected by changes in growth conditions. No PCR product was obtained from region IV (data not shown). None of the DNA fragments was amplified in samples pulled down by the normal mouse serum (data not shown).
FIG. 6.
Differential promoter entries by Myb2 and Myb3 in T. vaginalis. (A) The organization of the entire ap65-1 promoter is depicted. The MRE-1-like (ovals), MRE-2r-like (left-pointing triangles), and MRE-2f-like (right-pointing triangles) elements in the ap65-1 promoter are depicted in the top panel. Small arrows indicate the primer pairs used in the qPCR to amplify regions I, II, III, and IV as defined previously (25). Samples from HA-Myb3 overexpressing cells (B) or nontransfected cells (C) under iron-depleted (open bars) and iron-replete (closed bars) conditions for 8 h (left) and 18 h (right) were evaluated by ChIP using the anti-HA (B) or anti-Myb3 and anti-Myb2 antibodies (C). Relative levels of DNA pulled down by each antibody versus input DNA were determined by qPCR. In panel B, the lowest level of HA-Myb3 promoter entry under iron depletion for 18 h was considered 1 to illustrate relative promoter entry levels under other conditions. In panel C, the lowest levels of Myb3 and Myb2 promoter entry under iron depletion for 18 and 8 h, respectively, were considered 1 to illustrate the relative promoter entry levels under other conditions. The results are the average ± the standard error from three separate experiments.
Concomitant promoter entries by Myb2 and Myb3 were examined in nontransfected cells by ChIP using the anti-Myb2 and anti-Myb3 antibodies, respectively (Fig. 6C). Myb3 was associated with region I at similarly low levels under iron repletion for 8 h and iron depletion for 18 h. The entry level was four- or ∼3.5-fold higher in cells under iron depletion for 8 h or iron repletion for 18 h, respectively. Myb3 selected region III with a similar preference, but with less-than-twofold differences, and its selection of region II was refractory to changes in growth conditions. In contrast, Myb2's entry into all three regions reached similarly low levels under iron depletion for 8 h and iron repletion for 18 h but was 1.5- to threefold higher under 8 h of iron repletion and 18 h of iron depletion. No PCR product was obtained from region IV (data not shown). None of the DNA fragments was amplified in samples pulled down by normal rabbit serum (data not shown). Promoter entry by endogenous Myb1 could not be studied due to the incompetence of the anti-Myb1 antibody in ChIP.
Competitive Myb promoter entries.
Competitive entries by overall (overexpressed plus endogenous) Myb2 and Myb3 into region I of the ap65-1 promoter in transfected versus nontransfected cells were then examined by ChIP using the anti-Myb2 and anti-Myb3 antibodies, respectively (Fig. 7).
T. vaginalis was transfected with pAP65-2.2ha-myb1 (Fig. 7A) to overexpress HA-Myb1 in replacing the original unstable pFLPha-myb1 system (25). HA-Myb1 was detected in the nucleus of greater than 95% of transfected cells by IFA using the anti-HA-antibody (data not shown). Overexpression of HA-Myb1 also had little impact on expression levels of Myb2 and Myb3 (Fig. 7B, left); however, overall Myb2 promoter entry in transfected cells decreased by 50% or more (Fig. 7B, middle). A concurrent decrease in Myb3 promoter entry to less than 50% of the original level was detected in all cases, except under iron depletion for 18 h (Fig. 7B, right). Likewise, HA-Myb2 overexpression had little effect on Myb1 and Myb3 expressions (Fig. 7C, left), but a 1.5- to 3.5-fold increase in overall Myb2 promoter entry (Fig. 7C, middle) was detected in transfected cells, accompanied by a concurrent decrease by 50% or more in Myb3 promoter entry (Fig. 7C, right). In cells overexpressing HA-Myb3, the expression of Myb1 and Myb2 showed little variation (Fig. 7D, left). Under iron repletion for 8 h or iron depletion for 18 h, a fourfold increase in overall Myb3 promoter entry was detected in transfected cells (Fig. 7D, middle), while the concomitant Myb2 promoter entry diminished to 25% to 40% of the original level (Fig. 7D, right). Overall Myb3 promoter entry in transfected cells under iron depletion for 8 h or iron repletion for 18 h increased twofold (Fig. 7D, middle), but Myb2 promoter entry was not affected under these situations (Fig. 7D, right).
DISCUSSION
We previously demonstrated that Myb1 and Myb2 of the R2R3 subfamily and possibly a few more Myb proteins are involved in temporal and iron-inducible transcription of the ap65-1 gene in T. vaginalis (24, 25). How they coordinate in regulating transcription can just now be appreciated with the identification of Myb3, whose expression pattern, DNA-binding specificity, promoter selection mode, and potential functions are distinguishable from those of Myb1 and Myb2. These distinctions suggest that their actions may be monitored at multicellular checkpoints, which in ap65-1 transcription are likely interlaced to achieve a dynamic balance in accessing the same promoter sites.
Myb3 is expressed in T. vaginalis as ∼32-kDa doublet proteins with differential distributions in the cytoplasm and nucleus (Fig. 2), suggesting that Myb3 likely undergoes posttranslational modification(s) before its nuclear import. A similar phenomenon was observed for Myb1 and Myb2 (24, 25). Iron that slightly enhances the nuclear import of Myb1 exerts little of a similar effect on Myb2 under our test conditions, implying that the nuclear importation of respective Myb proteins is likely activated through different signaling pathways to control different phases of ap65-1 transcription or transcription of distinct gene subsets. The nuclear localization signal that functions in T. vaginalis has yet to be identified. In this regard, Myb3 is a suitable model to study how active nuclear import is achieved in this parasite as it has a putative nuclear localization signal in its protein sequence (Fig. 1), but one does not appear in that of Myb1 or Myb2 (24, 25). Myb3 expression in T. vaginalis varies within a much greater range than does Myb1 or Myb2 either under physiological conditions or in the transgenic systems (24, 25) (Fig. 2, 4, and 5), suggesting that the levels of Myb1 and Myb2, but not that of Myb3, in T. vaginalis must be maintained within a rather narrow window, perhaps to regulate some vital genes.
Unlike the dual recognition of both MRE-1/MRE-2r and MRE-2f by Myb1 and Myb2 (24, 25), Myb3 recognized only MRE-1, with a core sequence context closer to that of human c-Myb than to that of Myb1 or Myb2 (Fig. 1), perhaps due to the strict conservation between Myb3 and human c-Myb in all reputed base-contacting amino acids in DNA-binding domains (23). A search of the T. vaginalis genome (7) and cDNA databases (http://tvxpress.cgu.edu.tw/) revealed two myb3-like genes (those for 84370_m00120 and 88666_m00240), the open reading frames of which preserve all base-contacting amino acids found in Myb3. Analogous to Myb2-related proteins (25), which share similar base-contacting amino acids and display almost identical DNA-binding specificities (H. M. Hsu and J. H. Tai, unpublished data), proteins encoded by myb3-like genes may also have similar DNA-binding specificities. Whether such Myb proteins can select the same promoter sites under different situations, perhaps to suit various purposes, is one of the intriguing questions to be addressed in an effort to unravel how or whether they must be coordinated for proper function.
Myb3 shares little sequence homology with Myb1 or Myb2 outside of the DNA-binding domains (Fig. 1), suggesting that they may be composed of different functional motifs in the divergent transactivation domains in order to carry out distinct functions like the three Myb proteins in vertebrates (15, 22). In this regard, Myb2 is an activator for multifarious transcription of the ap65-1 gene under all test conditions (24), whereas Myb1 is a repressor in basal as well as iron-inducible transcription but may activate temporal transcription (25). Myb3 appears to regulate only basal and prolonged iron-inducible transcription as supported by the gene knockdown assay and its preferential promoter selections (Fig. 4 and 6). Although basal ap65-1 transcription was substantially increased when Myb3 was overexpressed, the synergistic effect was not seen in iron-inducible transcription (Fig. 5), suggesting that Myb3 may need a limiting coactivator to promote iron-inducible ap65-1 transcription. The role of Myb3 in temporal transcription as detected in transfected cells is negligible (Fig. 4 and 5), as also reflected by a substantial reduction in Myb3 promoter entry in cells with prolonged versus short-term iron depletion (Fig. 6). The roles of Myb3 as defined herein do not entirely conform to those of the cis-acting MRE-1 element, which was previously demonstrated to downregulate basal transcription but upregulate temporal as well as iron-inducible ap65-1 transcription (24). Whether this discrepancy is due to competition by other Myb3-related proteins for the same MRE-1 site remains to be determined.
The most crucial step in Myb-mediated transcription of the ap65-1 gene appears to be at the level of promoter selection, which is not solely determined by the DNA-binding specificity or gene expression pattern of respective Myb proteins (24, 25). For example, promoter selection by Myb2 can be up- or downregulated in a temporal manner by iron, which can also repress Myb2 expression (24). Myb2 seems to surpass Myb3 in its abilities to bind to and enter MRE-1/MRE-2r (Fig. 3 and 7), while Myb3 must also have an edge to enter this site (Fig. 6 and 7). Intriguingly, physiological conditions favoring Myb2 promoter entry are unfavorable to the concomitant entry of Myb3 into the same region and vice versa (Fig. 6). This phenomenon is explained only partially by a conditional competition between Myb2 and HA-Myb3 in gaining access to MRE-1/MRE-2r (Fig. 7). In conjunction with their DNA-binding specificities and expression profiles (24) (Fig. 2 and 3), the conditional competition suggests that Myb2 may constitutively select MRE-2f as the entry site, but it may additionally enter MRE-2r as depicted in Fig. 7E to block the access of the repressive Myb1 and boost iron-inducible transcription when the Myb3 expression level is low in the early cell growth stage (Fig. 2D). Under prolonged iron depletion, which favors the expression of Myb2 (24), Myb2's selection of MRE-2r may also be responsible for growth-related transcription when Myb3 promoter entry is minimal (Fig. 6).
Apparently, promoter selections by Myb2 and Myb3 are compounded by Myb1, which may initially select promoter region I, but the selection may further extend to regions II and III (25). When overexpressed, Myb1 not only inhibited Myb2 promoter entry under all test conditions but also inhibited Myb3 promoter entry in most situations, except after prolonged iron depletion (Fig. 7B). In conjunction with Myb1 DNA-binding specificities (25), this exception implies that Myb1 likely selects MRE-1/MRE-2r as the main entry site to block the promoter entry of Myb2 or Myb3 and repress iron-inducible transcription when cells are exposed to iron for a short or long period, respectively, but Myb1 may preferentially select MRE-2f as depicted in Fig. 7E to modulate temporal transcription when cells are exposed to prolonged iron depletion (26). The conditional promoter entry of Myb1 and Myb2 may allow each of them to regulate transcription of different gene subsets upon changes of environment. In human p53-mediated transcriptional regulation, site-specific phosphorylation of p53 may determine its preferential selections between two distinct promoters (20). Whether differential and competitive promoter selections described herein involve site-specific posttranslational modifications of respective Myb proteins remains to be studied.
In summary, we demonstrate herein that temporal and iron-inducible transcription of the ap65-1 gene in T. vaginalis is crucially regulated by the synergistic or antagonistic actions of at least three Myb-like transcription factors, Myb1, Myb2, and Myb3, which differentially and competitively select MRE-1/MRE-2r and MRE-2f as entry sites in the ap65-1 promoter when growth conditions change.
Acknowledgments
This work was supported by a grant from the National Science Council (NSC95-2320-B-001-027-MY3) and IBMS, Academia Sinica.
We thank Dan Chamberlin for editing the English of the manuscript and Hsing-Wei Liu for technical assistance. The monoclonal anti-malic enzyme antibody 15D7 and anti-cytosolic malic enzyme antibody were obtained from Guy Brugerolle at Universite Blaise Pascal de Clermont-Ferrand, France, and Ivan Hrdy at Charles University, Czech Republic, respectively.
Footnotes
Published ahead of print on 16 January 2009.
REFERENCES
- 1.Alderete, J. F. 1999. Iron modulates phenotypic variation and phosphorylation of P270 in double-stranded RNA virus-infected Trichomonas vaginalis. Infect. Immun. 674298-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alderete, J. F., J. Nguyen, V. Mundodi, and M. W. Lehker. 2004. Heme-iron increases levels of AP65-mediated adherence by Trichomonas vaginalis. Microb. Pathog. 36263-271. [DOI] [PubMed] [Google Scholar]
- 3.Alderete, J. F., D. Provenzano, and M. W. Lehker. 1995. Iron mediates Trichomonas vaginalis resistance to complement lysis. Microb. Pathog. 1993-103. [DOI] [PubMed] [Google Scholar]
- 4.Alderete, J. F., J. L. O'Brien, R. Arroyo, J. A. Engbring, O. Musatovova, O. Lopez, C. Lauriano, and J. Nguyen. 1995. Cloning and molecular characterization of two adhesion proteins involved in Trichomonas vaginalis cytoadherence. Mol. Microbiol. 1769-83. [DOI] [PubMed] [Google Scholar]
- 5.Boulikas, T. 1993. Nuclear localization signals (NLS). Crit. Rev. Eukaryot. Gene Expr. 3193-227. [PubMed] [Google Scholar]
- 6.Brugerolle, G., G. Bricheux, and G. Coffe. 2000. Immunolocalization of two hydrogenosomal enzymes of Trichomonas vaginalis. Parasitol. Res. 8630-35. [DOI] [PubMed] [Google Scholar]
- 7.Carlton, J. M., R. P. Hirt, J. C. Silva, A. L. Delcher, M. Schatz, Q. Zhao, J. R. Wortman, S. L. Bidwell, U. C. Alsmark, S. Besteiro, T. Sicheritz-Ponten, C. J. Noel, J. B. Dacks, P. G. Foster, C. Simillion, Y. Van de Peer, D. Miranda-Saavedra, G. J. Barton, G. D. Westrop, S. Muller, D. Dessi, P. L. Fiori, Q. Ren, I. Paulsen, H. Zhang, F. D. Bastida-Corcuera, A. Simoes-Barbosa, M. T. Brown, R. D. Hayes, M. Mukherjee, C. Y. Okumura, R. Schneider, A. J. Smith, S. Vanacova, M. Villalvazo, B. J. Haas, M. Pertea, T. V. Feldblyum, T. R. Utterback, C. L. Shu, K. Osoegawa, P. J. de Jong, I. Hrdy, L. Horvathova, Z. Zubacova, P. Dolezal, S. B. Malik, J. M. Logsdon, Jr., K. Henze, A. Gupta, C. C. Wang, R. L. Dunne, J. A. Upcroft, P. Upcroft, O. White, S. L. Salzberg, P. Tang, C. H. Chiu, Y. S. Lee, T. M. Embley, G. H. Coombs, J. C. Mottram, J. Tachezy, C. M. Fraser-Liggett, and P. J. Johnson. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolezal, P., S. Vanacova, J. Tachezy, and I. Hrdy. 2004. Malic enzymes of Trichomonas vaginalis: two enzyme families, two distinct origins. Gene 32981-92. [DOI] [PubMed] [Google Scholar]
- 9.Ganter, B., S. T. Chao, and J. S. Lipsick. 1999. Transcriptional activation by the Myb proteins requires a specific local promoter structure. FEBS Lett. 460401-410. [DOI] [PubMed] [Google Scholar]
- 10.Garcia, A. F., T. H. Chang, M. Benchimol, D. J. Klumpp, M. W. Lehker, and J. F. Alderete. 2003. Iron and contact with host cells induce expression of adhesins on surface of Trichomonas vaginalis. Mol. Microbiol. 471207-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harlow, E., and D. Land. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Hochheimer, A., and R. Tjian. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 171309-1320. [DOI] [PubMed] [Google Scholar]
- 13.Hrdý, I., and M. Muller. 1995. Primary structure of the hydrigenosomal malic enzyme of Trichomonas vaginalis and its relationship to homologous enzymes. J. Eukaryot. Microbiol. 42593-603. [DOI] [PubMed] [Google Scholar]
- 14.Lange, A., R. E. Mills, C. J. Lange, M. Stewart, S. E. Devine, and A. H. Corbett. 2007. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 2825101-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei, W., J. J. Rushton, L. M. Davis, F. Liu, and S. A. Ness. 2004. Positive and negative determinants of target gene specificity in Myb transcription factors. J. Biol. Chem. 27929519-29527. [DOI] [PubMed] [Google Scholar]
- 16.Liston, D. R., J. C. Carrero, and P. J. Johnson. 1999. Upstream regulatory sequences required for expression of the Trichomonas vaginalis alpha-succinyl CoA synthetase gene. Mol. Biochem. Parasitol. 104323-329. [DOI] [PubMed] [Google Scholar]
- 17.Liston, D. R., and P. J. Johnson. 1999. Analysis of a ubiquitous promoter element in a primitive eukaryote: early evolution of the initiator element. Mol. Cell. Biol. 192380-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liston, D. R., A. O. Lau, D. Ortiz, S. T. Smale, and P. J. Johnson. 2001. Initiator recognition in a primitive eukaryote: IBP39, an initiator-binding protein from Trichomonas vaginalis. Mol. Cell. Biol. 217872-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Y., C. Kung, J. Fishburn, A. Z. Ansari, K. M. Shokat, and S. Hahn. 2004. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol. 241721-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayo, L. D., Y. R. Seo, M. W. Jackson, M. L. Smith, J. Rivera Guzman, C. K. Korgaonkar, and D. B. Donner. 2005. Phosphorylation of human p53 at serine 46 determines promoter selection and whether apoptosis is attenuated or amplified. J. Biol. Chem. 28025953-25959. [DOI] [PubMed] [Google Scholar]
- 21.Müller, F., and L. Tora. 2004. The multicoloured world of promoter recognition complexes. EMBO J. 232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ness, S. A. 2003. Myb protein specificity: evidence of a context-specific transcription factor code. Blood Cells Mol. Dis. 31192-200. [DOI] [PubMed] [Google Scholar]
- 23.Ogata, K., S. Morikawa, H. Nakamura, A. Sekikawa, T. Inoue, H. Kanai, A. Sarai, S. Ishii, and Y. Nishimura. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79639-648. [DOI] [PubMed] [Google Scholar]
- 24.Ong, S. J., H. M. Hsu, H. W. Liu, C. H. Chu, and J. H. Tai. 2007. Activation of multifarious transcription of an adhesion protein ap65-1 gene by a novel Myb2 protein in the protozoan parasite Trichomonas vaginalis. J. Biol. Chem. 2826716-6725. [DOI] [PubMed] [Google Scholar]
- 25.Ong, S. J., H. M. Hsu, H. W. Liu, C. H. Chu, and J. H. Tai. 2006. Multifarious transcriptional regulation of adhesion protein gene ap65-1 by a novel Myb1 protein in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell 5391-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong, S. J., S. C. Huang, H. W. Liu, and J. H. Tai. 2004. Involvement of multiple DNA elements in iron-inducible transcription of the ap65-1 gene in the protozoan parasite Trichomonas vaginalis. Mol. Microbiol. 521721-1730. [DOI] [PubMed] [Google Scholar]
- 27.Rushton, J. J., L. M. Davis, W. Lei, X. Mo, A. Leutz, and S. A. Ness. 2003. Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene 22308-313. [DOI] [PubMed] [Google Scholar]
- 28.Schumacher, M. A., A. O. Lau, and P. J. Johnson. 2003. Structural basis of core promoter recognition in a primitive eukaryote. Cell 115413-424. [DOI] [PubMed] [Google Scholar]
- 29.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72449-479. [DOI] [PubMed] [Google Scholar]
- 30.Sorvillo, F., L. Smith, P. Kerndt, and L. Ash. 2001. Trichomonas vaginalis, HIV, and African-Americans. Emerg. Infect. Dis. 7927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stracke, R., M. Werber, and B. Weisshaar. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4447-456. [DOI] [PubMed] [Google Scholar]
- 32.Tsai, C. D., H. W. Liu, and J. H. Tai. 2002. Characterization of an iron-responsive promoter in the protozoan pathogen Trichomonas vaginalis. J. Biol. Chem. 2775153-5162. [DOI] [PubMed] [Google Scholar]
- 33.Vanácová, S., W. Yan, J. M. Carlton, and P. J. Johnson. 2005. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 1024430-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker, M. D. 1996. Screening expression cDNA libraries, p. 63-66. In K. Docherty (ed.), Gene transcription: DNA binding proteins. Wiley Interscience, New York, NY.