Abstract
Toxoplasma gondii is a haploid protozoan parasite infecting about one in seven people in the United States. Key to the worldwide prevalence of T. gondii is its ability to establish a lifelong, chronic infection by evading the immune system, and central to this is the developmental switch between the two asexual forms, tachyzoites and bradyzoites. A library of mutants defective in tachyzoite-to-bradyzoite differentiation (Tbd−) was created through insertional mutagenesis. This library contains mutants that, compared to the wild type, are between 20% and 74% as efficient at stage conversion. Two mutants, TBD5 and TBD8, with disruptions in a gene encoding a putative pseudouridine synthase, PUS1, were identified. The disruption in TBD8 is in the 5′ end of the PUS1 gene and appears to produce a null allele with a 50% defect in differentiation. This is about the same switch efficiency as obtained with an engineered pus1 deletion mutant (Δpus1). The insertion in TBD5 is within the PUS1 coding region, and this appears to result in a more extreme phenotype of only ∼10% switch efficiency. Complementation of TBD8 with the genomic PUS1 allele restored wild-type differentiation efficiency. Infection of mice with pus1 mutant strains results in increased mortality during the acute phase and higher cyst burdens during the chronic infection, demonstrating an aberrant differentiation phenotype in vivo due to PUS1 disruption. Our results suggest a surprising and important role for RNA modification in this biological process.
Toxoplasma gondii is an apicomplexan parasite that is unique in its wide host range and global presence. In humans, the prevalence rate ranges from 15% to 75%, depending on geographic region (26). This high prevalence is due, in part, to the ability of the parasite to efficiently initiate an infection using either of two developmental stages, each with its own mode of transmission. Chronic infection is also a result of the ability of Toxoplasma to successfully evade clearance by the host immune system. All of these phenomena are dependent on the complex but well-described developmental biology of this coccidian parasite.
Toxoplasma disseminates within a host primarily through interconversion between two asexual stages, the tachyzoite and bradyzoite forms. One way that a new host can be infected is through ingestion of raw or undercooked tissue from an infected animal. Bradyzoite-containing cysts in the tissue break open due to the pepsin and acidic environment of the digestive tract, after which the bradyzoites within are released and invade through the intestinal epithelium. These bradyzoites quickly convert into tachyzoites, the rapidly dividing, disease-causing form of the parasite, which spread throughout the infected individual. During parasite expansion, adaptive immunity is triggered; this efficiently clears the tachyzoites but not the encysted bradyzoites. This persistence of bradyzoites results in a chronic, lifelong infection that allows further dispersal of Toxoplasma. Thus, parasite differentiation represents a key step in Toxoplasma persistence and disease.
In vitro methods that stimulate interconversion between the tachyzoite and bradyzoite forms allow laboratory manipulation of Toxoplasma development. Bradyzoites can be induced to develop by mimicking the “stress” of the host immune response through treatment with gamma interferon, high temperature (43°C), nitric oxide, high pH (pH = 8.1), and/or mitochondrial inhibitors (3, 4, 34, 37). Subsequent removal of the stress stimuli causes bradyzoites to revert back to the tachyzoite stage. A reproducible differentiation efficiency of 90% to 95% is obtained using a combination of carbon starvation, low serum, and high pH, with the appearance of bradyzoite-specific markers after 48 h of stress treatment (12).
Despite improvements in in vitro methods to produce pure bradyzoite populations, little is known about the differentiation process at the molecular level. Several authors have reported bradyzoite-specific genes (5, 20, 32, 39, 42, 43). Analysis of T. gondii cDNA microarrays during an in vitro tachyzoite-to-bradyzoite conversion time course has revealed large groups of genes expressed in a stage-specific manner and large sets of transcripts with similar expression patterns that appear to be coregulated (9). Monitoring of transcript synthesis by pulse-labeling RNA with 4-thiouracil confirmed induced transcription and active repression of gene sets during a differentiation time course (10). The gene sets identified as having different expression patterns in activation and repression reveal underlying regulatory networks controlling transcription, but little is known about how these networks are regulated.
Chemical mutagenesis has been used to produce tachyzoite-to-bradyzoite differentiation (Tbd−) mutants with consistent, reproducible phenotypes, but the genes responsible have not been identified (25, 33). These Tbd− mutants have a 50% to 85% drop in differentiation efficiency compared to the wild type (WT) and produce fewer cysts in mice during chronic infection, implying that the lower cyst burden may be due to an inability to differentiate and consequent clearance by the immune system. Insertional mutagenesis and reverse genetics have proven successful in creating Tbd− mutants with identifiable genetic disruptions. Disruption of genes encoding either zinc finger protein 1 (ZFP1) or plasma membrane ATPase 1 (PMA1) results in a 50% decrease in differentiation efficiency in vitro (18, 38). Neither of these mutants, however, has provided insight into the molecular mechanisms of T. gondii differentiation. We produced a new library of Tbd− mutants through insertional mutagenesis of Toxoplasma Pru strain parasites. The mutants produced had differentiation efficiencies ranging from ∼20% to 80% of that of the WT parental strain. One of the confirmed mutants from this library (TBD8) and one from a previously generated library (TBD5) have disruptions of the gene encoding a putative pseudouridine synthase (PUS1). This surprising result suggests that RNA processing plays an important and unexpected role in Toxoplasma differentiation.
MATERIALS AND METHODS
Selection of Toxoplasma differentiation mutants.
Clone A7 of the Pru strain of Toxoplasma was used as the parental strain in this work (38). A7 lacks the gene for hypoxanthine-xanthine-guanine phosphoribosyl transferase (HPT) (and is thus designated Δhpt) and was previously engineered to constitutively express green fluorescent protein (GFP) and firefly luciferase. Two groups of 2 × 107 A7 tachyzoites were transformed with 70 μg of a plasmid insertional vector, pT/230 (22) (pT/230 carries the chloramphenicol acetyltransferase selectable marker). During transformation, 10 U of DpnII was added to the cuvette to increase integration of the circular plasmid by restriction enzyme-mediated integration (1). The mutagenized population was passaged in 30 μM chloramphenicol (Boehringer Mannheim) in human foreskin fibroblast (HFF) tissue culture for 18 days to select for stable integration of the insertional vector (all culturing was essentially as previously described) (36). After selection, parasites were harvested by syringe lysis of the infected host cells and inoculated into new HFFs in T25 flasks at a multiplicity of infection (MOI) of 0.2. Four hours were allowed for the parasites to invade, after which the normal growth medium was removed and replaced with differentiation medium, i.e., RPMI 1640 lacking sodium bicarbonate (Gibco), supplemented with 1% fetal calf serum (FCS), and adjusted to a pH of 8.0 with HEPES. Differentiation cultures were kept at 37°C using ambient CO2 levels with the caps loosened for 4 days. After this time, parasites were obtained by scraping the culture and passage through a syringe (35), after which they were assessed for their ability to differentiate from tachyzoites to bradyzoites by incubation with antibodies to the bradyzoite-specific surface antigens SRS9 (mouse antibodies used at 1:500) and SAG2X/Y (rabbit antibodies used at 1:300) using the fluorescent secondary antibodies goat anti-mouse-phycoerythrin (Calbiochem) and goat anti-rabbit-Cy5.5 (Caltag Laboratories) (31). Parasites were sorted using fluorescence-activated cell sorting (FACS). To obtain parasites deficient in their ability to switch from tachyzoite to bradyzoite, the collected fraction was gated to be GFP positive, phycoerythrin negative, and Cy5.5 negative. These collected tachyzoites were recultured in tachyzoite conditions for 3 days, allowing the parasites to recover, and the differentiation and FACS protocol was repeated twice more. After the third FACS, parasites were cloned by limiting dilution.
In vitro characterization of differentiation mutants.
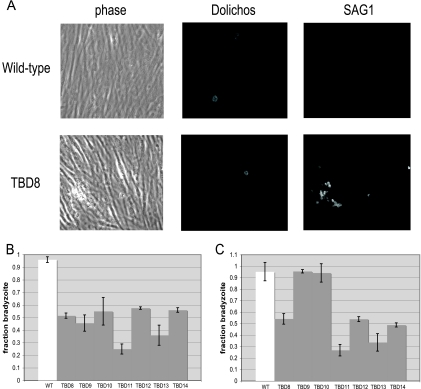
Clones were tested for a Tbd− phenotype by two in vitro differentiation protocols: with the differentiation medium used during selection or with the addition of atovaquone (Burroughs Wellcome Company) to the growth medium (37). For atovaquone-induced differentiation, HFFs were infected with Toxoplasma at an MOI of 0.2 in growth medium (Dulbecco modified Eagle medium containing 10% FCS and penicillin-streptomycin) containing 0.9 μM atovaquone and incubated at 37°C and 5.0% CO2 for 4 days. For both protocols, the infected monolayers were fixed with cold methanol after 4 days, permeabilized in 0.2% Triton X-100 (Sigma) in phosphate-buffered saline (PBS) for 20 min, and blocked with 3% (wt/vol) bovine serum albumin in PBS for 20 min (Sigma). Primary antibodies were diluted in PBS containing 0.2% Triton X-100 and 3% bovine serum albumin and incubated in the cell cultures for 1 hour. Rabbit anti-SAG1 was used at 1:5,000, fluorescein-conjugated Dolichos bifloros lectin (Vector laboratories) was used at 1:350, mouse anti-SRS9 was used at 1:5,000, and rabbit anti-SAG2X/Y was used at 1:2,000 (21). AlexaFluor 594-conjugated goat anti-mouse antibody was used to detect SAG1, and all bradyzoite antigens were targeted with AlexaFluor 488-conjugated goat anti-rabbit antibody (Invitrogen). To score bradyzoites and tachyzoites, Dolichos lectin and the antibody against the tachyzoite surface antigen SAG1 were used together. The presence of Dolichos staining and the absence or significantly reduced signal of SAG1 denoted bradyzoite cysts, and the presence of abundant SAG1 and no Dolichos indicated tachyzoites. Ten fields of view at a magnification of ×40 were quantified by this measure, with a sample size of >40 events, and averaged across three biological replicates, each with three technical replicates. Significance was determined by the Student t test distribution, comparing all Tbd− mutants to the parental WT strain.
Identification of the insertion site.
Genomic DNA from individual mutants was extracted using DNAzol (Invitrogen) according to the manufacturer's instructions. Twenty micrograms of DNA for TBD8 was digested overnight with 50 U MfeI, NotI, or SacII (New England Biosystems). The resulting digest was ligated in a 500-μl volume using high-concentration T4 ligase (NEB) according to the manufacturer's instructions. The reaction product was purified by phenol-chloroform extraction. Half of the extract was transformed into high-efficiency DH5α competent Escherichia coli cells (Invitrogen). The cells were plated onto ampicillin-LB agar plates, and plasmid from the picked colonies was sequenced to identify the flanking genomic sequence. The sequence obtained was subjected to BLAST analysis against the T. gondii genome database (http://www.toxodb.org/). Similarly, TBD5 was identified by digestion of genomic DNA with NotI. Two independent colonies were obtained for each of TBD5 and TBD8. Verification of the TBD5 and TBD8 insertion sites was performed using PCR. Primer pair 5′TAATTGTGGGTTTAGGGCCTG3′ (genomic) and 5′CAGTACGCCTGTTCTCTCGA3′ (vector) was used to amplify the TBD5 insertion specifically crossing the genome/vector boundary. Primer pair 5′GTCTTACCGGGTTGGACTCA3′ (vector) and 5′TCTGAAGAAACGCGTTCCGC3′ (genomic) was used to amplify the TBD8 genome/vector boundary. The disruption of the genomic site was also tested by attempting to PCR amplify the disrupted region using primers flanking the insertion site. For TBD5 and TBD8 the primer pairs were 5′CACAACGAGCCACAAAAAGA3′/5′CCTTCCCTTGCTATCCCTTC3′ and 5′CCAGTCGACGTTTATGGATC3′/5′GAAGACAGGAGAGGCAGGAA3′, respectively.
Characterization of the PUS1 locus.
To determine the full transcript and predicted coding regions of PUS1, reverse transcription-PCR (RT-PCR) was performed on RNA from in vitro-derived bradyzoites of the Pru strain. The full coding region was amplified using primers predicted to be in exonic regions for PUS1 (gene 20.m03736 in the draft 3 gene models of ToxoDB; www.Toxodb.org). The 3′ untranslated region (UTR) was identified by 3′ rapid amplification of cDNA ends (RACE) using oligo(dT) and primer 5′TGTCTTTCTTGAGCGGCATC3′, which is contained inside the predicted coding region. 5′ RACE was performed using terminal deoxynucleotidyl transferase poly(A) tract capping of cDNA and amplification of the 5′ end of the transcript with oligo(dT) and the primer 5′GGAAGAGACATCGAGGAGTC3′, which binds 100 bp upstream of the predicted initiator codon (ATG). The transcript sequence was translated using Biology Workbench (http://workbench.sdsc.edu/). The protein sequence was investigated for functional motifs using InterPro Scan (http://www.ebi.ac.uk/InterProScan/) and Pfam HMM (http://pfam.sanger.ac.uk/search?tab=searchSequenceBlock). Homology to other known genes was determined using BLAST methods at http://www.ncbi.nlm.nih.gov/blast/Blast.cgi/, http://www.yeastgenome.org/ and http://www.genome.wisc.edu/. The domain architecture of PUS1 was investigated using http://www.ncbi.nlm.nih.gov/sites/entrez?db=cdd. Analysis of the PUS1 protein sequence for a mitochondrial targeting sequence was performed using MitoProt (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html). Alignment of PUS1 with RluA-1 from Drosophila melanogaster, RPUSD2 from the human genome, PUS9 from Saccharomyces cerevisiae, RluC from Escherichia coli, and RluD from Arabidopsis thaliana was performed using ClustalW at the Biology Workbench v3.2 (http://workbench.sdsc.edu).
Creating a targeted Δpus1 Toxoplasma strain.
The regions 5′ and 3′ of the predicted PUS1 open reading frame (ORF) were amplified from genomic DNA and placed into the pmini-HPT vector flanking the HPT selectable marker (26). A 2.5-kb region at the 5′ end of the gene was amplified using primer pair 5′CCCTGGAGGAAACGCAGTC3′ and 5′AAGGAGACAGTGTGGGGTGA3′. A 2.5-kb region at the 3′ end of the gene was amplified using primer pair 5′GAGACTTCCTTTCTGCGCT3′ and 5′TGAGCGACAGACTAGGACTCG3′. The resulting construct is referred to as pPUS1KO. To increase the probability of homologous integration, clone A7 tachyzoites were partially induced to switch to bradyzoites by syringe-lysing infected HFF host cells and leaving the resulting parasites extracellular for 16 h before electroporation (6). After this overnight stressing of the parasites, 2 × 107 parasites were electroporated with 50 μg of linearized pPUS1KO. Parasites expressing the HPT marker were selected with 0.05 μM mycophenolic acid and 0.05 μM xanthine for 10 days to ensure stable integration of the knockout vector. Individual clones were screened by PCR with primers spanning the two genome/vector boundaries that should be created by double homologous recombination and replacement of PUS1 with HPT. The resulting mutant clone with complete deletion of the PUS1 ORF was named the Δpus1 strain. A strain in which insertion of the knockout vector occurred immediately 3′ of the PUS1 locus was obtained to serve as a control. This strain is named PUS1-shi (for single homologous integration).
Stress survival assay.
Toxoplasma tachyzoites were released from infected host cells by syringe passage, and 500 parasites were passed using one of the three following conditions: infect new cells in normal medium, infect new cells and stress with differentiation medium at 4 h postinfection, or maintain extracellular for 24 h and then infect new cells in normal medium. After application of each of the three conditions, the parasites were allowed to grow in T25 flasks and create plaques for 10 days undisturbed. After 10 days, the parasites were fixed and labeled with mouse anti-Toxoplasma serum at 1:1,000 followed by AlexaFluor 488-conjugated goat anti-mouse antibody (Invitrogen). The number of plaques in each flask was counted for each of two technical replicates and averaged for each strain. The fraction of plaques under each stress condition relative to no stress was determined to indicate survivability of the strain under that stress. This process was repeated three times independently for the parental type II and Δpus1, PUS1-shi, TBD5, and TBD8 strains. Significance was determined using Student's t test.
In vitro growth assay.
Parasites were syringe released from intact host cell monolayers and added to HFFs on coverslips at an MOI of 2. The parasites were allowed to invade and grow for 4, 12, 24, or 48 h before being fixed and labeled with mouse anti-Toxoplasma serum at 1:1,000 followed by AlexaFluor 488-conjugated goat anti-mouse antibody (Invitrogen). Coverslips for the 12-, 24-, and 48-hour time points were washed at 4 h to remove dead and unattached parasites. The number of vacuoles with two or more parasites was counted over 15 fields at a magnification of ×100, a sample size of over 100 vacuoles per experiment, and categorized by the number of parasites present in each vacuole (scored as 2, 4, 8, 16, 32, 64, 128, or 256) or lysed. At higher vacuole sizes not all vacuoles contained exact twofold increments of parasites, so vacuoles with 60 to 70 parasites are represented by the 64-parasite category, vacuoles with 115 to 140 parasites are represented by the 128-parasite category, and vacuoles with 240 to 280 parasites are represented by the 256-parasite category. Three biological replicates for each time point were performed blinded with two technical replicates. Significance was determined by a Student t test distribution comparing the complemented TBD8 mutant (TBD8comp), the pus1 knockout mutant (PUS1 KO), and TBD8 to the WT at each vacuole size for each time point.
Complementation of pus1 null mutant, TBD8.
To restore the WT differentiation phenotype to pus1 null mutants, the PUS1 genomic locus was amplified using Platinum Taq Hi-Fi polymerase (Invitrogen) from bp 906792 to 894632 of scaffold 995279 on chromosome VIIa encompassing the full PUS1 transcribed region identified through RT-PCR as well as 1,600 bp upstream of the transcription start site and 500 bp downstream of the 3′ UTR. The genomic locus was amplified in five pieces of roughly 2 kb, which were ligated together to create the full genomic locus. No nonsense mutations or frameshifts were found in the resulting genomic locus; there were, however, two missense mutations which were not expected to alter the function of the protein (as confirmed by the ability of the introduced DNA to complement a PUS1 mutant). The resulting full-length PUS1 gene was cloned into pHANA, and 10 μg of the resulting vector, pPUS1-HPT, or pHANA, lacking the PUS1 locus as a control, was transformed into 2 × 107 TBD8 tachyzoites each (2). The parasites were selected for 2 weeks with 0.05 μM mycophenolic acid and 0.05 μM xanthine and cloned by limiting dilutions. Individual clones containing pPUS1-HPT were screened by PCR for acquisition of the full genomic locus using primers that specifically amplify either end of the PUS1 locus in the pHANA vector. The clones transfected with pHANA were screened for retention of HPT, and a representative clone, TBD8pHANA, was chosen to serve as a control. These clones were then tested for restoration of a WT switch phenotype by the differentiation medium protocol with TBD8 and TBD8pHANA serving as negative controls and the parental A7 as a positive control. The scoring of differentiation efficiency was performed blinded by the method described for creation of the Tbd− mutants using SAG1 and cyst wall markers for tachyzoite and bradyzoite stages, respectively, over 15 fields of view at a magnification of ×40, a sample size of at least 50 vacuoles/cysts. Four biological replicates, each consisting of three technical replicates, were performed.
Infection of mice.
Eight-week-old BALB/c female mice (Jackson Laboratories) were infected with 500 tachyzoites by intraperitoneal injection in 0.2 ml sterile PBS as described previously (21). Tachyzoites were grown in vitro, mechanically lysed from host cells by passage through a 27-gauge needle, and washed twice with PBS supplemented with 1% FCS. Parasite numbers were determined by counting on a hemocytometer, and afterwards, injection of comparable numbers of viable parasites was verified by plaque assay and bioluminescence imaging (11, 17). Two groups of six mice were infected with each of the five Toxoplasma strains, WT (A7), PUS1-shi, TBD8comp, TBD8, and PUS1 KO. All animal studies have been reviewed and approved by Stanford University's Administrative Panel for Laboratory Animal Care.
Bioluminescence imaging.
Bioluminescence imaging procedures were performed using a Xenogen IVIS100 charge-coupled device system (Xenogen, Alameda, CA) as described previously (30). Briefly, mice were injected with 200 μl of luciferin (3 mg) and anesthetized in an oxygen-rich induction chamber with 1.5% isofluorane for 5 min. Mice were then imaged for 5 min dorsally and 5 min ventrally using a grayscale reference image and a photon emission image overlay from luciferase expression. Photon emission was pseudocolored to represent the light intensity collected from the animal, with blue as least and red as most intense. Region-of-interest measurements were collected for the mice and included the full body from the top of the skull to the base of the tail. Three region-of-interest measurements were taken for three of each set of mice in both experiments, and the standard deviations are shown.
Examination of brain cyst load and size.
Three mice infected with each strain were sacrificed at 4 weeks postinfection (wpi) and an additional three at 9 wpi. Brains were extracted from the mice, and one half of each brain was homogenized through a 100-μm cell strainer. The homogenate was washed three times in 25 ml of PBS supplemented with 3% FCS by centrifugation at 150 × g for 10 min. The homogenate was resuspended to 4 ml and incubated with rhodamine-conjugated Dolichos bifloros agglutinin (Vector Laboratories) at 1:150 for 2 h to stain the cyst wall. After staining, the homogenate was washed twice and resuspended in 1 ml PBS-3% FCS, and 50 μl was aliquoted into individual wells of a flat-bottom 96-well plate. Cysts were counted using an inverted fluorescence microscope (Nikon). To assess cyst size, photographs of brain cysts were captured using a camera attached to the microscope and used to measure the cyst diameter on an arbitrary scale.
Nucleotide sequence accession number.
The corrected sequence of the 20.m03736 gene has been submitted to GenBank with accession number FJ236805.
RESULTS
To identify genes necessary for tachyzoite-to-bradyzoite differentiation, we mutagenized Pru Δhpt tachyzoites engineered to constitutively express GFP and firefly luciferase (26). Mutagenesis was with an insertional vector containing the chloramphenicol acetyltransferase selectable marker and selection with chloramphenicol (36). Toxoplasma tachyzoites surviving this selection were induced to differentiate into bradyzoites in vitro for 4 days using a carbon starvation/low-serum/high-pH differentiation medium. Switch efficiency was monitored with antibodies specific to two bradyzoite-specific surface antigens, SAG2 X/Y and SRS9 (21). The cell population that was GFP positive but did not bind either bradyzoite-specific antibody was collected by FACS and recultured to repeat bradyzoite induction, antibody labeling, and FACS sorting twice more to enrich for parasites with a defect in tachyzoite-to-bradyzoite differentiation (Tbd−). Two independent mutagenesis procedures were performed, and a total of 114 Tbd− clones were selected.
Twenty-two of the individual mutant clones derived from the selected population by limiting dilution were tested for a Tbd− phenotype in vitro using alternative stimuli to induce differentiation: either the differentiation medium used during mutagenesis or the drug atovaquone, which has been shown previously to stimulate switching (37). These two methods of inducing Toxoplasma differentiation are thought to act by different mechanisms: atovaquone uncouples the electron transport chain in the parasites, whereas the differentiation medium acts through the host cells by relaying stress signals to the parasites (35). After being exposed to bradyzoite-inducing conditions (differentiation medium) for 4 days, 12 of the 22 clones were confirmed to have various degrees of Tbd− phenotype, ranging from a 20% to a 74% efficiency in differentiation relative to the WT (Fig. 1 and data not shown). Three of the confirmed Tbd− mutants with switch defects in differentiation medium had WT switch efficiencies when induced with atovaquone, indicating that switching is a complex response (Fig. 1B and C and data not shown). Although nine Tbd− mutants have consistent differentiation defects with both stress stimuli, these three mutants stand in contrast to previously published data in which Tbd− mutants, although obtained by high-pH selection, were also resistant to other inducers of bradyzoite formation (25, 33).
FIG. 1.
The Tbd− mutants show a range of phenotypes. (A) An immunofluorescence assay was used to score the ability of each mutant to differentiate in vitro. After 4 days of bradyzoite induction, the cultures were fixed and parasites were labeled with fluorescein Dolichos, which stains the cyst wall, and anti-SAG1, for the major tachyzoite surface antigen. Parasite cysts are identified by Dolichos staining and an absent or reduced SAG1 signal, while tachyzoite vacuoles lack Dolichos staining and express abundant SAG1. (B) The indicated Tbd− mutants were compared to the WT with respect to their ability to switch from the tachyzoite to bradyzoite form when grown for 96 h in differentiation medium (low CO2, low FCS, high pH). The fraction of each culture that switched to a bradyzoite pattern of gene expression was assessed by binding of the Dolichos bifloros lectin (specific for the cyst wall surrounding bradyzoites) or antibodies specific for the tachyzoite-specific antigen SAG1. Each bar represents two biological replicates with three technical replicates each, and error bars denote the standard deviation. (C) The same Tbd− mutants as in panel B were analyzed for differentiation in response to 900 nM atovaquone. Details are as described for panel B.
To identify the disrupted locus, plasmid rescue of the insertional vector was undertaken for two mutants with significant Tbd− phenotypes. One of these, TBD7, had an insertion within the region encoding the surface markers, SAG2C/D/X/Y, used in the selection and thus was not further pursued. The second, TBD8, has a single insertion of the mutagenesis vector 240 bp upstream of the predicted ORF designated 20.m03736 in ToxoDB 4.2 (http://www.toxodb.org) (Fig. 2A). PCR was used to confirm the insertion site (Fig. 2B). To determine if this disruption was within the 20.m03736 transcript, 5′ RACE was performed on bradyzoite cDNA. This indicated that the full 5′ UTR of this gene extends 1,030 bases upstream of the predicted start codon (data not shown), and thus the insertion site of the mutagenizing plasmid lies ∼790 nucleotides downstream of the transcription start site.
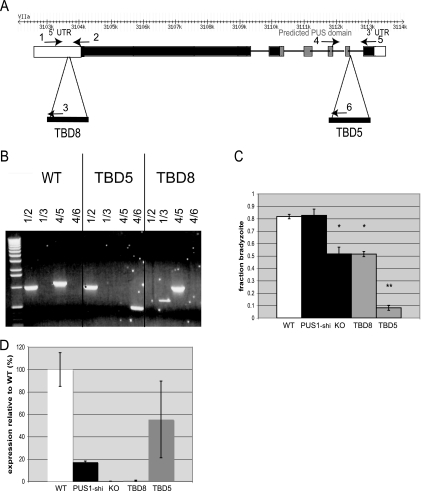
FIG. 2.
Two TBD mutants are the result of insertion in an apparent pseudouridine synthase gene. (A) The genomic locus of 20.m03736 comprises six exons located on chromosome VIIa and spanning 11 kb. The locus is comprised of a 1,030-bp 5′ UTR (white), a 2,121-amino-acid coding sequence (black) containing an RluD-like pseudouridine synthase (PUS) domain (gray), and a 350-bp 3′ UTR (white). TBD8 has a disruption 787 bp into the 5′ UTR, while TBD5 has a disruption immediately 3′ of the pseudouridine synthase domain in the indicated intron. (B) PCR from genomic DNA of the TBD5 and TBD8 mutants confirms the locations of the different mutations and distinguishes the PUS1 mutants. The numbers of the primers labeled on the genomic locus in panel A correspond to the number pairs for each lane. (C) Differentiation efficiency. The indicated mutants were exposed to high-pH switch conditions and assayed for switch efficiency as for Fig. 1. PUS1 KO is a knockout mutant in which the PUS1 locus has been deleted. PUS1-shi has a single, homologously integrated copy of the knockout vector but without deletion of the PUS1 locus. The bars represent three biological replicates, each with technical triplicates; error bars indicate standard deviations. *, P < 0.02; **, P < 0.005 (relative to WT). (D) Quantitative PCR was performed on cDNA from each strain to verify the molecular phenotype of each PUS1 disruption. TBD8 and PUS1 KO represent null alleles of PUS1, while TBD5 and PUS1-shi have less PUS1 transcript than WT parasites but are only slightly hypomorphic. The graph represents three independent experiments with technical triplicates normalized to a 0% to 100% range with the standard deviation of the cycle thresholds.
At the same time, investigation of a previously created mutant, TBD5, was undertaken (38). TBD5, which was also created by insertional mutagenesis of Pru strain Toxoplasma, has a differentiation efficiency of ∼10% relative to WT parasites, making it one of the strongest differentiation mutants of Toxoplasma yet described (38). Multiple attempts at plasmid rescue from this mutant have revealed only a single disruption with multiple, tandem copies of the mutagenesis vector inserted into the same ORF as in TBD8, i.e., 20.m03736. In the case of TBD5, the insertion occurred in the predicted final intron of the transcript (Fig. 2A and B). Thus, two independent mutants with a disruption of 20.m03736 had been selected with significant but varying defects in bradyzoite development, strongly indicating the importance of this gene in Toxoplasma differentiation.
To determine the full extent of the 20.m03736 gene, 5′ RACE, 3′ RACE, and RT-PCR were performed on bradyzoite cDNA from the WT parental strain (Fig. 2 and data not shown). The full 20.m03736 coding transcript is 6,363 nucleotides and spans six exons, producing a predicted 229-kDa protein of 2,121 amino acids; the 3′ UTR is 350 nucleotides. The predicted protein sequence matches the translation start and termination site of the 20.m03736 gene model, although two of the intron/exon boundaries were incorrect in that model. The PUS1 transcript was detectable by RT-PCR and quantitative RT-PCR in tachyzoites and parasites exposed to switch conditions in vitro for 2, 3, or 4 days; expression was low in all stages, with no significant difference seen between the tachyzoites and in vitro bradyzoites (data not shown).
BLAST analysis and motif finder algorithms identify an RluA family pseudouridine synthase domain (P = 4 × 10−15) at the carboxyl terminus of the 20.m03736 protein from amino acids 1864 to 2004. Pseudouridine synthases comprised all significant BLAST matches, and no other conserved motifs were found in the amino acid sequence. We have therefore designated this locus pseudouridine synthase 1 (PUS1). Pseudouridine synthases catalyze the conversion of the RNA base uridine to pseudouridine by isomerization of the uracil in RNA from a C1′-N1 linkage to a C1′-C5 linkage by rotation around the C3-C6 axis of the base. This protein contains all three conserved motifs found in pseudouridine synthases and the required catalytic aspartate of motif II that attacks the uracil being modified to remove the base from the nucleoside (Fig. 3) (6). Three other putative pseudouridine synthases exist in Toxoplasma; one of these proteins, 20.m03908, is a prototypical, short pseudouridine synthase, and the other two, 583.m05770 and 541.m02065, have a protein architecture similar to that of PUS1. The N terminus of the protein consists of ∼1,800 amino acids of unknown function. Using the Conserved Domain Database, four other proteins with architectures similar to that of PUS1 exist, and all are found in Apicomplexa, specifically Plasmodium and Cryptosporidium species. PUS1 is homologous to these other proteins only within the pseudouridine synthase domain and not across the extensive and as-yet-uncharacterized N-terminal region. The localization of PUS1 has not been investigated, and its predicted sequence does not provide a strong indication of where it might ultimately reside.
FIG. 3.
Alignment of the Toxoplasma PUS1 amino acid sequence with the sequences of five distinct RluD-like pseudouridine synthases. The five PUS proteins are RPUSD2 from the human genome, RluA-1 from Drosophila melanogaster, RluD from Arabidopsis thaliana, Pus9p from Saccharomyces cerevisiae, and RluC from Escherichia coli. Alignment analysis was performed using ClustalW. Alignment of weak amino acid groups is denoted by a period, conservation of a strong amino acid group is signified by a colon, and complete conservation of a residue is marked with an asterisk.
To understand the molecular phenotype of the mutations created in TBD5 and TBD8, a targeted Δpus1 knockout was created. A construct, pPUS1KO, containing the selectable marker HPT under the control of the DHFR promoter was used to replace PUS1 by flanking the HPT cassette with ∼2.5 kb of genomic sequence from either end of the PUS1 gene. This was electroporated into the parental Pru strain used to create TBD8. Only when parasites were transformed after having been kept extracellular overnight to promote bradyzoite gene expression was a Δpus1 clone found after HPT selection. These clones were identified by PCR to have experienced double homologous recombination and deletion of PUS1, as distinct from a single homologous insertion (PUS1-shi), which duplicates the PUS1 locus, leaving one copy completely intact. This serves as a useful control for the Δpus1 knockout strain. Utilizing stage-specific markers for tachyzoites and bradyzoites, the TBD8 and Δpus1 strains were found to have similar switch efficiencies (∼52% ± 5% and ∼52% ± 2%, respectively), compared to TBD5 (11% ± 2%) and the positive controls WT and PUS1-shi (∼82% ± 2% and 83% ± 5%, respectively) (Fig. 2C). Similarity in the switch phenotypes of the TBD8 and Δpus1 strains (Fig. 2C) suggests that TBD8 harbors a null mutation of PUS1 and TBD5 has a negative (antimorph) mutation that actively affects the parasites in an adverse way, analogous to a dominant negative mutation in a diploid system. Whether this negative phenotype is due to an aberrant product of the PUS1 locus in TBD5 or to an effect of one of the additional insertions of the mutagenizing vector is not yet known. Since the available data suggest that the additional insertions in TBD5 are all tandem and within the PUS1 locus, and since there is still a PUS1 transcript in this mutant (versus not in TBD8 or the Δpus1 strain [Fig. 2D]), we favor the antimorph explanation.
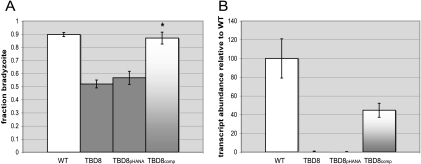
To ensure that it is the loss of a functional PUS1 in TBD8 rather than some artifactual result of the insertion that is responsible for creating a Tbd− phenotype, we complemented this strain with the full PUS1 genomic locus from position 3102000 to 3114000 of chromosome VIIa. Parasites were transformed with the complementation construct, pPUS1-HPT, or a control plasmid lacking PUS1, pHANA. The experiment was designed to achieve ectopic expression of the introduced gene rather than double homologous recombination and integration into the PUS1 locus. A complemented TBD8 mutant, TBD8comp, was obtained in this way and identified as having a stable, ectopic integration of the full PUS1 vector (data not shown). When exposed to in vitro switch conditions, the complemented strain reproduced WT levels of differentiation (Fig. 4A). The differentiation phenotype in the pHANA-transformed parasites was not significantly different from that in the TBD8 mutant itself (57% ± 5% versus 52% ± 3%, respectively). Transcript levels of PUS1 in TBD8comp appeared to be about half of WT levels, signifying that at least in regard to this phenotype, WT levels of PUS1 may normally be in modest excess (Fig. 4B). The restoration of WT differentiation in TBD8comp demonstrates that the absence of PUS1 is indeed responsible for the mutant Tbd− phenotype.
FIG. 4.
Complementation shows that PUS1 is indeed involved in the Tbd phenotype. (A) Clones of TBD8 parasites complemented with the genomic PUS1 locus, TBD8comp, or a control vector, TBD8pHANA were created. These parasites were induced to switch with differentiation medium for 4 days and labeled with the bradyzoite-specific cyst wall marker Dolichos bifluorescein and with the tachyzoite surface marker anti-SAG1. Fifteen fields were analyzed for tachyzoite vacuoles and bradyzoite cysts based on immunofluorescence. Three biological replicates with at least two technical replicates were performed and the data graphed with standard deviations. *, P < 0.005 relative to WT. (B) Quantitative PCR was performed on cDNA from each strain to verify the restoration of PUS1 transcription in TBD8comp. The graph represents two independent experiments with technical triplicates and the standard deviation of the cycle threshold.
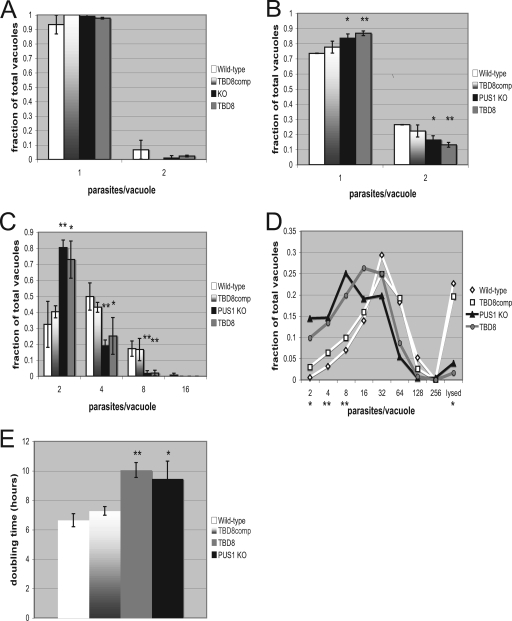
Mutation of individual pseudouridine synthases in Saccharomyces cerevisiae and Escherichia coli results in a slow-growth phenotype (13, 23, 28). To test for a slow-growth phenotype in Toxoplasma pus1 mutants, HFFs were infected with the WT, TBD8, TBD8comp, and Δpus1 strains at an MOI of 2 and allowed to grow for 4, 12, 24, and 48 h. The parasites were fixed, and the number of parasites per vacuole was counted in 15 fields. At 4 h postinfection, pus1 mutants had no noticeable growth difference compared to WT strains (Fig. 5). However, at 12 h postinfection, pus1 mutants began to show a delayed-growth phenotype, with significantly fewer vacuoles containing two parasites (P < 0.03 versus PUS1 KO and P < 0.01 versus TBD8) and significantly more vacuoles holding single parasites (P < 0.03 versus PUS1 KO and P < 0.01 versus TBD8). At 24 h there were significantly more two-parasite vacuoles in pus1 mutants (P < 0.01 versus PUS1 KO and P < 0.02 versus TBD8) and significantly fewer vacuoles containing four and eight parasites (P < 0.05 for TBD8 at four parasites/vacuole, P < 0.01 for PUS1 KO at four parasites/vacuole, and P < 0.01 for either strain at eight parasites/vacuole). At 48 h, this trend continued, with pus1 mutant Toxoplasma yielding significantly more vacuoles containing two, four, and eight parasites (P ≤ 0.02 for either pus1 mutant) and significantly fewer lysing vacuoles (P < 0.02 for either pus1 mutant). The doubling time calculated between the 12-hour time point and the 48-hour time point is 6.65 h per division for the WT and 7.28 h for TBD8comp. The doubling time is significantly increased for the TBD8 (10.06 h) and Δpus1 (9.46 h; P < 0.0004) strains. Calculations performed between the 12-hour and 24-hour time points or the 24-hour and 48-hour time points yield similar doubling rates and trends, indicating a consistently lower rate of replication in PUS1 mutants rather than just late invasion or a late start in replication upon invasion. Tbd− mutants have thus been characterized as having higher, equivalent, and, with these data, lower replication rates relative to WT (25, 38).
FIG. 5.
pus1 mutants replicate significantly more slowly than WT parasites. Cultures were infected and then analyzed at 4, 12, and 24 h postinfection (A, B, and C, respectively). The data represent three biological replicates done blinded in technical duplicates performed by counting the number of parasites per vacuole in 15 fields of view. Significance was calculated by grouping PUS1 WT strains versus pus1 mutant strains. *, P < 0.05; **, P < 0.01 (relative to the WT strain). (D) pus1 mutants continue to replicate significantly more slowly than WT parasites at 48 h. Parasites were counted and analyzed at 48 h postinfection as described for panels A, B, and C. Although the data do not represent a continuum between each increment (division within a given vacuole is synchronous), the data are presented as a line graph to allow easy comparison between the different strains over the full range of values. Significance was calculated by grouping PUS1 WT strains versus pus1 mutant strains at each vacuole size. *, P < 0.05; **, P < 0.01. (E) The doubling time between the 12- and 48-hour time points was calculated for each of the four strains. *, P < 0.02; **, P < 0.001.
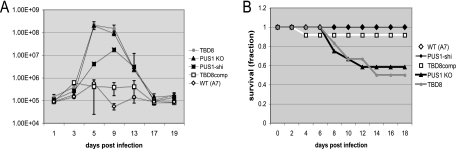
To test for an in vivo differentiation phenotype due to disruption of PUS1, mice were injected intraperitoneally with 500 Toxoplasma tachyzoites and the infection was tracked with bioluminescence imaging using Toxoplasma parasites constitutively expressing luciferase. For the five strains injected, A7, PUS1-shi, PUS1 KO, TBD8, and TBD8comp, the luminescence signal rose similarly up to day 3. By day 5 the signal intensity of the luminescence in the mice infected with PUS1 mutants had increased 100-fold over those in the WT and complemented control infections, indicating a massive increase in the parasite burden in the PUS1 mutant infections (Fig. 6A). The parasite burden in such mice peaked between day 5 and day 9 at just over 108 photons per second, compared to a peak intensity of ∼5 × 105 photons per second for WT-infected mice, and then began to decline steadily until the luminescence signal was no longer visible on day 21. The PUS1-shi-infected mice display an intermediate level of luminescence signal that may be due to a subtle effect of the single homologous integration at the PUS1 locus in this strain that leaves one intact copy of PUS1 but which does add DNA to the locus.
FIG. 6.
A more severe acute infection and lethality are observed in mice infected with PUS1 mutants. (A) Luminescence emitted from the body of the mouse, covering the head to the base of the tail, at the indicated days postinfection. The same three mice from each of two experiments, for a total of six mice per strain of parasite, were measured in this manner. Data are averages and standard deviations. (B) The survival of each infected mice was tracked daily during the infection and is shown as a Kaplan-Meier plot.
A strong luminescence signal from mice infected with the PUS1 mutant correlated strongly with increased mortality: using intraperitoneal infection and a dose of 500 tachyzoites, PUS1 mutant strains were significantly more lethal to mice than control strains (Fig. 6B), with TBD8 and PUS1 KO producing a lethal infection in approximately 50% of the mice between days 8 and 14 postinfection.
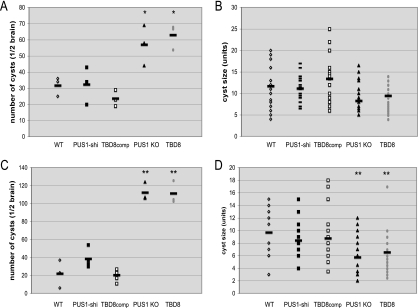
The effects of PUS1 mutagenesis on the chronic infection were investigated at 4 and 9 wpi. At 4 wpi there was a significantly increased number of cysts in mice infected with the PUS1 mutants compared to those infected with the WT or complemented mutants (P < 0.05 for TBD8 or PUS1 KO versus WT) (Fig. 7A). The sizes of the cysts were not significantly different in any strain used in the infections (Fig. 7B). At 9 wpi, the cyst burden in PUS1 mutant-infected mice was approximately 110 cysts per half mouse brain, a twofold increase over the number at 4 wpi, while the number of cysts in WT-infected mice did not increase, remaining at an average of ∼30 cysts per half brain (Fig. 7C). The difference in cyst burden between the WT and TBD8 or PUS1 KO is highly significant (P ≤ 0.01). Additionally, the cysts in mice infected with PUS1 mutants at 9 wpi were significantly smaller than those infected with WT or complemented parasites (Fig. 7D).
FIG. 7.
PUS1 mutants produce more and smaller cysts during the chronic infection. (A) Brains were harvested from infected mice at 4 wpi, and half of each brain was homogenized by passage through a 100-μm cell strainer. The homogenate was plated out, and cysts were counted only if they were labeled with the bradyzoite-specific cyst wall marker rhodamine-conjugated Dolichos bifloros agglutinin and expressed parasite-derived GFP. The results for the brains of three mice are represented as scatter plots for the mean cyst number. *, P < 0.05. (B) Images of the first seven cysts from each mouse analyzed at 4 wpi were captured, for a total of 21 images for each strain. The diameter of each cyst was measured using the obtained images, and results are represented as a scatter plot of the mean cyst diameter. (C) Cyst counts at 9 wpi. Details are as described for panel A. **, P ≤ 0.01. (D) Cyst size at 9 wpi. Details are as described for panel B. **, P ≤ 0.01.
DISCUSSION
Tachyzoite-to-bradyzoite conversion is a critical process in Toxoplasma gondii development, yet little is understood about the genetic mechanisms involved (41). Our results confirm that insertional mutagenesis is a potent method to create stable Tbd− mutants and identify genes involved in the developmental process. Interestingly, as with other studies, all of the mutants obtained here had a leaky phenotype, with at least some parasites still responding to the stimuli and differentiating to bradyzoites (18, 25, 33, 38). This suggests a branched and complex differentiation pathway. The ability of 3 of the 12 known TBD mutants (selected using high-pH conditions) to differentiate at WT levels when induced to differentiate with atovaquone further supports the idea of a complex differentiation pathway where distinct stimuli trigger different signaling cascades to initiate bradyzoite development.
Our results indicate that PUS1, a protein containing a pseudouridine synthase domain, is central to the process of Toxoplasma differentiation. PUS1 shows a high degree of conservation, including the key catalytic residues, in all three domains conserved in known pseudouridine synthases. No other enzymatic activity has ever been described for such a domain, and no sequence other than that from a pseudouridine synthase in the GenBank database showed significant homology to PUS1. Unfortunately, pseudouridine synthases are highly substrate specific (15, 19), and so establishing activity requires knowing the particular substrate that is acted upon. This can be determined only by in-depth biochemical analyses, leaving the question of whether PUS1 is indeed a pseudouridine synthase not yet proven. The arguments above, however, make this highly probable.
Pseudouridine synthases are found in all sequenced organisms from Archaea to chordates, where they catalyze the isomerization of uracil in uridine to create pseudouridine in RNA (8). Pseudouridine synthases modify RNAs whose structure is important for their function, such as tRNAs, rRNAs, snRNAs, and slRNAs (27). Pseudouridylation increases the stability of RNA folding by rigidifying the sugar-phosphate backbone, enhancing base stacking and coordinating water molecules in the RNA structure (40).
Previously described TBD mutants have demonstrated an equivalent or even accelerated growth compared to the WT parent under tachyzoite conditions (25, 33). In contrast, all Tbd− PUS1 mutants display a slow-growth phenotype under tachyzoite conditions. Mutation of various PUS genes in Saccharomyces cerevisiae and E. coli including mutations in rluA, the closest homologue to PUS1 in E. coli, also produce slow-growth phenotypes, consistent with PUS1 being a true pseudouridine synthase (7, 13, 14, 24). The cause for reduced growth of rluA mutants is unknown, although reduced translational capacity and a decrease in the rate of the aminoacyl-tRNA selection step during translation depending on the identity of the tRNA have been observed in rluD and truA E. coli mutants, respectively (7).
We have selected or engineered a total of three PUS1 mutants, which all display defects in tachyzoite-to-bradyzoite conversion. The Δpus1 and, apparently, TBD8 mutants are null mutants of PUS1 and show a 50% defect in differentiation. The phenotype of TBD5 is more severe, differentiating with only ∼10% efficiency. The suspected reason for this more extreme phenotype is that TBD5 appears to contain an antimorph mutation, where the resulting protein might bind but not release the RNA substrate, or a neomorph mutation, where the PUS1 protein now acts on novel RNAs or modifies different positions in the original RNA substrate. Such a new activity could be due to insertion into the 3′ end of the PUS1 gene of TBD5, producing a new carboxyl terminus of PUS1 that alters the protein's structure and thus its interaction with its RNA substrates. Alternatively, the presence of additional insertions in the TBD5 mutant could be playing a role in producing an enhanced phenotype. Regardless, the data presented here establish unequivocally a pivotal role for PUS1 in the tachyzoite-to-bradyzoite differentiation phenotype.
The role for PUS1 in differentiation was additionally confirmed in vivo. The PUS1 mutant-infected mice both showed a more intense acute infection and an increase in the number but not size of brain cysts (in fact, they were significantly smaller) during the chronic phase of infection. This could be due to the equilibrium of differentiation being shifted toward the more virulent tachyzoite form, producing more severe disease in the acute stages and greater numbers of parasites to establish cyst burdens in the chronic stages. The decreased size of the cysts in these mutants might be due to a slower growth in the bradyzoite stage and/or a tendency to switch back to tachyzoites during the chronic stages, yielding a consistently younger age of the cysts.
Mutations in a pseudouridine synthase, dyskerin, in mammals are associated with extreme cellular hyperproliferation, a defective differentiation response (16, 29). This is consistent with the identified role of PUS1 in cellular differentiation of Toxoplasma and suggests a broadly conserved biological role for these enzymes beyond the simple biochemical action of modifying a nucleobase.
Acknowledgments
We are grateful for the gifts of antibodies to SAG2X/Y and SRS9 from Jeroen Saeij and from Seon-Kyeong Kim, respectively; for the gift of anti-Toxoplasma mouse serum from Anita Koshy; for the assistance of Tim Knaack with fluorescence-activated cell sorting; and for crucial technical and theoretical assistance by Jon Boyle. We thank all the members of the Boothroyd lab for many helpful discussions and suggestions.
This work was supported by grants from the NIH (AI41014) to J.C.B. and the Stanford Genome Training Program (NIH 5 T32 HG00044) and by a Ford Foundation Predoctoral Diversity Fellowship from The National Academy of Sciences and a Stanford University DARE Fellowship to M.Z.A.
Footnotes
Published ahead of print on 5 January 2009.
REFERENCES
- 1.Black, M., F. Seeber, D. Soldati, K. Kim, and J. C. Boothroyd. 1995. Restriction enzyme-mediated integration elevates transformation frequency and enables co-transfection of Toxoplasma gondii. Mol. Biochem. Parasitol. 7455-63. [DOI] [PubMed] [Google Scholar]
- 2.Black, M. W., and J. C. Boothroyd. 1998. Development of a stable episomal shuttle vector for Toxoplasma gondii. J. Biol. Chem. 2733972-3979. [DOI] [PubMed] [Google Scholar]
- 3.Bohne, W., J. Heesemann, and U. Gross. 1993. Induction of bradyzoite-specific Toxoplasma gondii antigens in gamma interferon-treated mouse macrophages. Infect. Immun. 611141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bohne, W., J. Heesemann, and U. Gross. 1994. Reduced replication of Toxoplasma gondii is necessary for induction of bradyzoite-specific antigens: a possible role for nitric oxide in triggering stage conversion. Infect. Immun. 621761-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohne, W., U. Gross, D. J. Ferguson, and J. Heesemann. 1995. Cloning and characterization of a bradyzoite-specifically expressed gene (hsp30/bag1) of Toxoplasma gondii, related to genes encoding small heat-shock proteins of plants. Mol. Microbiol. 161221-1230. [DOI] [PubMed] [Google Scholar]
- 6.Boothroyd, J. C., M. Black, S. Bonnefoy, A. Hehl, L. J. Knoll, I. D. Manger, E. Ortega-Barria, and S. Tomavo. 1997. Genetic and biochemical analysis of development in Toxoplasma gondii. Philos. Trans. R. Soc. Lond. B 3521347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carbone, M. L., M. Solinas, S. Sora, and L. Panzeri. 1991. A gene tightly linked to CEN6 is important for growth of Saccharomyces cerevisiae. Curr. Genet. 191-8. [DOI] [PubMed] [Google Scholar]
- 8.Charette, M., and M. W. Gray. 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49341-351. [DOI] [PubMed] [Google Scholar]
- 9.Cleary, M. D., U. Singh, I. J. Blader, L. J. Brewer, and J. C. Boothroyd. 2002. Toxoplasma gondii asexual development: identification of developmentally regulated genes and distinct patterns of gene expression. Eukaryot. Cell 3329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleary, M. D., C. D. Meiering, E. Jan, R. Guymon, and J. C. Boothroyd. 2005. Biosynthetic labeling of RNA using uracil phosphoribosyltranferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat. Biotechnol. 23232-237. [DOI] [PubMed] [Google Scholar]
- 11.Dubey, J. P. 1997. Bradyzoite-induced murine toxoplasmosis: stage conversion, pathogenesis, and tissue cyst formation in mice fed bradyzoites of different strains of Toxoplama gondii. J. Eukaryot. Microbiol. 44592-602. [DOI] [PubMed] [Google Scholar]
- 12.Fux, B., J. Nawas, A. Khan, D. B. Gill, C. Su, and L. D. Sibley. 2007. Toxoplasma gondii strains defective in oral transmission are also defective in developmental stage differentiation. Infect. Immun. 752580-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutgsell, N. S., N. Englund, L. Niu, Y. Kaya, B. G. Lane, and J. Ofengand. 2000. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 61870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutgsell, N. D., M. D. Del Campo, S. Raychaudhuri, and J. Ofengand. 2001. A second function for pseudouridine synthases: a point mutant of RluD unable to form pseudouridines 1911, 1915, and 1917 in Escherichia coli 23S ribosomal RNA restores normal growth to an RluD-minus strain. RNA 7990-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamma, T., and A. R. Ferré-D'Amaré. 2006. Pseudouridine synthases. Chem. Biol. 131125-1135. [DOI] [PubMed] [Google Scholar]
- 16.Heiss, N. S., S. W. Knight, T. J. Vulliamy, S. M. Klauck, S. Wiemann, P. J. Mason, A. Poustka, and I. Dokal. 1998. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1932-38. [DOI] [PubMed] [Google Scholar]
- 17.Hitziger, N., N. I. Dellacasa, B. Albiger, and A. Barragan. 2005. Dissemination of Toxoplasma gondii to immunoprivileged organs and role of Toll interleukin-1 receptor signaling for host resistance assessed by in vivo bioluminescence imaging. Cell. Microbiol. 7837-848. [DOI] [PubMed] [Google Scholar]
- 18.Holpert, M., U. Gross, and W. Bohne. 2006. Disruption of the bradyzoite-specific P-type (H+)-ATPase PMA1 in Toxoplasma gondii leads to decreased bradyzoite differentiation after stress stimuli but does not interfere with mature tissue cyst formation. Mol. Biochem. Parasitol. 146129-133. [DOI] [PubMed] [Google Scholar]
- 19.Kaya, Y., and J. Ofengand. 2003. A novel unanticipated type of pseudourdine synthase with homologs in bacteria archaea and eukarya. RNA 9711-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, S. K., A. Karasov, and J. C. Boothroyd. 2007. Bradyzoite-specific surface antigen SRS9 plays a role in maintaining Toxoplasma gondii persistence in the brain and in host control of parasite replication in the intestine. Infect. Immun. 751626-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S. K., and J. C. Boothroyd. 2005. Stage-specific expression of surface antigens by Toxoplasma gondii as a mechanism to facilitate parasite persistence. J. Immunol. 1748038-8044. [DOI] [PubMed] [Google Scholar]
- 22.Knoll, L. J., and J. C. Boothroyd. 1998. Isolation of developmentally regulated genes from Toxoplasma gondii by a gene trap with the positive and negative selectable marker hypoxanthine-xanthine-guanine phosphoribosyltransferase. Mol. Cell. Biol. 18807-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecointe, F., O. Namy, I. Hatin, G. Simos, J. Rousset, and H. Grosjean. 2002. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 27634934-34940. [DOI] [PubMed] [Google Scholar]
- 24.Lecointe, F., G. Simos, A. Sauer, E. C. Hurt, Y. Motorin, and H. Grosjean. 1998. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J. Biol. Chem. 2731316-1323. [DOI] [PubMed] [Google Scholar]
- 25.Matrajt, M., R. G. Donald, U. Singh, and D. S. Roos. 2002. Identification and characterization of differentiation mutants in the protozoan parasite Toxoplasma gondii. Mol. Microbiol. 44735-747. [DOI] [PubMed] [Google Scholar]
- 26.Montoya, J., and O. Liesenfield. 2004. Toxoplasmosis. Lancet 3631965-1976. [DOI] [PubMed] [Google Scholar]
- 27.Nishikura, K., and E. M. De Robertis. 1981. RNA processing in microinjected Xenopus oocytes sequential addition of base modifications in a spliced transfer RNA. J. Mol. Biol. 145405-420. [DOI] [PubMed] [Google Scholar]
- 28.Raychaudhuri, S., J. Conrad, B. G. Hall, and J. Ofengand. 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 41407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruggero, D., S. Grisendi, F. Piazza, E. Rego, F. Mari, P. H. Rao, C. Cordon-Cardo, and P. P. Pandolfi. 2003. Dyskeratosis congenital and cancer in mice deficient in ribosomal RNA modification. Science 299259-262. [DOI] [PubMed] [Google Scholar]
- 30.Saeij, J. P., J. P. Boyle, M. E. Grigg, G. Arrizabalaga, and J. C. Boothroyd. 2005. Bioluminescence imaging of Toxoplasma gondii infection in living mice reveals cramatic differences between strains. Infect. Immun. 73695-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saeij, J. P., G. Arrizabalaga, and J. C. Boothroyd. 2008. A cluster of four surface antigen genes specifically expressed in bradyzoites, SAG2C/D/X/Y, plays an important role in Toxoplasma gondii persistence. Infect. Immun. 762402-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz, J. A., A. E. Fouts, C. A. Cummings, D. J. Ferguson, and J. C. Boothroyd. 2005. A novel rhoptry protein in Toxoplasma gondii bradyzoites and merozoites. Mol. Biochem. Parasitol. 144159-166. [DOI] [PubMed] [Google Scholar]
- 33.Singh, U., J. L. Brewer, and J. C. Boothroyd. 2002. Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction. Mol. Microbiol. 44721-733. [DOI] [PubMed] [Google Scholar]
- 34.Soete, M., and J. F. Dubremetz. 1996. Toxoplasma gondii: kinetics of stage-specific protein expression during tachyzoite-bradyzoite conversion in vitro. Curr. Top. Microbiol. Immunol. 21976-80. [PubMed] [Google Scholar]
- 35.Soete, M., B. Fortier, D. Camus, and J. F. Dubremetz. 1993. Toxoplasma gondii kinetics of bradyzoite interconversion in vitro. Exp. Parasitol. 76259-264. [DOI] [PubMed] [Google Scholar]
- 36.Soldati, D., and J. C. Boothroyd. 1993. Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science 260349-352. [DOI] [PubMed] [Google Scholar]
- 37.Tomavo, S., and J. C. Boothroyd. 1995. Interconnection between organellar functions, development and drug resistance in the protozoan parasite, Toxoplasma gondii. Int. J. Parasitol. 251293-1299. [DOI] [PubMed] [Google Scholar]
- 38.Vanchinathan, P., J. L. Brewer, O. S. Harb, J. C. Boothroyd, and U. Singh. 2005. Disruption of a locus encoding a nucleolar zinc finger protein decreases tachyzoite-to-bradyzoite differentiation in Toxoplasma gondii. Infect. Immun. 736680-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss, L. M., and K. Kim. 2000. The development and biology of bradyzoites of Toxoplasma gondii. Front. Biosci. 5391-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson, J. R. 2000. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 7834-837. [DOI] [PubMed] [Google Scholar]
- 41.Wong, S. Y., and J. S. Remington. 1993. Biology of Toxoplasma gondii. AIDS 7299-316. [DOI] [PubMed] [Google Scholar]
- 42.Yang, S., and S. F. Parmley. 1997. Toxoplasma gondii expresses two distinct lactate dehydrogenase homologous genes during its life cycle in intermediate hosts. Gene 1841-12. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, Y. W., S. K. Halonen, Y. F. Ma, M. Wittner, and L. M. Weiss. 2001. initial characterization of CST1, a Toxoplasma gondii cyst wall glycoprotein. Infect. Immun. 69501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reference deleted.