Abstract
The relationships among gene regulatory mechanisms in the malaria parasite Plasmodium falciparum throughout its asexual intraerythrocytic developmental cycle (IDC) remain poorly understood. To investigate the level and nature of transcriptional activity and its role in controlling gene expression during the IDC, we performed nuclear run-on on whole-transcriptome samples from time points throughout the IDC and found a peak in RNA polymerase II-dependent transcriptional activity related to both the number of nuclei per parasite and variable transcriptional activity per nucleus over time. These differential total transcriptional activity levels allowed the calculation of the absolute transcriptional activities of individual genes from gene-specific nuclear run-on hybridization data. For half of the genes analyzed, sense-strand transcriptional activity peaked at the same time point as total activity. The antisense strands of several genes were substantially transcribed. Comparison of the transcriptional activity of the sense strand of each gene to its steady-state RNA abundance across the time points assayed revealed both correlations and discrepancies, implying transcriptional and posttranscriptional regulation, respectively. Our results demonstrate that such comparisons can effectively indicate gene regulatory mechanisms in P. falciparum and suggest that genes with diverse transcriptional activity levels and patterns combine to produce total transcriptional activity levels tied to parasite development during the IDC.
Plasmodium falciparum, the most deadly human malaria parasite, undergoes many developmental changes and thrives in both human and mosquito hosts. Since the genome sequence of P. falciparum became available (17), microarray studies of its intraerythrocytic developmental cycle (IDC) (3, 37) and other asexual and bloodborne stages (34) have demonstrated that differential gene expression is integral to the parasite's development. The stage specificity of steady-state RNA expression supports previous observations that the morphological changes of the IDC correspond to many less-apparent changes in physiology, such as protein and nucleic acid metabolism (10, 19) and sensitivity to small molecules (14, 22, 61). The Plasmodium repertoire of gene regulatory mechanisms is comparable to those of other eukaryotes (7, 23), implying that stage-specific gene expression may be regulated at many levels, including chromatin structure, transcriptional activation, and posttranscriptional modulation of translation and stability (11, 39, 41, 59). However, the relationship between such mechanisms during the IDC, specifically whether regulation of primary transcriptional activity determines the steady-state RNA level, remains uncertain for the vast majority of genes.
P. falciparum's steady-state expression patterns during the IDC have been hypothesized to result from cis-acting regulatory DNA sequences (e.g., promoters and enhancers), given its canonical basal transcriptional machinery (4, 5, 55). Numerous studies of individual putative transcription factors or of individual target genes have found relationships between cis-acting DNA sequences and the expression of individual loci or small groups of related genes (23, 38, 44, 47, 53), in some cases demonstrating the activity of putative regulatory proteins on the motifs (18, 20, 50, 62). Recently, de Silva et al. (12) established the functionality of two members of the ApiAP2 family of putative transcription factors and their binding motifs (2). Bioinformatic approaches have predicted many conserved motifs associated with coregulation (65, 72, 75), but in few resulting cases have physiological relevance to regulation, sufficiency of the motif alone, and biochemical function all been demonstrated jointly.
With limited direct evidence that the observed steady-state patterns of most Plasmodium genes are driven by promoter-based transcriptional regulation during the IDC, complementary mechanisms of widespread posttranscriptional regulation have received increasing attention. RNA-binding proteins were predicted to be abundant in P. falciparum (8), a temporal offset in the accumulation of protein and RNA of some genes suggested differential translational regulation during the IDC (24, 33), regulatory RNA binding was observed in both Plasmodium berghei (41) and P. falciparum (40), and RNA half-lives were found to be regulated in a functionally specific manner during the P. falciparum IDC (59). Although plasmodia do not encode the components of the canonical RNA interference pathway, despite their conservation in some other protozoan parasites (64), abundant steady-state antisense RNA (21, 51) and individual examples of cRNA (29) have triggered speculation that antisense RNA could be involved in regulation at the transcriptional or posttranscriptional levels (46). Together, such studies highlight the enigmatic relationships between gene regulatory mechanisms in P. falciparum.
In this study, we investigate the role that transcriptional activity plays in the expression of genes during the IDC in P. falciparum, independent of downstream modulation. Total transcriptional activity changes during developmental transitions in systems as disparate as tubers and nematodes (68, 70) due to the activation state of the RNA polymerase (RNAP) enzymes themselves, the availability of the DNA template, or both. The physiological changes of the P. falciparum IDC incorporate many aspects of cellular development, and differential overall transcriptional activity has been suggested by early observations (19, 26). Several previous studies have employed nuclear run-on to directly assay transcriptional activity—nascent RNA production—at individual loci (26, 27, 29, 31, 32, 58) and compared changes in relative transcriptional activity to changes in steady-state RNA levels. Yet, cases of posttranscriptional regulation implied by previous comparisons remain unverified by matched RNA half-life studies, and the possibility of stage-specific patterning of overall transcriptional activity could fundamentally affect the interpretation of the changes observed at specific loci across morphological stages by nuclear run-on.
Here we test the hypothesis that differential total transcriptional activity of the composite transcriptome is an intrinsic component of P. falciparum stage-specific physiology during the IDC, using nuclear run-on. We report that the activity of RNAP II varies significantly and reproducibly with the morphological stage, due both to increases in the number of nuclei during schizogony and to temporal variability in transcriptional activity per nucleus. We use this differential trend in total transcriptional activity during the IDC to compare the absolute transcriptional activities of both sense and antisense strands of a set of individual genes across four time points and to compare changes in their transcriptional activities to changes in their steady-state RNA expression levels through the IDC. We demonstrate that this analysis offers unique insights into the mechanisms controlling the RNA levels of genes during the P. falciparum IDC and can consistently identify known targets of posttranscriptional regulation.
MATERIALS AND METHODS
Parasite culture and nuclear harvest.
Plasmodium falciparum strain 3D7 was cultured by standard methods (63) in RPMI-HEPES medium supplemented with 5% human serum (O+) and 5% Albumax II (Gibco). Parasites were synchronized using 5% sorbitol solution (30) during three generations and assayed at 4% hematocrit and <2% parasitemia. These highly synchronized populations were then harvested for transcriptionally active nuclei as previously described (45). Upon harvest, thin-smear slides of the infected red blood cell (iRBC) culture were made, and parasitemia, number of nuclei, and morphological stage were evaluated by modified Wright stain (Diff-Quik) (see Fig. S1 in the supplemental material), utilizing the guidelines in Silamut and White (60). Briefly, iRBCs were centrifuged at ∼1,800 rpm in a Sorvall RT6000B, washed once in phosphate-buffered saline, pH 7.4, and then lysed in 0.2% saponin-phosphate-buffered saline. The liberated parasites were washed once in solution A {20 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 7.5, 15 mM NaCl, 60 mM KCl, 0.5 mM EGTA, 4 mM EDTA, 0.15 mM spermine, 0.5 mM spermidine, 0.125 mM phenylmethylsulfonyl chloride, 14 mM β-mercaptoethanol} and then lysed in a final concentration of 0.64% Nonidet P-40 substitute with ∼10 strokes with a Dounce homogenizer (B pestle). Nuclei were pelleted at 1,000 × g in a microcentrifuge, a force which yields incorporation identical to that of pelleting at the range of forces (see Fig. SA5 in the supplemental material) utilized in the literature (26, 45). Nuclei were then washed once with solution A, suspended in glycerol storage buffer (50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 0.1 mM EDTA, 40% glycerol), and then flash frozen and stored at −80°C. For total-incorporation nuclear run-on, 25 ml culture (1 ml packed RBCs) was harvested; for nuclear run-on for gene-specific hybridization blots, 100 ml culture (4 ml packed RBCs) was harvested. Samples prepared by these methods yield membrane-bound nuclei containing DNA, stainable by DAPI (4′,6-diamidino-2-phenylindole) or Sybr green (data not shown).
Total incorporation nuclear run-on.
Frozen nuclei prepared from 1 ml packed RBCs were thawed on ice and incubated for 15 min at 4°C with either 100 μg/ml α-amanitin or an equivalent volume of water. A concentration of 100 μg/ml α-amanitin fully abolishes RNAP II activity (35), inhibits RNAP III to <10% activity (71), and is effective on nuclei isolated from P. falciparum (31, 45). Nuclei were heated to 37°C for 30 min in the following reaction buffer: 50 mM HEPES, pH 7.9, 50 mM NaCl, 10 mM MgCl2, 1.2 mM dithiothreitol, 15% glycerol, 10 mM creatine phosphate, 0.2 mg/ml creatine kinase, 0.1 U/μl RNasin (Promega), 4 mM ATP, 1 mM CTP, 1 mM GTP, and 1.5 mCi/ml [α-32P]UTP (∼0.5 μM; Perkin Elmer). Reactions were stopped by the addition of an ice-cold solution of 5% trichloroacetic acid, 100 mM sodium pyrophosphate (NaPP), and 20 μg/ml sheared salmon sperm DNA (Ambion). Stopped reaction mixtures were allowed to incubate on ice for ∼1 h before macromolecules were collected on fiberglass filter disks (Whatman GF/C) by vacuum manifold, washed with cold 5% trichloroacetic acid-100 mM NaPP, and precipitated with cold 95% ethanol. Filter disks were dried at room temperature, and then scintillation was counted in a Beckman LS 1801 liquid scintillation system.
Gene-specific nuclear run-on.
Single-stranded DNA probes for the slot blots were obtained by the M13 helper-phage method (56). Briefly, sequence fragments from genes of interest (see Table S2 in the supplemental material) were PCR cloned into the TOPO TA pCR2.1 vector (Invitrogen), XL2-blue Escherichia coli were transformed with these constructs, and cultures were infected with M13K07 helper phage (New England Biolabs) according to the manufacturer's instructions. Phage was suspended in Tris-EDTA buffer and lysed by extraction with phenol and chloroform. DNA was precipitated with sodium acetate and ethanol and then dissolved in nuclease-free water. A 3-μg amount of each probe was applied to NytranN nylon membranes (Schleicher & Schuell) in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) by using a 48-well Bio-Dot slot-blotting apparatus (Bio-Rad), cross-linked in a Bio-Rad GS GeneLinker with 120 mJ of 254-nm light, and allowed to dry. Just prior to hybridization with the [32P]RNA generated in the nuclear run-on reaction (see below), filters were prehybridized [50 mM HEPES, pH 7.4, 0.3 M NaCl, 10 mM EDTA, 0.2% sodium dodecyl sulfate, 1 mg/ml poly(A) (GE Lifesciences), 1 mg/ml yeast RNA (Ambion), 1% NaPP (Sigma), 5× Denhardt's solution without BSA] at 65°C for 6 h.
Gene-specific nuclear run-on was performed on samples of nuclei harvested from 4 ml packed RBCs, stored identically to those for total incorporation, and heated in the same composition reaction mixture to 37°C for 30 min. Following transcription, these samples were treated with 0.1 U/μl RQ1 DNase (Promega) for 5 min at room temperature and then digested by the addition of SET buffer (0.5% SDS, 5 mM EDTA, 1 mM Tris, pH 7.4), proteinase K (Qiagen), and 0.25 mg/ml yeast RNA (Ambion) at 37°C for an additional 30 min. The resultant RNA was extracted with TriReagent LS and alcohol precipitated according to the manufacturer's protocol. The radiolabeled RNA samples were resuspended in 100 μl nuclease-free H2O and then added to 4 ml hybridization solution [50 mM HEPES, pH 7.4, 0.3 M NaCl, 10 mM EDTA, 0.2% SDS, 0.1 mg/ml poly(A), 0.1 mg/ml yeast RNA, 0.1% NaPP, 1× Denhardt's solution without bovine serum albumin] and incubated with the prepared slot-blotted membranes for 48 h at 60°C. Membranes were given 20-min washes twice in 6× SSC-0.1% SDS at 25°C and then in 2× SSC-0.1% SDS at 50°C. A 10-min wash in 2× SSC and 10 μg/ml RNaseA at 37°C was used to degrade RNA not bound to the single-stranded DNA probes. These membranes were then exposed to a phosphorimager screen for 14 days. Images were detected by using a Storm 820 phosphorimager and analyzed with ImageQuant software (Molecular Dynamics).
Quantification of morphological stages and nuclear copy number.
Thin-smear slides of the blood cultures were methanol fixed at each time point and stained with Diff-Quik (Wright-Giemsa). Twelve or fewer fields per slide were imaged by using an Olympus BX 41 microscope with a Qcolor 5 camera and a 100× oil immersion objective, and parasitemias of six morphological stages (early rings, late rings, early trophozoites, mid-trophozoites, late trophozoites, and schizonts) were calculated based on these images, according to the criteria outlined by Silamut and White (60). These images were also used to approximate the number of nuclear bodies per parasite.
DNA synthesis.
In a single-culture, short-exposure adaptation of the [3H]hypoxanthine incorporation assay (13), complete RPMI-HEPES medium was removed from 25 ml of P. falciparum culture (described above) 3 h prior to time points outlined in Results. This was replaced with medium supplemented with only 3H-labeled hypoxanthine to a final concentration of 33 to 100 μM. At each time point, iRBCs were harvested by centrifugation at 1,800 rpm, the medium removed, and the pellet frozen at −80°C. These cells were thawed, and DNA isolated with a Qiagen blood mini DNA kit according to the manufacturer's protocol. A portion of the [3H]DNA was quantified by measuring the absorption at 260 nm in a Beckman DU540 spectrophotometer, and the remainder was scintillation counted to generate the 3H cpm/(μg/ml) ratios (see Fig. S2A in the supplemental material).
Stage specificity analysis.
We hypothesized that each morphological stage has a characteristic level of transcriptional activity and that the observed peak in the total transcriptional activity of the population could be attributed to the presence of stages possessing higher levels of activity. Using the observed values for total transcriptional activity (from the total-incorporation nuclear run-on) and the morphological stages (from slides made of each time point) to describe the hypothesis in terms of the normal equations in the form Ax = b, we used an inequality-constrained linear least squares algorithm (home-written in Python) to solve for the best-fit vector (x) of characteristic values representing the total transcriptional activity of each morphological stage, given the parasitemias of those stages (the matrix A) and the total transcriptional activity at each time point (the vector b), with the constraint that all values be nonnegative.
To test the fit of the stage-specific model to the observed data, the vector resulting from our algorithm was multiplied by the experimentally measured matrix of morphological-stage parasitemias. The resulting R2 values were calculated in Microsoft Excel.
Reverse transcription and quantitative real-time PCR.
RNA was harvested directly from iRBCs at each time point by using TriReagent (MRC) according to the manufacturer's instructions. RNA from each time point was quantified by measuring the absorption at 260 nm, treated with Turbo DNase (Ambion), and reverse transcribed at a final concentration of 30 ng/μl by using a SuperScript first-strand synthesis kit for reverse transcriptase PCR (Invitrogen), primed with a mixture of (oligo)dT and random hexamers. The resulting cDNA was amplified in triplicate (0.1 μl per reaction) by quantitative real-time PCR (Applied Biosystems 7300 and ABI Sybr green PCR master mix), using the same primer pairs as were used to generate the DNA probes for the nuclear run-on blots (see Table S2 in the supplemental material), except in the cases of PF07_0029, PFI0755c, PFB0100c, PF10_0345, PF14_0323, and 18S rRNA. Primers amplifying PF08_0085 were used to compare the time point RNA samples to a mixed-stage cDNA sample; subsequently, the absolute quantity of each gene-specific transcript in each time point sample (and in the control sample) was compared to that of PF08_0085 by using standard methods and accounting for primer efficiency by the Pfaffl method (52). The absolute steady-state RNA levels of the genes assayed were obtained through an operation analogous to that used to calculate their absolute transcriptional activity: all values were normalized by the total RNA yield of the sample and then multiplied by coefficients reflecting the difference in ideal RNA yield (43) for time points closely matched for morphological-stage composition.
RESULTS
Total transcriptional activity peaks late in the IDC.
In order to assess the total activity of the RNAPs throughout the IDC, independent of posttranscriptional regulators, P. falciparum 3D7 was synchronized with sorbitol and nuclei were harvested at regular time points for one full developmental cycle (48 h). We performed nuclear run-on with [α-32P]UTP and quantified the total transcriptional activity, proportional to polymerase occupancy at a locus (25), at each time point by measuring the incorporation of the radiolabeled nucleotide into these samples by precipitation and scintillation counting.
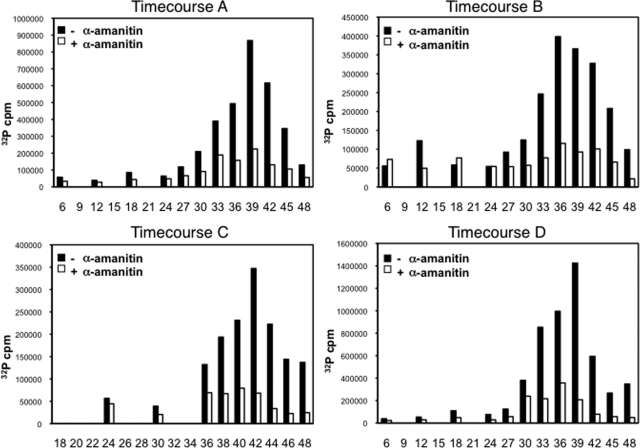
Over several independent time course experiments, we observed a robust peak in total transcriptional activity late in the IDC (Fig. 1), although the exact peak time point varied from time point (in hours) 36 (T36) to T42 between time courses due to minor differences in the synchronized populations (see Fig. S1 in the supplemental material). Activity during the first part of the IDC was significantly lower (see Table S1 in the supplemental material), although a detectable level of transcriptional activity was observed at early time points. Nuclear run-on labeling in the presence of α-amanitin, which inhibits RNAP II with high specificity (35, 71), resulted in significantly lower levels of incorporation (see Table S1 in the supplemental material), implying that the major component of the peak in total transcriptional activity is RNAP II dependent. Total incorporation by α-amanitin-treated samples did increase during approximately the second half of the IDC but did not correlate well with the RNAP II incorporation (R2 = 0.425, averaged for time courses A to D), indicating differential regulation of these two transcriptional activities.
FIG. 1.
Total transcriptional activity across the 48-h IDC. Transcriptional activity was measured in counts per minute (cpm) of total 32P incorporation by nuclei harvested during four independent time courses. Nuclei were treated with RNAP II inhibitor α-amanitin (+) or an equal volume of water (−) prior to incubation at 37°C for 30 min in the presence of [α-32P]UTP. x axes represent hours postreinvasion. Time points during time course C occur at 2 h resolution during the final 10 h.
Increased transcriptional activity occurs in multinucleate morphological stages.
Given the specific, singular occurrence of elevated total transcriptional activity during the IDC, we hypothesized that elevated transcriptional activity was associated with a particular morphological stage or stages. Thus, we sought to test the association of characteristic levels of transcriptional activity with the morphological stages observable by thin smear (see Fig. S1 in the supplemental material). The hypothesis that each of the six morphological stages (see Materials and Methods) has a characteristic level of transcriptional activity gives rise to the following description of the total transcriptional activity, as measured in the total-incorporation nuclear run-on assay:
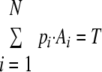 |
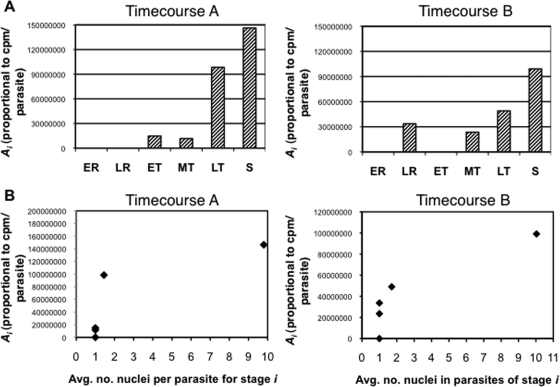
where, for any time point, the transcriptional activity of the population (T) can be described as the characteristic level of transcriptional activity for a stage i (Ai), multiplied by the number of parasites in that morphological stage (pi), summed over all morphological stages (1 − N). We solved for a best-fit vector for this overdetermined system (see Materials and Methods), which assigned transcriptional activity levels to each of the morphological stages (Fig. 2; see the supplemental material). To assess the degree to which a stage-specific model described the data, the output vector for each time course was multiplied with the morphological-stage data to render the transcriptional activity at each time point predicted by the model. These values fit the observed nuclear run-on data well (for time course A, R2 = 0.920, and for time course B, R2 = 0.914), validating that the assignment of characteristic levels of transcriptional activity to the six morphological stages of the IDC describes the observed trend. Furthermore, the output vectors implicated late trophozoites and schizonts, the multinucleate stages, in the observed elevated activity.
FIG. 2.
Fit of stage-specific model to observed transcriptional activity. (A) Total transcriptional activity per parasite for each stage as calculated by fitting algorithm. The vectors representing the best-fit characteristic transcriptional activity levels of the six morphological stages as output by the algorithm for time courses A and B are graphed. ER, early rings; LR, late rings; ET, early trophozoites; MT, mid-trophozoites; LT, late trophozoites; S, schizonts. (B) Relationship between model's output for transcriptional activity per parasite and number of nuclei per parasite. The number of nuclei per parasite was counted for parasites of each of the six stages (x axis) and compared to the characteristic transcriptional activity level for that stage (from panel A, y axis).
Transcriptional activity of the population results from both the number of nuclei per parasite and variable transcriptional activity per nucleus.
The morphological criteria by which these parasites were counted specify that late trophozoites contain one or two nuclei, while parasites with more than two nuclei are considered schizonts (60). The assignment of heightened transcriptional activity to late trophozoites and even greater activity to schizonts implicated additional genomic templates in the elevation of transcriptional activity. In order to pinpoint the timing of the transcriptional upregulation relative to DNA synthesis, radiolabeled [3H]hypoxanthine was added to cultures 3 h prior to each harvest to allow incorporation into freshly synthesized DNA. The proportion of 3H-labeled DNA was predicted to peak when the radiolabeling interval had included the first round of DNA replication. This peak in the proportion of 3H radioactivity occurred at the T30 time point in time course B (see Fig. S2A in the supplemental material), which coincided approximately with the beginning of upregulation of RNAP II transcriptional activity leading up to the peak.
The observation that the peak in transcriptional activity occurred after the onset of DNA replication led us to characterize the relationship between the multiple genome copies and transcriptional activity quantitatively. The same Wright-stained slides that provided morphological data for each trial (see Fig. S1 in the supplemental material) were used to count the number of nuclei per parasite. Although the correlation between the number of nuclei per parasite and RNAP II-dependent transcriptional activity was strong (see Fig. S2B in the supplemental material), the transcriptional activity assigned to a given stage increased monotonically, but not linearly, with the average number of nuclei in parasites of that stage (Fig. 2B). For all the time course experiments, the total transcriptional activity at each time point (Fig. 1), divided by the number of nuclei in the culture, was not constant (see Fig. S3A in the supplemental material), though the variable activity per nucleus could not be modeled by the six morphological stages (see Fig. S3B in the supplemental material). Together, the assignment of the highest level of transcriptional activity to the most-nucleated stage (schizonts) and the variable transcriptional activity per nucleus suggest that increased transcriptional activity of the synchronized population results from both the availability of template DNA and variable transcriptional activity per nucleus.
Transcriptional activity of specific genes.
The observed trend in total transcriptional activity during the IDC motivated investigation of whether the same pattern was recapitulated at the level of individual genes. The stage-specific peak of RNAP II-dependent total transcriptional activity during the late stages of the IDC could reflect a coinciding peak activity of the majority of genes or only that of a minority of high-activity genes. To address whether the coincident transcriptional activity peak was shared by genes with different steady-state RNA expression levels, a set was chosen for their variety with regard to (i) steady-state RNA profiles (3, 9, 34), (ii) gene ontology group, and (iii) adjacent genes in a chromosomal region of interest with respect to both genomic diversity and steady-state RNA expression in clinical isolates (9, 67) (Table 1).
TABLE 1.
Genes probed by nuclear run-on slot blot
| Description of gene group | Gene | Function | Relevant gene ontology term(s) |
|---|---|---|---|
| Constitutively high steady-state RNA levels by | PF07_0029 | Heat shock protein 86 (PfHsp90) | Response to heat, response to unfolded protein |
| microarray | PF13_0268 | 60S ribosomal protein L17 | Translation, cytosolic large ribosomal subunit |
| PFE0810c | 40S ribosomal protein S14 | Translation, cytosolic small ribosomal subunit | |
| Stage-specific steady-state | PF14_0187 | Glutathione S-transferase | Glutathione transferase activity |
| RNA levels | PFB0100c | Knob-associated histidine-rich protein | Host cell modification |
| PF14_0323 | Calmodulin | Calcium ion binding, cellular component, enzyme binding | |
| PF10_0345 | Merozoite surface protein 3 | Entry into host cell, cellular component | |
| Large-amplitude changes | PFI1755c | Hypothetical, conserved | None |
| in steady-state RNA | PFI1740c | Hypothetical | None |
| levels or putative | PFB0120w | Early-transcribed membrane protein | Membrane, apicoplast, molecular function |
| promoter | PF14_0097 | CDP diacylglycerol synthetase | Phospholipid biosynthetic process, phosphatidate cytidylyltransferase activity |
| Gene expression or chromatin-related gene | PF11_0062 | Histone H2B | Nucleosome, DNA binding, chromosome organization and biogenesis |
| products | PF07_0054 | Histone H2B variant | Chromatin assembly or disassembly, DNA binding, nucleus, nucleosome |
| PFC0915w | RNA DEAD/H box helicase | ATP-dependent RNA helicase activity, nucleic acid binding | |
| High steady-state RNA | PFD0695w | Hypothetical, conserved | None |
| levels in vivo, low | PF14_0134 | Hypothetical | None |
| expression in vitro | PFI0735c | NADH dehydrogenase | Electron transport, disulfide oxidoreductase activity |
| Chromosome 13 region of interest | MAL13P1.220 | Lipoate synthase, putative | Lipoic acid biosynthetic process, catalytic activity, mitochondrion, Fe-ion binding |
| PF13_0251 | DNA topoisomerase III | DNA unwinding during replication, DNA modification, DNA topoisomerase type 1 | |
| PF13_0252 | Nucleotide transporter 1 | Transport, membrane, nucleoside transporter activity | |
| PF13_0253 | Ethanolamine-P cytidylyltransferase, putative | Phospholipid biosynthetic process, catalytic activity, nucleotidyltransferase activity | |
| PF13_0254 | Hypothetical, conserved | None |
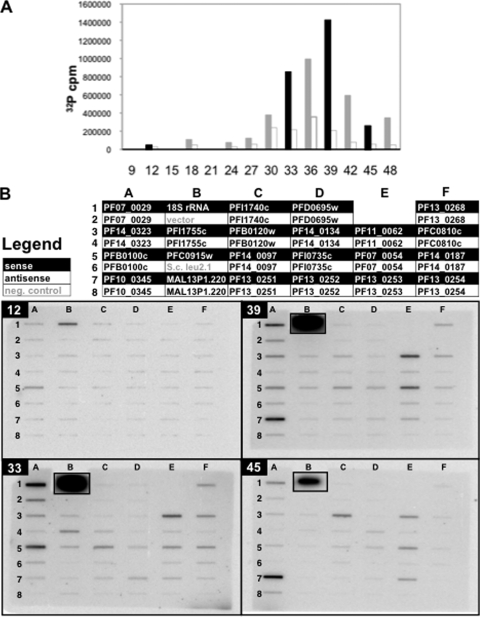
Four time points from time course D were chosen (Fig. 3A), representing ring stage (T12), late trophozoites (T33), schizonts (T39), and schizonts/rings (T45) (see Fig. S1 in the supplemental material). Nuclei from these time points, harvested from the same culture as those in the corresponding total-incorporation experiment, were labeled by [α-32P]UTP nuclear run-on. The resulting [32P]RNA (see Table S5 in the supplemental material) was hybridized to nylon filters (Fig. 3B) with immobilized single-stranded DNA probes for the sense and antisense strands of the genes. Consistent with the total incorporation data, very little transcriptional activity was detected among the genes assayed at T12. At later time points, both sense and antisense RNA were detected for all genes assayed, in agreement with previous results (45). Although α-amanitin-resistant activity was not the major component of total incorporation, unlike that of RNAP II, it is concentrated on a single locus during the IDC (69); accordingly, transcriptional activity of rRNA was detected more strongly than the RNAP II-transcribed genes on all blots.
FIG. 3.
Nuclear run-on gene-specific hybridization. (A) Total-incorporation nuclear run-on of time course D is depicted as in Fig. 1, with time points which were assayed by gene-specific nuclear run-on (T12, T33, T39, and T45) shown in black. x axes represent hours postreinvasion. (B) Single-stranded DNA probes were slot blotted onto membranes, which were hybridized with run-on-labeled [32P]RNA from time points T12, T33, T39, and T45 and imaged by exposure of a phosphorimager screen. The legend describes which RNA strand was detected. rRNA slots were cut from their filters and exposed separately to avoid interference between their signals and those of adjacent slots but are shown inset in their original positions.
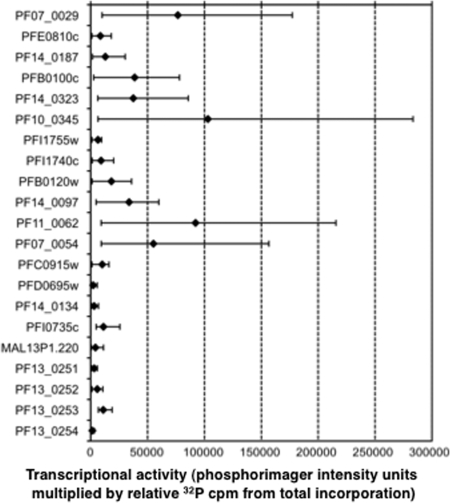
The normalized gene-specific nuclear run-on intensities were multiplied by the total transcriptional activity level of the population for each of the four time points assayed, allowing quantitative analysis of the absolute transcriptional activity of both sense and antisense strands of genes. The means and ranges of values for the sense strand of each gene across the four time points are plotted in Fig. 4. Sense-strand transcriptional activity for most genes varied by more than 10-fold for some genes and by 2- to 10-fold for others (e.g., PFI0735c, PF13_0252, and PF13_0253). The high sensitivity of radioactive nuclear run-on allows the detection of basal transcriptional activity not resulting from activation of the polymerase complex (1), and genes with a mean transcriptional activity level of the 10th percentile or below (PF14_0134, PFD0695w, and PF13_0254) were not included in further analysis.
FIG. 4.
Ranges of transcriptional activity across time points for sense strands of specific genes. The signal from the sense strand of each gene (shown in the same order as in Table 1) as detected by phosphorimager (see Table S4 in the supplemental material), corrected for single-stranded DNA probe purity and [32P]U content of the target RNA sequence, was normalized by the total radioactivity applied to its filter during hybridization and then multiplied by the total incorporation for that time point (relative to that of T12), yielding the absolute intensity value for that strand. The mean sense-strand signal for each gene across all four time points (diamonds) is shown between the maximum and minimum values (whiskers) among the four time points.
Overall, the timing of peak transcriptional activity for individual genes was distributed across the four time points assayed (Fig. 5; see Table S5 in the supplemental material). High mean transcriptional activity was not restricted to genes peaking at the same time point as the total transcriptional activity (T39). Nine of the 18 genes peaked in sense-strand transcriptional activity at T39 and represented a diversity of mean transcriptional activity. The five genes whose activity peaked at T33 encode proteins known to be expressed early in the IDC, such as PF07_0029 (33), PFB0100c (66), and PFE0810c (15). Of the three genes with peak transcriptional activities at T12, two are hypothetical, while MAL13P1.220 encodes a putative lipoate synthase, an important biosynthetic enzyme (73). The only gene with peak transcriptional activity at T45 was PFB0120w, a member of the early-transcribed membrane protein family, annotated for high steady-state RNA levels early in the IDC. Although this small gene set and the limited number of time points cannot prove the composition of the total transcriptional activity peak, these data are consistent with a model in which (i) many genes of diverse transcriptional activities share the peak time point with the total activity curve and (ii) the times of peak transcriptional activity for genes are distributed, though possibly not evenly, throughout the IDC.
FIG. 5.
Relationship between transcriptional activity and steady-state RNA levels. The corrected, normalized transcriptional activity of each strand, detected by gene-specific nuclear run-on in the slot blots depicted in Fig. 3 and multiplied by the total transcriptional activity for each time point, is displayed with respect to the maximum value for the gene among the four time points (bars: S, sense; AS, antisense). Levels of steady-state RNA (RNA) assayed by reverse transcription and real-time PCR are graphed with respect to their maximum values among the time points. All y axes represent the fraction of maximum for both transcriptional activity and steady-state RNA. Panels A to F represent the six gene groups assayed, as described in Table 1. Asterisks denote genes for which not all data were available; genes of low mean value were not included (see Table S3 in the supplemental material).
Transcriptional activity of antisense strands.
Consistent with previous nuclear run-on data (45), antisense transcription was detectable for most genes at most time points (Fig. 5; see Table S4 in the supplemental material). Sense strands of all genes across all time points constituted 83% of the RNAP II activity observed (a comparison of the sum of intensities from sense- and antisense-strand slots on the four blots), which is in approximate agreement with the results of steady-state RNA studies of mixed-stage parasites (21, 51). Accordingly, sense strands generally exhibited higher activity than their complementary strands by individual comparison of loci at time points. However, in the cases of PFI1755c and PFB0120w, inversion of this pattern occurs at T33. For both genes, T33 was both the peak of antisense-strand transcriptional activity and the time of minimum sense-strand transcriptional activity, suggesting that regulation of the transcriptional activities of these complementary strands may be related.
Relationship between transcriptional activity and steady-state RNA.
To characterize the relationship between transcriptional activity and steady-state RNA for these genes and thereby address whether transcriptional activation could be responsible for modulation of gene expression, steady-state RNA was quantified. Real-time RT-PCR was performed on whole-parasite RNA samples harvested concurrently with the nuclei used in gene-specific nuclear run-on during time course D (Fig. 3A). These quantities were corrected for recovery artifacts to reflect the absolute steady-state RNA level of each gene (see the supplemental material) and are graphed alongside the transcriptional activity of each strand according to nuclear run-on, all with respect to their maximum values, in Fig. 5.
Since four time points were included in the assay, we compared the three transitional changes occurring between these time points. For both transcriptional activity and steady-state RNA level for each gene, each change was described as the difference between a value and that of the previous time point. The correlation coefficient between the changes in steady-state RNA levels and those in transcriptional activities was then calculated for each gene (see Fig. S4 in the supplemental material). This comparison revealed a strong positive correlation (R > 0.95) for 7 of the 18 genes analyzed and a continuum of partial, poor, and negative correlation for the remainder. Those with the highest correlations, and therefore the most direct relationship between transcriptional activity and steady-state RNA level, included RNA DEAD/H box helicase PFC0915w (R = 0.998), CDP diacylglycerol synthase PF14_0097 (R = 0.989), and calmodulin PF14_0323 (R = 0.988). Lower values reflected cases where some but not all changes were correlated. To investigate whether discrepancies between the changes in the transcriptional activities and steady-state RNA levels of a gene could be used to predict regulation at the level of RNA stability, we compared the changes seen in our results to previously reported changes in RNA half-life (59) for those genes during those transitions (Table 2). For the genes and transitions for which both transcriptional activity and RNA stability datasets were available, we found that most discrepancies between changes in transcriptional activity and steady-state RNA accumulation could be accounted for by an opposing change in RNA stability. For example, the decrease in the transcriptional activity of PF10_0345 (msp3) between T39 and T45 and the contrasting increase in steady-state RNA was, as predicted, concurrent with an increase in its RNA half-life.
TABLE 2.
Comparison of increases and decreases in transcriptional activity, steady-state RNA level, and RNA half-lifea
| Gene | Time point transitionb | Transcriptional activity | Steady-state RNA | RNA stability (half-life)c |
|---|---|---|---|---|
| PFE0810c | 33 to 39 | − | + | + |
| PF10_0345 | 12 to 33 | (−) | (+) | − |
| 39 to 45 | − | + | +/− | |
| PFI0735c | 12 to 33 | − | + | +/+ |
| MAL1P13.220 | 12 to 33 | − | + | + |
| PF13_0253 | 12 to 33 | (−) | (+) | (−) |
| 39 to 45 | − | + | + |
+ and − indicate direction of change, and parentheses indicate changes that were not large (<5% of the maximum value for the gene). RNA half-life data are as reported by Shock et al. (59).
Numbers represent hourly time points.
Genes for which data from two probes were available in the data of Shock et al. have both changes noted, separated by a slash.
DISCUSSION
We have demonstrated that the reproducible peak in total transcriptional activity during the P. falciparum IDC is stage specific and dependent on RNAP II activity, which in turn varies with both the number of nuclei per parasite and a changing level of transcriptional activity per nucleus. Applying the total transcriptional activity level of the population, which is differential among time points, to gene-specific nuclear run-on data from those time points allowed the calculation of the absolute transcriptional activities of both sense and antisense strands of genes. Overall, the changes between time points in the transcriptional activity of each gene correlated well with the changes in its steady-state RNA level, and individual discrepancies were strong indicators of changes in RNA stability observed in previous studies. Comparison of transcriptional activity and steady-state RNA level, a strategy used to infer posttranscriptional mechanisms of gene regulation in other organisms (16), offers insight into the competing influences of transcriptional activity and RNA stability on the expression of individual genes and the physiology underpinning the morphological stages of the IDC.
The absolute transcriptional activity levels obtained through the combination of total-incorporation and gene-specific hybridization run-on revealed three subsets of genes among our set: (i) those with high correlation between changes in transcriptional activity and steady-state RNA level, (ii) those with major discrepancies suggestive of posttranscriptional regulation, and (iii) those with very low transcriptional activity. For the first subset, excellent correlation implied that steady-state expression was regulated primarily at the level of transcriptional activity throughout the IDC. We corroborated several cases in which transcriptional activity has been proposed as the main mechanism of regulation (PF14_0097 [50], PF14_0323 [54], PF07_0029 [44], and PFB0100c [31]). Among the second group, the direction of previously observed changes in RNA stability between these time points (59) was consistent with the change in steady-state RNA levels (Table 2; see the supplemental material), validating the idea that discrepancies between sense-strand transcriptional activities and steady-state RNA levels indicate the influence of changes in RNA stability during the IDC. These findings support the model of regulated RNA stability proposed by Shock et al. (59) and provide motivation for the use of other discrepancies in the flow of gene expression (33) to diagnose targets of regulation during the P. falciparum IDC. The third group, whose low signal may represent only basal transcriptional activity, was populated by genes chosen for their low steady-state expression level in cultured P. falciparum but high expression level in samples taken directly from patients (9). Their low transcriptional activities in this assay implies that their differential regulation in some in vivo parasites may occur at the level of transcriptional activity.
Antisense transcriptional activity was observed for all genes during at least one time point, though in some cases (e.g., for PFI0735c) it was constitutively very low. In two cases, antisense activity was not only substantial but exceeded that of sense-strand transcription during one time point assayed (PFB0120w and PFI1755c). The peak in antisense transcriptional activity of both genes occurs at T33 and coincides with their lowest levels of steady-state RNA. This finding highlights the importance of investigating the physiological role of P. falciparum's abundant antisense transcripts (21), particularly whether they regulate their complementary mRNAs at the transcriptional or posttranscriptional level.
Total RNAP II activity during the cell cycle in metazoan cells becomes repressed during S, G2, and M phases (74). Our observation of the opposite pattern during the P. falciparum IDC suggests that the regulation of transcriptional activity intrinsic to the morphological stages is not simply an overlay of the canonical cell cycle onto this complex cell but instead resembles developmental modulation of RNAP activity by modification of the enzyme itself or by template accessibility (36, 68, 70). The proteins responsible for phosphorylation/activation of RNAP II are encoded in the P. falciparum genome and give rise to a functional enzyme complex (6), but their expression and regulation during the IDC are yet uncharacterized in living parasites. The positive relationship between total transcriptional activity and number of nuclei implies transcription from multiple templates, the uniformity of which varies greatly among multinucleate systems (48, 49), while the variable transcriptional activity per nucleus implies regulation of bulk RNAP activity.
Similarities between the P. falciparum IDC and cellular development beyond mitotic cycling suggest that the parasite may rely on a combination of mechanisms to regulate the expression of specific genes (28, 36, 57). Regarding transcriptional activity, although the peak activities of genes were distributed among the four time points assayed, several of the genes with the closest correlation between transcriptional activity and steady-state RNA level share a T39 peak with the total transcriptional activity curve. Agreement with the composite transcriptome coincided with transcriptional activity being the dominant regulatory step in gene expression. This observation hints that the primary role of cis-acting DNA sequences might be to regulate only certain genes whose expression must deviate from a driving trend of RNAP II activity: that is, to repress activity at a locus during the increase in RNAP II activity or induce it at an alternative time. Genes assayed by nuclear run-on in this study and previously (26, 27, 29, 31, 32, 58) have exhibited a diversity of patterns in transcriptional activity across the IDC, supporting a model of stage-specific total transcriptional activity arising from the collective patterns of many individual genes which are nonidentical but cumulatively reflect both bulk RNAP II activity and template availability. Thus, future studies which consider total transcriptional activity in concert with that of specific genes and their steady-state RNA levels promise to provide insight into mechanisms of regulation at the transcriptional and posttranscriptional levels.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation graduate research fellowships (J.S.S. and V.P.P.), NIH postdoctoral fellowship AI050303-01 (K.T.M.), and NIH grant GM61351-03 (D.F.W.).
We thank Simon Dove and his lab members for use of the Storm 820 phosphorimager, as well as Dan Milner and the Pathology Department of Brigham and Women's Hospital for use of their microscope and camera. We thank Gilberto Ramirez, David Rosen, Nira Mahesh, and Amanda Lukens for technical support and Johanna Daily, Michelle LeRoux, and Carolyn Dong for valuable discussion of methods and the manuscript. Special thanks to Manuel Llinás and Eric Rubin for critical reading of the manuscript.
Footnotes
Published ahead of print on 16 January 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Akhtar, A., G. Faye, and D. L. Bentley. 1996. Distinct activated and non-activated RNA polymerase II complexes in yeast. EMBO J. 154654-4664. [PMC free article] [PubMed] [Google Scholar]
- 2.Balaji, S., M. M. Babu, L. M. Iyer, and L. Aravind. 2005. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 333994-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bzik, D. J. 1991. The structure and role of RNA polymerases in Plasmodium. Parasitol. Today 7211-214. [DOI] [PubMed] [Google Scholar]
- 5.Callebaut, I., K. Prat, E. Meurice, J. P. Mornon, and S. Tomavo. 2005. Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: conserved features and differences relative to other eukaryotes. BMC Genomics 6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y., D. Jirage, D. Caridha, A. K. Kathcart, E. A. Cortes, R. A. Dennull, J. A. Geyer, S. T. Prigge, and N. C. Waters. 2006. Identification of an effector protein and gain-of-function mutants that activate Pfmrk, a malarial cyclin-dependent protein kinase. Mol. Biochem. Parasitol. 14948-57. [DOI] [PubMed] [Google Scholar]
- 7.Coleman, B. I., and M. T. Duraisingh. 2008. Transcriptional control and gene silencing in Plasmodium falciparum. Cell. Microbiol. 101935-1946. [DOI] [PubMed] [Google Scholar]
- 8.Coulson, R. M., N. Hall, and C. A. Ouzounis. 2004. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 141548-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daily, J. P., D. Scanfeld, N. Pochet, K. Le Roch, D. Plouffe, M. Kamal, O. Sarr, S. Mboup, O. Ndir, D. Wypij, K. Levasseur, E. Thomas, P. Tamayo, C. Dong, Y. Zhou, E. S. Lander, D. Ndiaye, D. Wirth, E. A. Winzeler, J. P. Mesirov, and A. Regev. 2007. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 4501091-1095. [DOI] [PubMed] [Google Scholar]
- 10.Deans, J. A., A. W. Thomas, P. M. Inge, and S. Cohen. 1983. Stage-specific protein synthesis by asexual blood stage parasites of Plasmodium falciparum. Mol. Biochem. Parasitol. 845-51. [DOI] [PubMed] [Google Scholar]
- 11.Deitsch, K., M. Duraisingh, R. Dzikowski, A. Gunasekera, S. Khan, K. Le Roch, M. Llinas, G. Mair, V. McGovern, D. Roos, J. Shock, J. Sims, R. Wiegand, and E. Winzeler. 2007. Mechanisms of gene regulation in Plasmodium. Am. J. Trop. Med. Hyg. 77201-208. [PubMed] [Google Scholar]
- 12.De Silva, E. K., A. R. Gehrke, K. Olszewski, I. Leon, J. S. Chahal, M. L. Bulyk, and M. Llinas. 2008. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc. Natl. Acad. Sci. USA 1058393-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desjardins, R. E., C. J. Canfield, J. D. Haynes, and J. D. Chulay. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieckmann, A., and A. Jung. 1986. Stage-specific sensitivity of Plasmodium falciparum to antifolates. Z. Parasitenkd. 72591-594. [DOI] [PubMed] [Google Scholar]
- 15.Florens, L., M. P. Washburn, J. D. Raine, R. M. Anthony, M. Grainger, J. D. Haynes, J. K. Moch, N. Muster, J. B. Sacci, D. L. Tabb, A. A. Witney, D. Wolters, Y. Wu, M. J. Gardner, A. A. Holder, R. E. Sinden, J. R. Yates, and D. J. Carucci. 2002. A proteomic view of the Plasmodium falciparum life cycle. Nature 419520-526. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Martinez, J., A. Aranda, and J. E. Perez-Ortin. 2004. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell 15303-313. [DOI] [PubMed] [Google Scholar]
- 17.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gissot, M., S. Briquet, P. Refour, C. Boschet, and C. Vaquero. 2005. PfMyb1, a Plasmodium falciparum transcription factor, is required for intra-erythrocytic growth and controls key genes for cell cycle regulation. J. Mol. Biol. 34629-42. [DOI] [PubMed] [Google Scholar]
- 19.Gritzmacher, C. A., and R. T. Reese. 1984. Protein and nucleic acid synthesis during synchronized growth of Plasmodium falciparum. J. Bacteriol. 1601165-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gunasekera, A. M., A. Myrick, K. T. Militello, J. S. Sims, C. K. Dong, T. Gierahn, K. Le Roch, E. Winzeler, and D. F. Wirth. 2007. Regulatory motifs uncovered among gene expression clusters in Plasmodium falciparum. Mol. Biochem. Parasitol. 15319-30. [DOI] [PubMed] [Google Scholar]
- 21.Gunasekera, A. M., S. Patankar, J. Schug, G. Eisen, J. Kissinger, D. Roos, and D. F. Wirth. 2004. Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Mol. Biochem. Parasitol. 13635-42. [DOI] [PubMed] [Google Scholar]
- 22.Gupta, S. K., S. Schulman, and J. P. Vanderberg. 1985. Stage-dependent toxicity of N-acetyl-glucosamine to Plasmodium falciparum. J. Protozool. 3291-95. [DOI] [PubMed] [Google Scholar]
- 23.Hakimi, M. A., and K. W. Deitsch. 2007. Epigenetics in Apicomplexa: control of gene expression during cell cycle progression, differentiation and antigenic variation. Curr. Opin. Microbiol. 10357-362. [DOI] [PubMed] [Google Scholar]
- 24.Hall, N., M. Karras, J. D. Raine, J. M. Carlton, T. W. Kooij, M. Berriman, L. Florens, C. S. Janssen, A. Pain, G. K. Christophides, K. James, K. Rutherford, B. Harris, D. Harris, C. Churcher, M. A. Quail, D. Ormond, J. Doggett, H. E. Trueman, J. Mendoza, S. L. Bidwell, M. A. Rajandream, D. J. Carucci, J. R. Yates III, F. C. Kafatos, C. J. Janse, B. Barrell, C. M. Turner, A. P. Waters, and R. E. Sinden. 2005. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 30782-86. [DOI] [PubMed] [Google Scholar]
- 25.Hirayoshi, K., and J. T. Lis. 1999. Nuclear run-on assays: assessing transcription by measuring density of engaged RNA polymerases. Methods Enzymol. 304351-362. [DOI] [PubMed] [Google Scholar]
- 26.Horrocks, P., M. Jackson, S. Cheesman, J. H. White, and B. J. Kilbey. 1996. Stage specific expression of proliferating cell nuclear antigen and DNA polymerase delta from Plasmodium falciparum. Mol. Biochem. Parasitol. 79177-182. [DOI] [PubMed] [Google Scholar]
- 27.Horrocks, P., and M. Lanzer. 1999. Mutational analysis identifies a five base pair cis-acting sequence essential for GBP130 promoter activity in Plasmodium falciparum. Mol. Biochem. Parasitol. 9977-87. [DOI] [PubMed] [Google Scholar]
- 28.Kuersten, S., and E. B. Goodwin. 2003. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 4626-637. [DOI] [PubMed] [Google Scholar]
- 29.Kyes, S., Z. Christodoulou, R. Pinches, and C. Newbold. 2002. Stage-specific merozoite surface protein 2 antisense transcripts in Plasmodium falciparum. Mol. Biochem. Parasitol. 12379-83. [DOI] [PubMed] [Google Scholar]
- 30.Lambros, C., and J. P. Vanderberg. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65418-420. [PubMed] [Google Scholar]
- 31.Lanzer, M., D. de Bruin, and J. V. Ravetch. 1992. A sequence element associated with the Plasmodium falciparum KAHRP gene is the site of developmentally regulated protein-DNA interactions. Nucleic Acids Res. 203051-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanzer, M., D. de Bruin, and J. V. Ravetch. 1992. Transcription mapping of a 100 kb locus of Plasmodium falciparum identifies an intergenic region in which transcription terminates and reinitiates. EMBO J. 111949-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Roch, K. G., J. R. Johnson, L. Florens, Y. Zhou, A. Santrosyan, M. Grainger, S. F. Yan, K. C. Williamson, A. A. Holder, D. J. Carucci, J. R. Yates III, and E. A. Winzeler. 2004. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 142308-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, P. De La Vega, A. A. Holder, S. Batalov, D. J. Carucci, and E. A. Winzeler. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 3011503-1508. [DOI] [PubMed] [Google Scholar]
- 35.Lindell, T. J., F. Weinberg, P. W. Morris, R. G. Roeder, and W. J. Rutter. 1970. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science 170447-449. [DOI] [PubMed] [Google Scholar]
- 36.Lis, J. T. 2007. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature 450198-202. [DOI] [PubMed] [Google Scholar]
- 37.Llinas, M., Z. Bozdech, E. D. Wong, A. T. Adai, and J. L. DeRisi. 2006. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 341166-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Estrano, C., A. M. Gopalakrishnan, J. P. Semblat, M. R. Fergus, D. Mazier, and K. Haldar. 2007. An enhancer-like region regulates hrp3 promoter stage-specific gene expression in the human malaria parasite Plasmodium falciparum. Biochim. Biophys. Acta 1769506-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez-Rubio, J. J., L. Riviere, and A. Scherf. 2007. Shared epigenetic mechanisms control virulence factors in protozoan parasites. Curr. Opin. Microbiol. 10560-568. [DOI] [PubMed] [Google Scholar]
- 40.Loyevsky, M., F. Mompoint, E. Yikilmaz, S. F. Altschul, T. Madden, J. C. Wootton, J. Kurantsin-Mills, O. O. Kassim, V. R. Gordeuk, and T. A. Rouault. 2003. Expression of a recombinant IRP-like Plasmodium falciparum protein that specifically binds putative plasmodial IREs. Mol. Biochem. Parasitol. 126231-238. [DOI] [PubMed] [Google Scholar]
- 41.Mair, G. R., J. A. Braks, L. S. Garver, J. C. Wiegant, N. Hall, R. W. Dirks, S. M. Khan, G. Dimopoulos, C. J. Janse, and A. P. Waters. 2006. Regulation of sexual development of Plasmodium by translational repression. Science 313667-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reference deleted.
- 43.Martin, R. E., R. I. Henry, J. L. Abbey, J. D. Clements, and K. Kirk. 2005. The “permeome” of the malaria parasite: an overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 6R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Militello, K. T., M. Dodge, L. Bethke, and D. F. Wirth. 2004. Identification of regulatory elements in the Plasmodium falciparum genome. Mol. Biochem. Parasitol. 13475-88. [DOI] [PubMed] [Google Scholar]
- 45.Militello, K. T., V. Patel, A. D. Chessler, J. K. Fisher, J. M. Kasper, A. Gunasekera, and D. F. Wirth. 2005. RNA polymerase II synthesizes antisense RNA in Plasmodium falciparum. RNA 11365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Militello, K. T., P. Refour, C. A. Comeaux, and M. T. Duraisingh. 2008. Antisense RNA and RNAi in protozoan parasites: working hard or hardly working? Mol. Biochem. Parasitol. 157117-126. [DOI] [PubMed] [Google Scholar]
- 47.Myrick, A., A. Munasinghe, S. Patankar, and D. F. Wirth. 2003. Mapping of the Plasmodium falciparum multidrug resistance gene 5′-upstream region, and evidence of induction of transcript levels by antimalarial drugs in chloroquine sensitive parasites. Mol. Microbiol. 49671-683. [DOI] [PubMed] [Google Scholar]
- 48.Newlands, S., L. K. Levitt, C. S. Robinson, A. B. Karpf, V. R. Hodgson, R. P. Wade, and E. C. Hardeman. 1998. Transcription occurs in pulses in muscle fibers. Genes Dev. 122748-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newport, J., and M. Kirschner. 1982. A major developmental transition in early Xenopus embryos. II. Control of the onset of transcription. Cell 30687-696. [DOI] [PubMed] [Google Scholar]
- 50.Osta, M., L. Gannoun-Zaki, S. Bonnefoy, C. Roy, and H. J. Vial. 2002. A 24 bp cis-acting element essential for the transcriptional activity of Plasmodium falciparum CDP-diacylglycerol synthase gene promoter. Mol. Biochem. Parasitol. 12187-98. [DOI] [PubMed] [Google Scholar]
- 51.Patankar, S., A. Munasinghe, A. Shoaibi, L. M. Cummings, and D. F. Wirth. 2001. Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Mol. Biol. Cell 123114-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porter, M. E. 2001. The DNA polymerase delta promoter from Plasmodium falciparum contains an unusually long 5′ untranslated region and intrinsic DNA curvature. Mol. Biochem. Parasitol. 114249-255. [DOI] [PubMed] [Google Scholar]
- 54.Robson, K. J., and M. W. Jennings. 1991. The structure of the calmodulin gene of Plasmodium falciparum. Mol. Biochem. Parasitol. 4619-34. [DOI] [PubMed] [Google Scholar]
- 55.Ruvalcaba-Salazar, O. K., M. del Carmen Ramirez-Estudillo, D. Montiel-Condado, F. Recillas-Targa, M. Vargas, and R. Hernandez-Rivas. 2005. Recombinant and native Plasmodium falciparum TATA-binding-protein binds to a specific TATA box element in promoter regions. Mol. Biochem. Parasitol. 140183-196. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 57.Schaner, C. E., and W. G. Kelly. 24 January 2006. Germline chromatin. In The C. elegans Research Community, WormBook. doi: 10.1895/wormbook.1.73.1. [DOI] [PMC free article] [PubMed]
- 58.Schieck, E., J. M. Pfahler, C. P. Sanchez, and M. Lanzer. 2007. Nuclear run-on analysis of var gene expression in Plasmodium falciparum. Mol. Biochem. Parasitol. 153207-212. [DOI] [PubMed] [Google Scholar]
- 59.Shock, J. L., K. F. Fischer, and J. L. DeRisi. 2007. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intra-erythrocytic development cycle. Genome Biol. 8R134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silamut, K., and N. J. White. 1993. Relation of the stage of parasite development in the peripheral blood to prognosis in severe falciparum malaria. Trans. R. Soc. Trop. Med. Hyg. 87436-443. [DOI] [PubMed] [Google Scholar]
- 61.Skinner, T. S., L. S. Manning, W. A. Johnston, and T. M. Davis. 1996. In vitro stage-specific sensitivity of Plasmodium falciparum to quinine and artemisinin drugs. Int. J. Parasitol. 26519-525. [DOI] [PubMed] [Google Scholar]
- 62.Sunil, S., V. S. Chauhan, and P. Malhotra. 2008. Distinct and stage specific nuclear factors regulate the expression of falcipains, Plasmodium falciparum cysteine proteases. BMC Mol. Biol. 947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trager, W., and J. B. Jensen. 1976. Human malaria parasites in continuous culture. Science 193673-675. [DOI] [PubMed] [Google Scholar]
- 64.Ullu, E., C. Tschudi, and T. Chakraborty. 2004. RNA interference in protozoan parasites. Cell. Microbiol. 6509-519. [DOI] [PubMed] [Google Scholar]
- 65.van Noort, V., and M. A. Huynen. 2006. Combinatorial gene regulation in Plasmodium falciparum. Trends Genet. 2273-78. [DOI] [PubMed] [Google Scholar]
- 66.Vernot-Hernandez, J. P., and H. G. Heidrich. 1984. Time-course of synthesis, transport and incorporation of a protein identified in purified membranes of host erythrocytes infected with a knob-forming strain of Plasmodium falciparum. Mol. Biochem. Parasitol. 12337-350. [DOI] [PubMed] [Google Scholar]
- 67.Volkman, S. K., P. C. Sabeti, D. DeCaprio, D. E. Neafsey, S. F. Schaffner, D. A. Milner, Jr., J. P. Daily, O. Sarr, D. Ndiaye, O. Ndir, S. Mboup, M. T. Duraisingh, A. Lukens, A. Derr, N. Stange-Thomann, S. Waggoner, R. Onofrio, L. Ziaugra, E. Mauceli, S. Gnerre, D. B. Jaffe, J. Zainoun, R. C. Wiegand, B. W. Birren, D. L. Hartl, J. E. Galagan, E. S. Lander, and D. F. Wirth. 2007. A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 39113-119. [DOI] [PubMed] [Google Scholar]
- 68.Walker, A. K., P. R. Boag, and T. K. Blackwell. 2007. Transcription reactivation steps stimulated by oocyte maturation in C. elegans. Dev. Biol. 304382-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Waters, A. P., C. Syin, and T. F. McCutchan. 1989. Developmental regulation of stage-specific ribosome populations in Plasmodium. Nature 342438-440. [DOI] [PubMed] [Google Scholar]
- 70.Wechselberger, M., B. Wielgat, and G. Kahl. 1979. Rhythmic changes in transcriptional activity during the development of potato tubers. Planta 147199-204. [DOI] [PubMed] [Google Scholar]
- 71.Weil, P. A., and S. P. Blatti. 1975. Partial purification and properties of calf thymus deoxyribonucleic acid dependent RNA polymerase III. Biochemistry 141636-1642. [DOI] [PubMed] [Google Scholar]
- 72.Wu, J., D. H. Sieglaff, J. Gervin, and X. S. Xie. 2008. Discovering regulatory motifs in the Plasmodium genome using comparative genomics. Bioinformatics 241843-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeh, I., T. Hanekamp, S. Tsoka, P. D. Karp, and R. B. Altman. 2004. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res. 14917-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yonaha, M., T. Chibazakura, S. Kitajima, and Y. Yasukochi. 1995. Cell cycle-dependent regulation of RNA polymerase II basal transcription activity. Nucleic Acids Res. 234050-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Young, J. A., J. R. Johnson, C. Benner, S. F. Yan, K. Chen, K. G. Le Roch, Y. Zhou, and E. A. Winzeler. 2008. In silico discovery of transcription regulatory elements in Plasmodium falciparum. BMC Genomics 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.