Abstract
We report characterization of the gene encoding putative transcription factor PRO1, identified in transcriptional profiling studies as being downregulated in the chestnut blight fungus Cryphonectria parasitica in response to infection by virulence-attenuating hypoviruses. Sequence analysis confirmed that pro1 encodes a Zn(II)2Cys6 binuclear cluster DNA binding protein with significant sequence similarity to the pro1 gene product that controls fruiting body development in Sordaria macrospora. Targeted disruption of the C. parasitica pro1 gene resulted in two phenotypic changes that also accompany hypovirus infection, a significant reduction in asexual sporulation that could be reversed by exposure to high light intensity, and loss of female fertility. The pro1 disruption mutant, however, retained full virulence. Although hypovirus CHV1-EP713 infection was established in the pro1 disruption mutant, infected colonies continually produced virus-free sectors, suggesting that PRO1 is required for stable maintenance of hypovirus infection. These results complement the recent characterization of the hypovirus-responsive homologue of the Saccharomyces cerevisiae Ste12 C2H2 zinc finger transcription factor gene, cpst12, which was shown to be required for C. parasitica female fertility and virulence.
Reduced virulence (hypovirulence) of the chestnut blight fungus, Cryphonectria parasitica, caused by viruses in the family Hypoviridae (hypoviruses), is accompanied by reduced conidiation and loss of female fertility (reviewed in references 3, 21, and 28). These deficiencies in the reproductive capacity of hypovirus-infected C. parasitica strains restrict transmission of hypoviruses through fungal populations and thus reduce the efficacy of hypovirus-mediated biological control of chestnut blight (discussed in reference 24). It has been possible to partially uncouple hypovirulence from reduced conidiation by creating chimeric hypoviruses from mild and severe virus isolates (10). However, a basic understanding of the regulatory pathways that govern C. parasitica asexual and sexual reproduction would provide additional tools for enhancing hypovirus-mediated biological control potential.
Our laboratory recently reported that the C. parasitica homologue of the Saccharomyces cerevisiae Ste12 C2H2 zinc finger transcription factor gene, cpst12, identified by microarray analysis as downregulated following hypovirus infection (1, 2), was required for female fertility and virulence (16). A gene encoding a second putative transcription factor with sequence similarity to the Zn(II)2Cys6 transcription factor PRO1 of Sordaria macrospora was also found to be downregulated following infection by mild and severe hypovirus isolates CHV1-Euro7 and CHV1-EP713, respectively (2). Interestingly, the S. macrospora pro1 gene is required for female fertility (22, 23), and the Neurospora crassa pro1 homologue was able to rescue the female infertility phenotype when introduced into the S. macrospora pro1 deletion mutant (23).
Thus, it was of interest to further characterize the hypovirus-responsive C. parasitica pro1 gene to determine whether this putative transcription factor is also involved in C. parasitica virulence and sexual reproduction. Targeted disruption of the C. parasitica pro1 gene confirmed that it is required for female fertility but not for virulence. Surprisingly, the disruption mutant also exhibited a light-reversible defect in asexual spore development and a propensity to escape hypovirus infection. These results are discussed in terms of the role of host transcription factors in the elaboration of hypovirus-mediated alterations of host gene expression and phenotype.
MATERIALS AND METHODS
Fungal strains, culture conditions, and phenotypic measurements.
The C. parasitica strains used in this study (Table 1) were maintained on potato dextrose agar (PDA; Difco, Detroit, MI) at 22 to 24°C with a 12-h/12-h light/dark cycle and a light intensity of 1,300 to 1,600 lx. Cultures used for RNA preparations were grown for 7 days under similar conditions on cellophane overlaying PDA (PDA-cellophane). Potato dextrose broth (Difco, Detroit, MI) was used for liquid cultures, essentially as described previously (12), except that cultures were incubated for 48 h on an orbital shaker set at a rotation speed of 110 rpm. Hyphal anastomosis (hyphal fusion assay) for examination of virus transmission, susceptibility, and stability (31) was performed by inoculating strain EP713, which contains hypovirus CHV1-EP713, and virus-free strain EP155 or the pro1 disruption mutant strains, spaced approximately 2 cm apart on PDA plates, and incubating them under the conditions described above. As the two fungal colonies grow, the vegetatively compatible hyphae merge and fuse, resulting in cytoplasmic exchange and transmission of the CHV1-EP713 hypovirus to the virus-free strain. The transmitted virus spreads into the rapidly growing hyphae at the expanding edge of the recipient strain colony. Since the presence of hypovirus CHV1-EP713 results in phenotypic changes in host colony morphology, e.g., a loss of orange pigmentation and reduced aerial hyphae, the virus-infected (converted) mycelia of the recipient strain can be easily distinguished and isolated from the unconverted, virus-free mycelia. Asexual sporulation measurements, virulence assays with dormant chestnut tree stems, and statistical analysis were performed as described by Deng et al. (16), with the modification that sporulation was determined as the number of spores per ml instead of per cm2. Images of fungal colonies were captured using a Discovery V12 stereomicroscope (Carl Zeiss SMT, Inc., Thornwood, NY).
TABLE 1.
Cryphonectria parasitica strains used in this study
| Strain | Characteristic(s) | Source or reference |
|---|---|---|
| EP155 | Wild type; mating type Mat-2 | ATCC 38755 |
| EP713 | EP155 infected with hypovirus CHV1-EP713 | ATCC 52571 |
| EP146 | Wild type; mating type Mat-1 | ATCC 64671 |
| Δpro1T3 | pro1 disruption mutant no. 3 in EP155 genetic background | This study |
| Δpro1T6 | pro1 disruption mutant no. 6 in EP155 genetic background | This study |
| Δpro1T6/pG1 | pro1 disruption mutant Δpro1T6 strain complemented with pro1 gene plasmid pGPRO1 | This study |
| Δcpst12 strain | cpst12 disruption mutant in EP155 genetic background | 16 |
Identification and disruption of the C. parasitica pro1 gene.
A portion of the C. parasitica pro1 gene was amplified with the primer set comprising pro1-5 (5′-GCCATACGAGATTGACGTCAA-3′) and pro1-3 (5′-GTTGAAATGAAGGGTTAGTC-3′), based on the sequence of the C. parasitica expressed sequence tag cDNA clone CEST-36-H-03, identified in microarray analyses as a hypovirus-responsive gene (1), and a homologue of the S. macrospora pro1 gene involved in fruiting body formation (23). The entire pro1 gene was assembled with additional sequencing of PCR products and the sequence information from the C. parasitica draft genome produced by the Joint Genome Institute (http://genome.jgi-psf.org/Crypa1/Crypa1.home.html). The predicted intron/exon junctions were confirmed by sequencing PCR-amplified pro1 cDNA generated from a C. parasitica total RNA preparation. Phylogenetic analysis was performed using the CLUSTALW online program (http://align.genome.jp/), and the amino acid sequences of PRO1 and closely related Zn(II)2Cys6 transcription factors were aligned with Lasergene (DNASTAR, Inc., Madison, WI).
Disruption of the C. parasitica pro1 gene was performed by the PCR-based strategy described by Kuwayama et al. (19). The putative DNA activation domain was replaced with a gene cassette conferring G418 resistance in a process involving three rounds of PCR. The 5′ and 3′ ends of the pro1 gene were amplified by the primer set comprising Pro1-KF1 (5′-CCGTCACTGATGGCTTTCC-3′) and Pro1-KR1 (5′-TTCGCCCTATCCAACATGGTGATCTGCTCCGTCCATTCGCC-3′) for the 5′ terminus and the primer set comprising Pro1-KF2 (5′-AGTGTCTACTGCTGGCGTCGACGGCTCCGATCTCATCCTGC-3′) and Pro1-KR2 (5′-GTTGAAATGAAGGGTTAGTC-3′) for the 3′ terminus, using EP155 genomic DNA as a template. Primers Neo-F (5′-TCACCATGTTGGATAGGGCGAA-3′) and Neo-R (5′-GTCGACGCCAGCAGTAGACACT-3′) were used to amplify the G418 resistance gene cassette, consisting of the neomycin phosphotransferase gene flanked by the Cochliobolus heterostrophus glyceraldehyge-3-phosphate dehydrogenase gene promoter and the N. crassa β-tubulin gene terminator, from pSK666 (a kind gift from Seogchan Kang at the Pennsylvania State University). The portions of primers Pro1-KR1 and Pro1-KF2 corresponding to Neo-F and Neo-R are underlined. The amplified products were gel purified with a QIAquick gel extraction kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. A second round of PCR was performed (10 cycles consisting of denaturation at 94°C for 20 s, annealing at 55°C for 10 min, and extension at 68°C for 10 min) to assemble the three fragments. Primers Pro1-KF1 and Pro1-KR2 were used to amplify the full-length disruption construct in a third round of PCR (30 cycles consisting of denaturation at 94°C for 20 s, annealing at 58°C for 30 s, and extension at 72°C for 6 min). This full-length disruption construct was gel purified and used to transform spheroplasts of C. parasitica strain EP155 according to the method of Churchill et al. (14), followed by selection of putative transformants in the presence of 20 μg/ml of G418. Putative disruptants were placed under intense constant-light conditions (∼4,000 lx) to promote asexual sporulation (18), followed by selection of uninuclear single conidial isolates on G418-containing PDA to eliminate heterokaryons. Disruption of the pro1 gene in single-spored transformants was confirmed by Southern blot hybridization and PCR analyses.
DNA isolation and Southern blot analysis.
Cultures grown in 30 ml potato dextrose broth for 2 days were harvested, frozen in liquid nitrogen, and ground into a fine powder by using a mortar and pestle. Total genomic DNA was extracted with phenol-chloroform and then precipitated with ethanol. Ten micrograms of genomic DNA was digested with SacI (New England Biolabs, Ipswich, MA) and separated by electrophoresis in 0.8% agarose gel. After electrophoresis, the gel was denatured and DNA was transferred to a Hybond-N+ nylon membrane (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). The transferred DNA was hybridized with a 0.4-kb pro1-specific probe that was amplified by PCR from EP155 genomic DNA, using a PCR DIG probe synthesis kit (Roche Diagnostics, Mannheim, Germany). The entire procedure from prehybridization to signal detection was performed with a digoxigenin detection kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's protocols.
Functional complementation of the pro1 disruption mutant.
A genomic DNA copy of the full-length pro1 gene, the entire protein-coding sequence plus the single intron, was amplified from C. parasitica genomic DNA by using Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA). The PCR product was digested with KpnI, and the digested fragment (2,431 bp) was cloned into the C. parasitica expression vector pCPXHY1 (13) to generate pGPRO1. In this plasmid, the pro1 gene open reading frame is under the control of the promoter and terminator elements of the C. parasitica glyceraldehyde-3-phosphate dehydrogenase gene (gpd1), and the hygromycin resistance gene cassette is derived from plasmid pUCDH25 (15). Plasmid pGPRO1 was used to transform pro1 disruption mutant 6 (Δpro1T6), resulting in the selection of the complemented Δpro1T6/pG1 strain.
Mating and ascospore germination analysis.
Mating was performed on autoclaved twigs of American chestnut tree embedded in 2% water agar as previously described (4). The pro1 disruption mutant Δpro1T3 and Δpro1T6 strains (mating type Mat-2 [7]) were tested as both the male and the female in crosses with strain EP146 (mating type Mat-1 [7]). Reciprocal EP155 × EP146 crosses served as controls. The bark of autoclaved chestnut tree twigs was cut longitudinally multiple times with a scalpel prior to inoculation to ensure effective colonization by the strains serving as female parents in the cross. The colonized twigs were incubated at 20° to 22°C following spermatization with conidia from the male parent until perithecia could be observed in the stroma.
Mature perithecia were detached from the stroma with a dissecting needle. Each perithecium was rolled over 4% water agar to remove stromatic debris. Perithecia were then transferred to microscope slides, washed with sterile, distilled water, and squashed with a blunt glass rod. Ascospores were carefully collected and transferred to a centrifuge tube and counted with the aid of a hemacytometer. Serial dilutions of the spore suspensions were plated on 2% water agar and incubated at 28°C. Spore germination was monitored with a dissecting microscope (Bausch & Lomb, Rochester, NY) after 24 to 36 h.
RNA isolation and semiquantitative real-time RT-PCR.
Total RNA was prepared from cultures grown on PDA-cellophane as described previously (30). For samples used for sexual stage RNA preparation, the entire stroma, including perithecia formed from crosses between strains EP155 (as the female) and EP146 (as the male), were removed from the chestnut tree twigs with a scalpel. Real-time reverse transcription-PCR (RT-PCR) analysis of pro1 gene transcript accumulation was performed with TaqMan reagents (Eurogentec, Seraing, Belgium) and a model 7300 real-time PCR system (Applied Biosystems, Foster City, CA) as described previously (30). Probes and oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Real-time RT-PCR was performed in triplicate for each of at least two independent RNA preparations with primers and probes specific for cDNAs of 18S rRNA and the target gene. Transcript abundance relative to the amount of 18S rRNA in the sample was calculated by using the comparative threshold cycle method with primers and conditions described by Parsley et al. (30).
Nucleotide sequence accession numbers.
The pro1 and pro41 nucleotide sequences and predicted amino acid sequences were submitted to GenBank under accession no. FJ348264 and FJ348265, respectively.
RESULTS
Sequence analysis and expression of the C. parasitica pro1 gene.
Sequencing of overlapping DNA fragments of the pro1 gene amplified by PCR from C. parasitica genomic DNA and from corresponding mRNA revealed a 2,163-bp open reading frame interrupted by a single 259-bp intron. The coding domain was predicted to encode a polypeptide consisting of 720 amino acids with a molecular mass of 80 kDa and a high level of sequence identity (67%) with PRO1 of S. macrospora (22), which included the typical GAL4-like Zn(II)2Cys6 binuclear cluster DNA-binding domain and an activation domain within the N-terminal region.
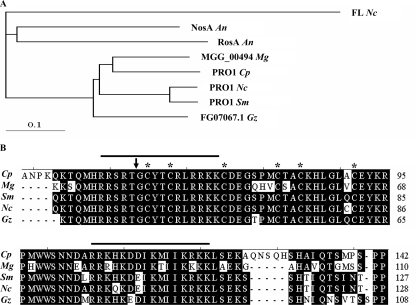
An alignment of the PRO1 Zn(II)2Cys6 binuclear cluster domain with that of putative orthologues from closely related fungal species showed a high level of sequence conservation that included the six cysteine residues involved in coordination of two zinc ions and two putative nuclear localization signal domains (Fig. 1B). Comparative analysis of the PRO1 amino acid sequence with orthologues previously implicated in fungal reproduction and from fungal species closely related phylogenetically to C. parasitica revealed the highest level of conservation with orthologues from the plant pathogenic fungal species Gibberella zeae and Magnaporthe grisea and the nonpathogen N. crassa (Fig. 1A), consistent with conclusions drawn in a recent large-scale C. parasitica expressed sequence tag analysis (32). The identical location of the single conserved intron in the pro1 orthologue in each of five related ascomycetes (Fig. 1B) suggests a common ancestry.
FIG. 1.
Comparative sequence analysis of the predicted amino acid sequences of the C. parasitica pro1 gene and homologous genes reported to function in fungal sexual development or from closely related plant pathogenic fungi. (A) Phylogram of predicted amino acid sequences of pro1 gene orthologues. The tree was generated by CLUSTALW (http://align.genome.jp/) with the default parameter setting, using the full-length amino acid sequence. FL Nc, N. crassa fluffy protein (GenBank accession no. AAB80932); NosA An, A. nidulans NosA protein (GenBank accession no. CAJ76908); RosA An, A. nidulans RosA protein (GenBank accession no. CAD58393); MGG_00494 Mg, M. grisea hypothetical protein MGG_00494 (GenBank accession no. XP_368750); PRO1 Cp, C. parasitica PRO1 protein (GenBank accession no. FJ348264); PRO1 Nc, N. crassa PRO1 protein (GenBank accession no. CAB89819); PRO1 Sm, S. macrospora PRO1 protein (GenBank accession no. CAB52588); FG07067.1 Gz, G. zeae hypothetical protein FG07067.1 (GenBank accession no. XP_387243). The 0.1 scale bar shows 10% sequence aberration. (B) Alignment of the Zn(II)2Cys6 fungal binuclear cluster motif for five clustered PRO1 orthologues. The alignment was performed with MegAlign in Lasergene, using the default setting. Cp, C. parasitica PRO1 protein; Gz, G. zeae hypothetical protein FG07067.1; Mg, M. grisea hypothetical protein MGG_00494; Nc, N. crassa PRO1 protein; Sm, S. macrospora PRO1 protein. The cysteine residues involved in the coordination of the Zn2+ atoms are highlighted with an asterisk. The position of the conserved intron is indicated by an arrowhead. The two predicted bipartite nuclear localization signals are indicated with a line above the sequence.
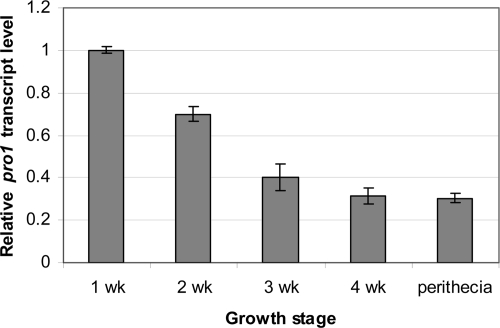
Semiquantitative real-time RT-PCR analysis showed that pro1 expression levels are highest during the early growth phase and decrease with increasing time of culturing (Fig. 2). A modest level of pro1 transcript accumulation was also observed in sexual fruiting body tissues (Fig. 2). We also confirmed earlier reports (1, 2) that pro1 transcript accumulation is reduced severalfold following hypovirus infection (data not shown).
FIG. 2.
Growth and tissue-specific accumulation of pro1 gene transcripts. Real-time RT-PCR analysis was performed as described in Materials and Methods for RNA isolated from wild-type C. parasitica strain EP155 cultures harvested from PDA-cellophane plates at weekly intervals and from perithecia collected from chestnut tree twigs. The values were normalized to the pro1 transcript accumulation level measured for 1-week-old PDA-cellophane culture (value set to 1), with the standard deviations, based on three independent measurements of two independent RNA preparations, indicated by the error bars.
Targeted disruption of pro1 and phenotypic characterization.
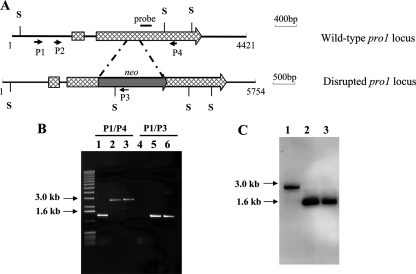
Gene disruption analysis was employed to examine whether the sequence conservation between C. parasitica pro1 and pro1 of S. macrospora extended to conservation of functional roles. A PCR-based strategy was used to generate a disruption construct consisting of a neomycin selectable marker flanked by pro1 gene-specific sequences (Fig. 3A) that was used to transform C. parasitica wild-type strain EP155. Disruption of pro1 was confirmed by PCR and Southern analysis (Fig. 3B and C). As indicated in Fig. 3B, a 1,350-bp fragment amplified from EP155 genomic DNA with primers P1 and P4 (Fig. 3A) was replaced with a fragment of 2,694 bp for the two pro1 disruption mutants, while a 1,367-bp fragment produced with primers P1 and P3 for the two pro1 disruption mutants was absent for strain EP155. Southern blot analysis using a pro1-specific probe (Fig. 3A) detected a 1,669-bp fragment for SacI-digested pro1 disruption mutant genomic DNA that contrasted with the 2,767-bp fragment detected for SacI-digested wild-type strain EP155 genomic DNA (Fig. 3C), consistent with the presence of a SacI restriction site both in the neomycin resistance gene cassette and in the pro1 gene (Fig. 3A). Additionally, pro1 mRNA was undetectable for the pro1 disruption mutant strains by real-time RT-PCR analysis using primers located in the 3′ portion of the pro1 gene at a position past the neomycin resistance gene cassette insertion site (data not shown).
FIG. 3.
Disruption of the C. parasitica pro1 gene. (A) Cartoon of pro1 gene organization and disruption construct. A pro1 gene disruption construct was generated by replacing a 200-bp fragment that encodes most of the putative DNA activation domain with a neomycin resistance gene cassette, using a PCR-based strategy as described in Materials and Methods. (B) PCR analysis of pro1 disruption mutants. Templates used for PCR analysis included genomic DNA isolated from wild-type strain EP155 (lanes 1 and 4) and pro1 disruption mutants Δpro1T3 (lanes 2 and 5) and Δpro1T6 (lanes 3 and 6). (C) Southern blot analysis of the EP155 (lane 1), Δpro1T3 (lane 2), and Δpro1T6 (lane 3) strains. Genomic DNA was digested with SacI and hybridized with a pro1-specific probe as shown in panel A.
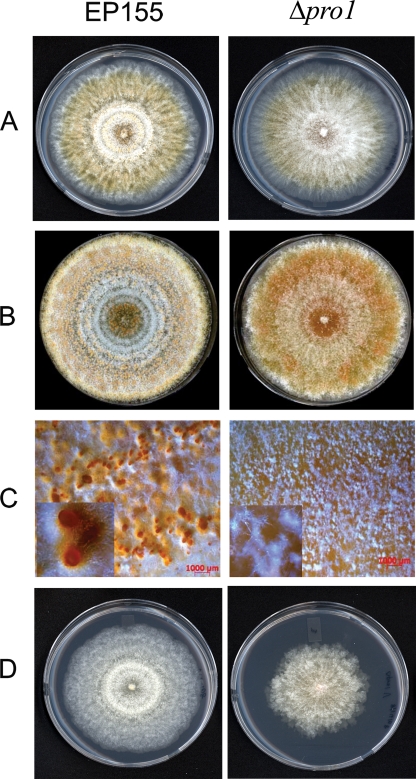
The growth characteristics and colony morphology exhibited by the pro1 disruption mutant strains were very similar to those exhibited by the EP155 parental strain under standard laboratory conditions (22 to 24°C, 12-h/12-h light/dark cycle with a light intensity of 1,300 to 1,600 lx) during the first 7 days of culturing on PDA medium, with the exception that the mutant strains produced more aerial hyphae (Fig. 4A). However, significant differences were observed during the second week of culturing (Fig. 4B). The disruption mutants failed to produce normal orange, asexual-spore-forming structures, resulting in a brown colony with little to no conidium production (Fig. 4C and Table 2). However, the conidiation defect observed for the pro1 mutants was reversible upon exposure of the growing colonies to high light intensity (Table 2). In this regard, our laboratory previously reported that high-light-intensity exposure also reversed hypovirus-mediated suppression of host conidium production (18).
FIG. 4.
Colony morphology of the pro1 disruption mutant strain. Colonies of wild-type strain EP155 (left) and pro1 disruption mutant Δpro1T6 (right) after 7 and 14 days of culturing are shown in panels A and B, respectively. Photographs were taken of cultures incubated on PDA at 22 to 24°C under standard light conditions (12-h/12-h light/dark cycle with a light intensity of 1,300 to 1,600 lx). The pro1 disruption mutant formed small putative proto-pycnidia (C, right) rather than mature conidium-producing pycnidia as produced by strain EP155 (C, left). The defect in conidium production in the pro1 disruption mutant was restored to wild-type levels in the complemented Δpro1T6/pG1 strain (Table 2). Panel D shows the colonies formed by the hypovirus CHV1-EP713-infected EP155 (left) and Δpro1T6 (right) strains after 7 days of culturing under the conditions described above.
TABLE 2.
Conidiation by pro1 disruption mutants under high-intensity and standard light conditions
| Strain | No. of conidiospores/mla
|
|
|---|---|---|
| Standard laboratory lightb | High lightc | |
| EP155 | (5.65 ± 1.54) × 108 | (9.36 ± 1.87) ×108 |
| Δpro1T3 | <1 × 104d | (10.87 ± 3.07) × 108 |
| Δpro1T6 | <1 × 104 | (8.36 ± 2.21) × 108 |
| Δpro1T6/pG1 | (6.08 ± 3.2) × 108 | (17.6 ± 4.5) × 108 |
Conidiation was measured after 2 weeks of culturing on PDA by using a hemacytometer. Data are shown as means ± standard deviations for four independent replicates.
Twelve hours of light and 12 h of darkness at 1,300 to 1,600 lx.
Twenty-four hours of continuous light at ∼4,000 lx.
Ten thousand conidiospores/ml is the lower threshold for spore detection with this quantification method.
To ensure that the sporulation deficiency phenotype was caused by the disruption of pro1, a complementation vector expressing the full-length of pro1 gene under the direction of the C. parasitica gpd1 promoter region, and conferring resistance to hygromycin, was constructed and used to transform the pro1 disruption mutant Δpro1T6. Hygromycin resistance transformants produced conidiospores at a level comparable to that of the wild-type strain EP155 under standard light conditions (Table 2).
The pro1 disruption mutants were also inoculated onto American chestnut tree stems to test whether disruption of this gene affected virulence. The mean canker areas ± standard deviations (cm2) from the virulence assay for pro1 disruption mutant strains on dormant chestnut tree stems were as follows: for EP155, 47.5 ± 8.4; for Δpro1T3, 37.2 ± 10.2; for Δpro1T6, 48.9 ± 8.7; and for EP713, 6.3 ± 2.2. No differences were found between the mutants and the wild-type strain, indicating that the disruption of the pro1 gene in C. parasitica did not affect fungal virulence.
Disruption of pro1 results in female infertility.
The ability of the pro1 disruption mutants (mating type Mat-2 [7]) to serve as either a male or a female in sexual crosses was tested in mating experiments with strain EP146 (mating type Mat-1 [7]). Spermatization of strain EP146 with conidia from either wild-type strain EP155 or the two pro1 disruption mutants resulted in a similar number of perithecia (sexual fruiting body structures) containing viable ascospores (data not shown). In contrast, spermatization of the two pro1 disruption mutants with conidia derived from strain EP146 resulted in no perithecium production, whereas large numbers of perithecia were produced when the same batch of EP146 conidia were used to spermatize strain EP155 (Fig. 5). Female fertility was restored in the complemented Δpro1T6/pG1 strain. We conclude that pro1, as was reported (16) for the transcription factor gene cpst12, is required for C. parasitica female fertility.
FIG. 5.
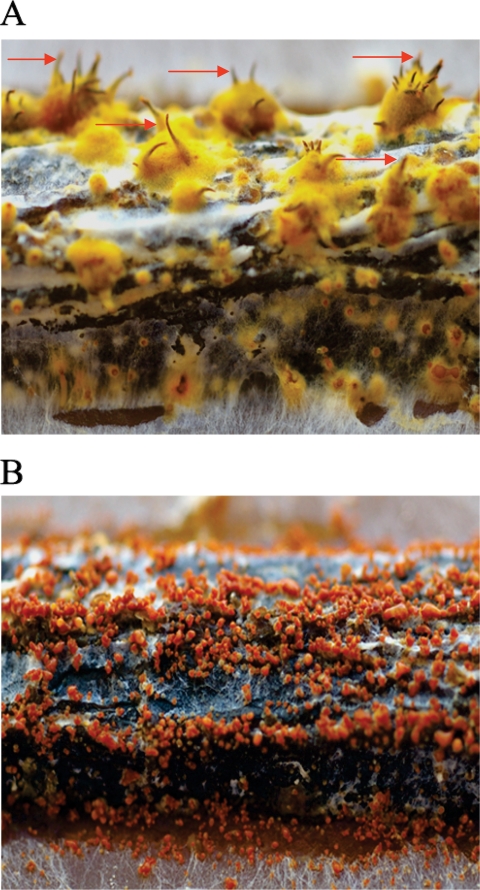
The pro1 disruption mutants are female sterile on American chestnut tree twigs. Mating experiments were performed with pro1 disruption mutant or EP155 strains as the female and strain EP146, of the opposite mating type, as the male. Photographs were captured at 6 months after initiation of the mating, using a Pentax K100D digital SLR camera equipped with a macro lens at a magnification of ×4. Mature perithecia with protruding ostiolar necks (arrows) were present in abundance for the control EP146(♂) × EP155(♀) crosses (A) but were completely absent in the EP146(♂) × Δpro1T6(♀) crosses (B).
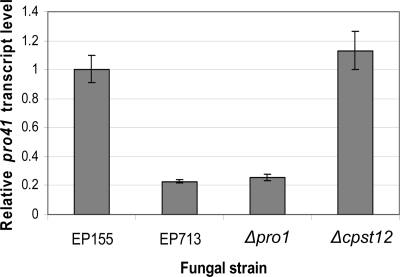
The developmental mutant screen that identified the S. macrospora pro1 (Smpro1) gene also identified an endoplasmic reticulum membrane protein gene, pro41 (27). The Smpro41 mutant was blocked at the same stage of development as the Smpro1 mutant. Moreover, Smpro41 transcript accumulation was greatly reduced in Smpro1 mutant strains, and microarray analysis of Smpro1/Smpro41 double mutants supported the conclusion that Smpro41 is epistatic to Smpro1. In view of these results, we used the Smpro41 amino acid sequence in a BLAST analysis of the draft C. parasitica genome assembly to identify the C. parasitica orthologue. Only one credible match was obtained, with an E value of 1 × 10−37 (the next-highest hit had an E value of 6.5). Sequence alignment analysis revealed 72% identity at the predicted amino acid sequence level for the two genes (Cppro41; GenBank accession no. FJ348265). As shown in Fig. 6, Cppro41 transcript accumulation was significantly reduced in the ΔCppro1 mutant and in the hypovirus CHV1-EP713-infected strain compared to the level found in wild-type strain EP155. It is interesting to note that Cppro41 expression is not reduced in the cpst12 disruption mutant strain (Fig. 6), even though both Δpro1 and Δcpst12 mutant strains are female infertile.
FIG. 6.
Semiquantitative real-time RT-PCR analysis of pro41 expression in the EP155, EP713, Δpro1T6, and Δcpst12 strains. Relative transcript levels were measured using cDNA of 18S rRNA generated in the same RT reaction for normalization, with standard deviations, based on three independent measurements of two independent RNA preparations, indicated by the error bars.
Disruption of pro1 alters maintenance of hypovirus CHV1-EP713 infection.
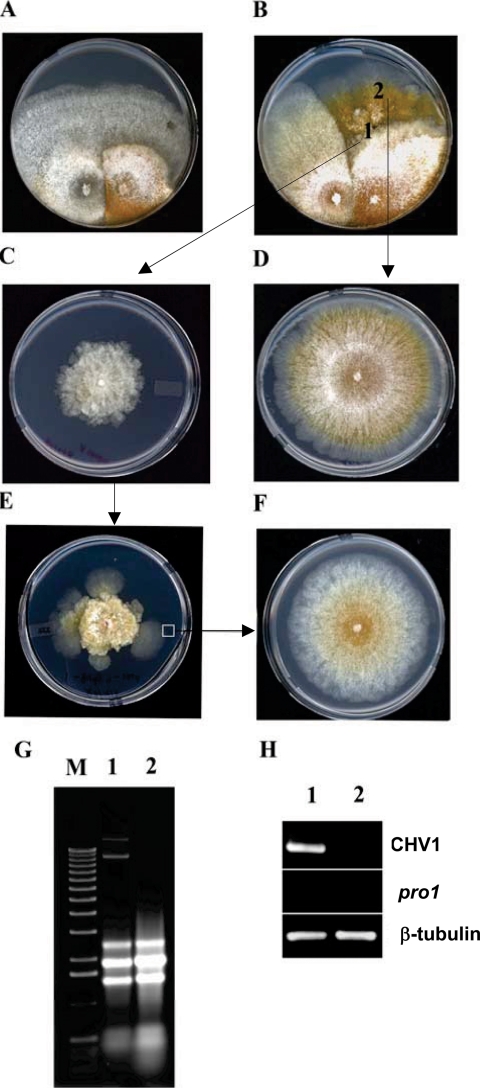
The pro1 disruption mutant strains were infected by hypovirus CHV1-EP173 introduced either by anastomosis-mediated transfer of virus from strain EP713 (see Materials and Methods) or by transfection with the viral coding-strand RNA (8). However, the infected pro1 disruption mutants differed from the infected wild-type strain in several properties. As shown in Fig. 7B, when infected by anastomosis-mediated transfer of hypovirus CHV1-EP713 from strain EP713, the newly infected (converted) pro1 disruption mutant strain displayed a severe morphology, with slower growth and fewer aerial hyphae, than the newly converted wild-type strain (Fig. 7A). This severe growth phenotype was invariably accompanied by the appearance of faster-growing sectors.
FIG. 7.
Requirement of the pro1 gene for CHV1-EP713 virus maintenance. (A and B) Anastomosis-mediated transfer of hypovirus CHV1-EP713 from strain EP713 (left side of each panel) to wild-type strain EP155 (A) and pro1 disruption mutant Δpro1T6 (B). (C) Subcultures taken from the converted, CHV1-EP713-infected pro1 disruption mutant culture at a point near the initial contact between the paired colonies (position 1) exhibited virus infection symptoms that included loss of orange pigment production and altered colony morphology (Fig. 4D). (D) Subcultures taken from rapidly growing sectors originating from the converted pro1 disruption mutant (position 2) exhibited a virus-free colony phenotype. (E) With successive subculturing (transfer every 7 days), the CHV1-EP713-infected Δpro1T6 mycelia produced rapidly growing sectors at a high frequency and the colonies derived from those sectors always exhibited a virus-free phenotype (F). (G) Total RNA was extracted from colonies originating from mycelia taken from positions 1 and 2 (C and D, respectively) and analyzed on an agarose gel. The slowly migrating bands present in lane 1 but absent in lane 2 are full-length and defective CHV1-EP713 replicative double-stranded RNAs. (H) RT-PCR analysis detected a virus-specific product (lane 1) from colonies originating from position 1 (C) but not from position 2 (D). Primers specific for CHV1-EP713 genome RNA and pro1 mRNA were used along with primers specific for β-tubulin mRNA as reaction and loading controls.
CHV1-EP713-infected mycelia that were subcultured from the slower-growing portion of the converted pro1 disruption mutant (position marked “1” in Fig. 7B) resembled strains infected with wild-type EP155 in the loss of orange pigmentation but exhibited much slower growth and an irregular colony margin (compare the colonies in the left and right panels of Fig. 4D). In contrast, the mycelia recovered from the rapidly growing regions (position marked “2” in Fig. 7B) resembled the uninfected Δpro1 mutant strain during the early stage of colony growth (Fig. 7D). Agarose gel and RT-PCR analysis of RNA extracted from colonies derived from the rapidly growing mycelia confirmed the absence of CHV1-EP713 viral RNA and the Δpro1 genetic background (Fig. 7G and H).
As shown in Fig. 7E, colonies subcultured from CHV1-EP713-infected Δpro1 mycelia recovered from anastomosis plates (Fig. 7C) also developed rapidly growing sectors at a high frequency. Colonies derived from these sectors were also virus free (Fig. 7F). In contrast, virus-free sectors were not observed for the CHV1-EP713-infected, complemented Δpro1T6/pG1 strain. These combined results suggest a role for pro1 in stable maintenance of hypovirus infection.
DISCUSSION
Many of the mycoviruses that attenuate virulence of phytopathogenic fungal hosts also cause additional symptoms that can compromise the utility of these agents for biological control of fungal disease (reviewed in reference 28). For example, the loss of female fertility and reduced asexual sporulation levels caused by hypovirus infections reduce the ecological fitness and spread of hypovirulent C. parasitica strains (reviewed in references 3, 21, and 25). Recent studies have shown that hypovirus infections persistently modify host transcriptional profiles and alter cellular signaling pathways that are required for conidiation and sexual reproduction, e.g., G-protein (9, 11, 17) and mitogen-activated protein kinase (29) signaling pathways. The list of hypovirus-responsive genes identified by microarray analysis (1, 2) included two genes that encode putative transcription factors: cpst12, an orthologue of yeast C2H2 zinc finger transcription factor ste12 (16), and the subject of this report, pro1, a Zn(II)2Cys6 binuclear cluster transcription factor gene. Remarkably, characterization of cpst12 (16) and pro1 (this study) revealed that both transcription factors are required for female fertility, that pro1 is required for conidiation, and that cpst12 is required for full virulence, three important biological processes that are altered by hypovirus infection. Paradoxically, pro1 is also required for stable maintenance of hypovirus replication in C. parasitica.
The Zn(II)2Cys6 binuclear cluster protein family, to which pro1 belongs, comprises a large class of fungus-specific transcription factors, e.g., the largest class of transcription factor genes in N. crassa (6). Although most of the characterized members of this family participate in regulation of primary and secondary metabolic pathways, several have been shown to regulate fungal developmental processes (33). Gene deletion and complementation studies have shown that Smpro1 is required for sexual development and that the N. crassa pro1 (Ncpro1) orthologue can complement the Smpro1 deletion mutant (22, 23). In A. nidulans, the nosA gene is required for sexual development (35), while the rosA gene appears to repress sexual development (34). In contrast, the N. crassa fluffy gene (5) is required for conidiation. As indicated in Fig. 1A, the predicted C. parasitica pro1 (Cppro1) amino acid sequence clusters with the Zn(II)2Cys6 orthologues implicated in regulating sexual development in S. macrospora (67%) and N. crassa (67%) and related pro1 orthologues from the plant pathogenic fungal species M. grisea (69%) and G. zeae (67%). Additionally, each of the five orthologues contains a single intron at the same position in the coding region just before the conserved Zn(II)2Cys6 binuclear cluster domain (Fig. 1B). Thus, the requirement of Cppro1 for female fertility is consistent with its phylogenetic position relative to the other pro1 orthologues.
The genetic relationship reported for Smpro1 and Smpro41 in S. macrospora (27) also appears to be conserved for the corresponding orthologues in C. parasitica. Transcript accumulation is reduced for both pro41 orthologues in the corresponding pro1 disruption mutant strains (Fig. 6) (27). The observed reduction in pro41 transcript accumulation in the CHV1-EP713-infected strain could be explained as a result of an epistatic relationship with pro1, as has been proposed for S. macrospora (27). However, additional studies are required to confirm the nature of this relationship in C. parasitica and to identify the complement of pro1-regulated genes. The fact that pro41 transcript accumulation is not altered by disruption of cpst12 (Fig. 6), a transcription factor also essential for female fertility, indicates that female fertility in C. parasitica is subject to regulation by multiple pathways.
The functional role of pro1 in C. parasitica is distinctive among characterized Zn(II)2Cys6 binuclear cluster proteins in that it is required for both sexual development and asexual sporulation (Fig. 5 and Table 2). A requirement for conidiation has been demonstrated for the Zn(II)2Cys6 transcription factor gene fluffy (5) in the closely related fungus N. crassa (Fig. 1A). Surprisingly, we were unable to identify an orthologue of the N. crassa fluffy gene in the draft C. parasitica genome sequence assembly. The absence of a fluffy orthologue and the requirement of pro1 for conidiation in C. parasitica suggest considerable functional divergence within the fungal Zn(II)2Cys6 binuclear cluster protein family. Interestingly, the defect in conidiation observed in the pro1 disruption mutant is reversible by exposure to high light intensity (Table 2), similar to the hypovirus-mediated suppression of conidiation (18).
An additional distinctive functional role for pro1 is its apparent requirement for stable maintenance of hypovirus infection. The Δpro1 mutant strain responds to CHV1-EP713 infection by growing at a much lower rate than the CHV1-EP713-infected parental strain and by producing rapidly growing, virus-free sectors at a high frequency. The observation that a virus downregulates a host transcription factor gene that is also required for virus maintenance presents an intriguing paradox. However, it is important to note that pro1 expression is significantly reduced by CHV1-EP713 but not eliminated as occurs in the Δpro1 mutant strains. Polashock et al. (31) previously reported the isolation and characterization of a spontaneous C. parasitica mutant, NB58F, which was deficient in hypovirus maintenance. Although the colony morphologies of the NB58F and Δpro1 mutant strains are different, the NB58F mutant, like Δpro1, was deficient in conidium production, but not to the same extent. The role of pro1 in maintenance of hypovirus infection and its relevance to the NB58F mutant warrant further investigation.
The fact that both the cpst12 and the pro1 disruption mutants are female sterile indicates that these two transcription factors, one of the C2H2 zinc finger class and the other of the Zn(II)2Cys6 class, are individually necessary but insufficient for female fertility in C. parasitica. Mature perithecia fail to form in crosses in which either the cpst12 or the pro1 disruption mutants are spermatized by conidia from a strain of the opposite mating type (reference 16 and this study, respectively). This contrasts with the defects in fruiting body formation observed for the pro1 and ste12 orthologue mutants of S. macrospora, where the Smpro1 mutant fails to form mature perithecia (22, 23) while the Smste12 mutant forms mature perithecia, but these perithecia contain nonviable ascospores (26). These differences may reflect the differences in the homothallic and heterothallic life cycles employed by S. macrospora and C. parasitica, respectively.
Although cpst12 and pro1 appear to have coordinating roles in the regulation of C. parasitica female fertility, each transcription factor is independently required for the proper regulation of other very distinct processes (Table 3):the cpst12 disruption mutant exhibits significantly reduced virulence, and the pro1 deletion mutant does not produce asexual spores under standard culture conditions. These observations suggest that these two transcription factors are located at positions well up in the transcriptional regulatory network and that modulation of the expression of one or both of the genes for these transcription factors would likely result in pleiotropic phenotypic changes.
TABLE 3.
Comparison of C. parasitica pro1 and cpst12 gene disruption mutant strains
| Characteristic | Result for:
|
|
|---|---|---|
| pro1 disruption mutant | cpst12 disruption mutantb | |
| Vegetative growth | Same as the wild-type level, with increased aerial hyphae | Same as the wild-type level |
| Asexual sporulation | No or few conidia produced under standard laboratory conditionsa | Significantly more conidia produced than for the wild-type strain |
| Sexual sporulation | Female sterile | Female sterile |
| Virulence | Same as the wild-type level | Significantly reduced |
It is premature to conclude that hypovirus-mediated downregulation of cpst12 and pro1 expression is responsible for the reduced virulence, female infertility, and reduced condiation observed in hypovirus CHV1-EP713-infected C. parasitica strains. However, it is remarkable that two of three transcription factor genes that were identified, through transcriptional profiling analysis of 2,200 of the estimated 11,184 C. parasitica genes, as being hypovirus responsive (1, 2) turned out to be required for regulation of three major biological processes that are altered as a result of hypovirus infection. Inspection of the C. parasitica draft genome sequence (http://genome.jgi-psf.org/Crypa1/Crypa1.home.html) reveals over 80 open reading frames with a Zn(II)2Cys6 fungal binuclear cluster motif and over 20 open reading frames comprising putative C2H2 zinc finger transcription factor genes. The recent development of a highly efficient C. parasitica gene disruption system (20) and the draft genome sequence will allow a systematic analysis of the role of host transcription factors in the elaboration of hypovirus-mediated symptom expression as well as hypovirus replication, persistence, and transmission.
Acknowledgments
This work was supported in part by Public Health Service grant GM55981 to D.L.N.
Footnotes
Published ahead of print on 29 December 2008.
REFERENCES
- 1.Allen, T. D., A. L. Dawe, and D. L. Nuss. 2003. Use of cDNA microarrays to monitor transcriptional responses of the chestnut blight fungus Cryphonectria parasitica to infection by virulence-attenuating hypoviruses. Eukaryot. Cell 21253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. D., and D. L. Nuss. 2004. Specific and common alterations in host gene transcript accumulation following infection of the chestnut blight fungus by mild and severe hypoviruses. J. Virol. 784145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anagnostakis, S. L. 1982. Biological control of chestnut blight. Science 215466-471. [DOI] [PubMed] [Google Scholar]
- 4.Anagnostakis, S. L. 1984. Nuclear gene mutations in Endothia (Cryphonectria) parasitica that affect morphology and virulence. Phytopathology 74761-765. [Google Scholar]
- 5.Bailey, L. A., and D. J. Ebbole. 1998. The fluffy gene of Neurospora crassa encodes a Gal4p-type C6 zinc cluster protein required for conidial development. Genetics 1481813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner, N. D. Read, S. Seiler, D. Bell-Pedersen, J. Paietta, N. Plesofsky, M. E. Nargang, A. Radford, C. Selitrennikoff, J. E. Galagan, J. C. Dunlap, J. J. Loros, D. Catcheside, H. Inoue, R. Aramayo, M. Polymenis, E. U. Selker, M. S. Saks, G. A. Marzluf, I. Paulsen, R. Davis, D. J. Ebbole, A. Zelter, E. R. Kalkman, R. O'Rourke, F. Bowring, Y. Yeadon, C. Ishii, K. Suzuki, W. Sakai, and R. Pratt. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 681-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, B., G. H. Choi, and D. L. Nuss. 1993. Mitotic stability and nuclear inheritance of integrated viral cDNA in engineered hypovirulent strains of the chestnut blight fungus. EMBO J. 122991-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, B., G. H. Choi, and D. L. Nuss. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 2641762-1764. [DOI] [PubMed] [Google Scholar]
- 9.Chen, B., S. Gao, G. H. Choi, and D. L. Nuss. 1996. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc. Natl. Acad. Sci. USA 937996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, B., L. M. Geletka, and D. L. Nuss. 2000. Using chimeric hypoviruses to fine-tune the interaction between a pathogenic fungus and its plant host. J. Virol. 747562-7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, G. H., B. Chen, and D. L. Nuss. 1995. Virus-mediated or transgenic suppression of a G-protein alpha subunit and attenuation of fungal virulence. Proc. Natl. Acad. Sci. USA 92305-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, G. H., T. G. Larson, and D. L. Nuss. 1992. Molecular analysis of the laccase gene from the chestnut blight fungus and selective suppression of its expression in an isogenic hypovirulent strain. Mol. Plant-Microbe Interact. 5119-128. [DOI] [PubMed] [Google Scholar]
- 13.Choi, G. H., and D. L. Nuss. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257800-803. [DOI] [PubMed] [Google Scholar]
- 14.Churchill, A. C. L., L. M. Ciufetti, D. R. Hansen, H. D. Van Etten, and N. K. Van Alfen. 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 1725-31. [Google Scholar]
- 15.Cullen, D., S. A. Leong, L. J. Wilson, and D. J. Henner. 1987. Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene 5721-26. [DOI] [PubMed] [Google Scholar]
- 16.Deng, F., T. D. Allen, and D. L. Nuss. 2007. Ste12 transcription factor homologue CpST12 is down-regulated by hypovirus infection and required for virulence and female fertility of the chestnut blight fungus Cryphonectria parasitica. Eukaryot. Cell 6235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, S., and D. L. Nuss. 1996. Distinct roles for two G protein alpha subunits in fungal virulence, morphology and reproduction revealed by targeted gene disruption. Proc. Natl. Acad. Sci. USA 9314122-14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillman, B. I., R. Shapira, and D. L. Nuss. 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80850-956. [Google Scholar]
- 19.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushihara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan, X., Z. Yao, Y. Zhou, J. Shang, H. Lin, D. L. Nuss, and B. Chen. 2008. Deletion of the cpku80 gene in the chestnut blight fungus, Cryphonectria parasitica, enhances gene disruption efficiency. Curr. Genet. 5359-66. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald, W. L., and D. W. Fulbright. 1991. Biological control of chestnut blight: use and limitations of transmissible hypovirulence. Plant Dis. 75651-661. [Google Scholar]
- 22.Masloff, S., S. Poggeler, and U. Kuck. 1999. The pro1+ gene from Sodaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152191-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masloff, S., S. Jacobsen, S. Poggeler, and U. Kuck. 2002. Functional analysis of the C6 zinc finger gene pro1 involved in fungal sexual development. Fungal Genet. Biol. 36107-116. [DOI] [PubMed] [Google Scholar]
- 24.Milgroom, M. G. 1995. Population biology of the chestnut blight fungus, Cryphonectria parasitica. Can. J. Bot. 73(Suppl. 1)S311-S319. [Google Scholar]
- 25.Milgroom, M. G., and P. Cortesi. 2004. Biological Control of chestnut blight with hypovirulence: a critical analysis. Annu. Rev. Phytopathol. 42311-338. [DOI] [PubMed] [Google Scholar]
- 26.Nolting, N., and S. Poggeler. 2006. A STE12 homologue of the homothallic ascomycete Sordoria macrospora interacts with the MADS box protein MCM1 and is required for ascosporogenesis. Mol. Microbiol. 62853-868. [DOI] [PubMed] [Google Scholar]
- 27.Nowrousian, M., S. Frank, S. Koers, P. Strauch, T. Weitneer, C. Ringelberg, J. C. Dunlap, J. J. Loros, and U. Kuck. 2007. The novel ER membrane protein PRO41 is essential for sexual development in the filamentous fungus Sordoria macrospora. Mol. Microbiol. 64923-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nuss, D. L. 2005. Hypovirulence: mycoviruses at the fungal-plant interface. Nat. Rev. Microbiol. 3632-642. [DOI] [PubMed] [Google Scholar]
- 29.Park, S.-M., E.-S. Choi, M.-J. Kim, B.-J. Cha, M.-S. Yang, and D.-K. Kim. 2004. Characterization of HOG1 homologue CpMK1 from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol. Microbiol. 511267-1277. [DOI] [PubMed] [Google Scholar]
- 30.Parsley, T. B., B. Chen, L. M. Geletka, and D. L. Nuss. 2002. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell 1401-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polashock, J. J., S. L. Anagnostakis, M. G. Milgroom, and B. I. Hillman. 1994. Isolation and characterization of a virus-resistant mutant of Cryphonectria parasitica. Curr. Genet. 26528-534. [DOI] [PubMed] [Google Scholar]
- 32.Shang, J., X. Wu, X. Lan, Y. Fan, H. Dong, Y. Deng, D. L. Nuss, and B. Chen. 2008. Large-scale expressed sequence tag analysis for the chestnut blight fungus Cryphonectria parasitica. Fungal Genet. Biol. 45319-327. [DOI] [PubMed] [Google Scholar]
- 33.Todd, R. B., and A. Andrianopoulos. 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21388-405. [DOI] [PubMed] [Google Scholar]
- 34.Vienken, K., M. Scherer, and R. Fischer. 2005. The Zn(II)2Cys6 putative Aspergillus nidulans transcription factor repressor of sexual development inhibits development under low-carbon conditions and in submerged culture. Genetics 169619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vienken, K., and R. Fischer. 2006. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol. Microbiol. 61544-554. [DOI] [PubMed] [Google Scholar]