Abstract
The metazoan cytosolic fatty acid synthase (FAS) contains all of the enzymes required for de novo fatty acid biosynthesis covalently linked around two reaction chambers. While the 3D architecture of FAS has been mostly defined, it is unclear how reaction intermediates can transfer between distant catalytic domains. Using single-particle electron microscopy we have identified a near continuum of conformations consistent with remarkable flexibility of FAS. The distribution of conformations was influenced by the presence of substrates and altered by different catalytic mutations suggesting a direct correlation between conformation and specific enzymatic activities. 3D reconstructions were interpreted by docking high-resolution structures of individual domains and illustrate that the substrate loading and condensation domains dramatically swing and swivel to access substrates within either reaction chamber. Concomitant rearrangement of the β-carbon processing domains synchronizes acyl-chain reduction in one chamber with acyl-chain elongation in the other.
The synthesis de novo of long-chain fatty acids universally involves a suite of enzymes that catalyze the iterative elongation and processing of the carbon chain followed by product release (Fig. 1). In all fatty acid synthase (FAS) systems, covalently bound reaction intermediates are translocated between active sites by an acyl carrier protein (ACP). In prokaryotes, chloroplasts and mitochondria, the constituent enzymes are freestanding proteins but in the cytosol of eukaryotes they are integrated into giant multifunctional polypeptide chains1. Interest in the structure and mechanism of action of the mammalian FAS has been stimulated by the realization that the protein is a potential target for treatment of obesity and cancer, since FAS inhibitors are effective appetite suppressants and can selectively target several types of cancer cells2,3.
Figure 1.
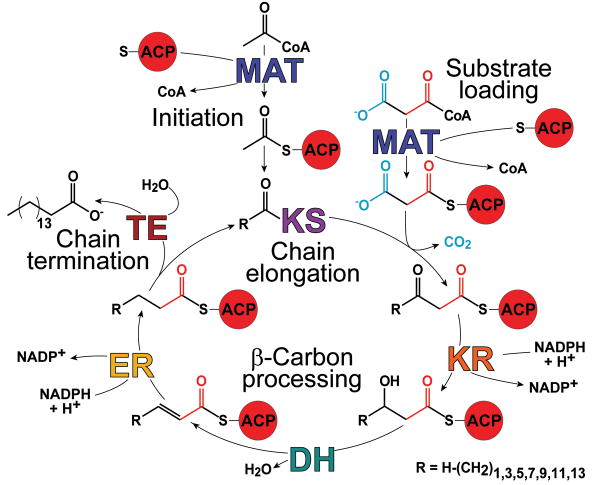
Structural and functional organization of the metazoan FAS. (a) Fatty acid biosynthesis reaction cycle initiates with transfer of the acetyl moiety to the KS via an ACP-bound intermediate (Initiation). The malonyl thioester is similarly transferred to an ACP (Substrate Loading) and then condensed with the KS-bound acyl chain (Chain Extension). The resulting β-ketone is then reduced and dehydrated yielding a saturated acyl chain (β-Carbon Processing) that is delivered to the KS initiating the next cycle. After 7 cycles the 16 carbon acyl chain, palmitate, is released (Chain Termination).
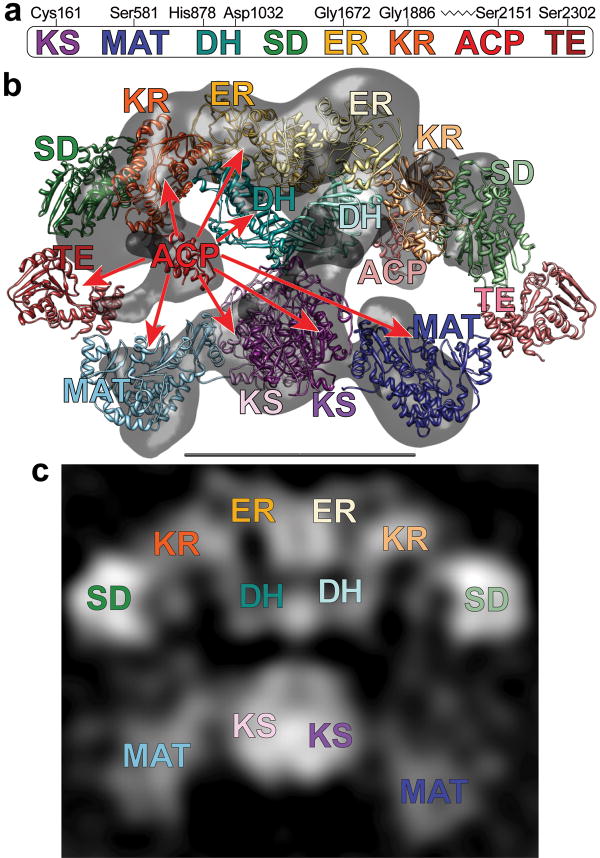
Surprisingly, the evolution of the eukaryotic megasynthases has followed two radically different architectural themes. In the 2.6-megadalton fungal FAS constituent enzymes are all embedded in the interior wall of a rigid barrel-shaped structure. Access of the enzymes to their ACP-bound substrates is facilitated entirely by movement of the ACP domains about their attachment points in the center of the barrel4,5. By contrast, the homodimeric 0.54-megadalton metazoan FAS (Fig. 2a) is an extremely flexible macromolecule6. A model of FAS derived by fitting high resolution structures of individual prokaryotic enzymes into a 4.5 Å resolution FAS crystallographic density map7, revealed that the FAS subunits come together to form a central interface comprised of dimeric β-ketoacyl synthase (KS) and enoyl reductase (ER) domains, and a pair of pseudodimeric dehydratase (DH) domains (Fig. 2b). The ER and DH domains in the upper portion of the FAS structure are flanked by appended pairs of monomeric ketoreductase (KR) domains. In the lower section, the KS domains are positioned between monomeric malonyl/acetyl transferase (MAT) domains. Upper and lower sections are joined by a narrow connection formed by the distal end of the linker connecting the MAT and DH domains. The resolution of the X-ray density map was insufficient to determine whether the FAS subunits were in a back-to-back or crossed-over arrangement. An additional section of the structure must comprise the ACP and the thioesterase (TE) domain that catalyzes the chain termination step. Extensive flexibility in their flanking linker regions prevented imaging of these domains in the X-ray crystallography electron density map, but we surmise (by virtue of the covalent linkage between the ACP and the carboxyl terminus of the KR domain) that an ACP-sized density observed below the KR domains in the cryo-electron microscopy (EM) structure marks the position of the ACP domain in the upper section of the EM reconstruction.
Figure 2.
(a) The domains of FAS are linearly arranged along the FAS polypeptide. Key active-site residues for the KS, MAT, DH and TE domains, the location of the glycine-rich motifs of the nucleotide-binding sites in the ER and KR domains, and the site of posttranslational phosphopantetheinylation in the ACP domain are marked (rat FAS numbering). (b) Crystal and cryo-EM structures capture different conformations of FAS. Atomic structures of individual catalytic domains were positioned according to the intermediate-resolution crystal structure7. ACP domains36 were fitted into remaining densities located below the KR domains in the cryo-EM structure (gray)6. Densities corresponding to the TE domains were not apparent in either the crystal structure or the cryo-EM structure but the domains are positioned near the outer edge of the two reaction chambers based on evidence presented in this article. Subunits in the FAS homodimer are depicted in a crossed-over arrangement with the domains of one subunit in faded colors. Catalytic contacts made by the ACP of one subunit are indicated by arrows. Flexibility of the KR-ACP linker and mobility of the phosphopantetheine are insufficient to explain contacts with the distant KS and MAT active sites. (c) A 2D class average calculated from images of FAS molecules preserved in stain has recognizable structural elements and shows good correspondence with the X-ray and cryo-EM 3D structures. The scale bar represents 100 Å.
The FAS structure defined by the 4.5 Å crystallographic density map neatly compartmentalizes constituent domains. Domains involved in chain extension (KS and MAT) grouped in the lower portion of the structure and domains responsible for β-carbon processing (KR, DH, and ER) in the upper portion alternately engage the ACP during each catalytic cycle and are arranged around two discrete reaction chambers in which each ACP has access to only one set of catalytic domains7. However, a substantial body of biochemical evidence, including mutant complementation analyses8 and site-specific cross-linking9, indicates that the ACP domains are able to make functional contacts with the KS and MAT domains of either subunit (red arrows in Fig. 2b). Clearly, substantial flexibility of the megasynthase would be required to permit functional contacts between these domains that, in the crystal structure, appear distantly located. In this study, we used single-particle macromolecular electron microscopy (EM) to characterize a wide range of conformations that enable functionality of a mammalian (Rattus norvegicus) FAS by facilitating all required catalytic interactions.
Results
Structural Analysis of FAS using single particle EM
Derivation of structural information from noisy EM images of single macromolecules relies on averaging of properly aligned images that, in principle, originate from identical particles. If particles are heterogeneous in conformation the analysis becomes considerably more complex, especially for the unstained specimens in which biological macromolecules are best preserved. Preservation of particles in stain can result in some distortion but the relatively high signal-to-noise ratio in the resulting images, in combination with suitable statistical analysis, can allow for a quantitative description of different molecular conformations10. Earlier EM reconstructions of FAS calculated from molecules preserved in stain or ice were remarkably similar6 and both resembled the intermediate resolution X-ray structure7 in overall shape and size (Fig. 2b), indicating that FAS is fairly resistant to stain-induced deformation. Moreover, upon adsorption to the amorphous carbon support film used to prepare stained samples, FAS showed a strongly preferred orientation, so that image alignment and classification can be used to sort out changes in molecular conformation without the complication of having to determine particle orientation parameters. Therefore, images of FAS from stained samples could be used to calculate 2D class averages (Fig. 2c) and corresponding 3D reconstructions that would reveal relative domain positions in different conformational states.
FAS structural pliability
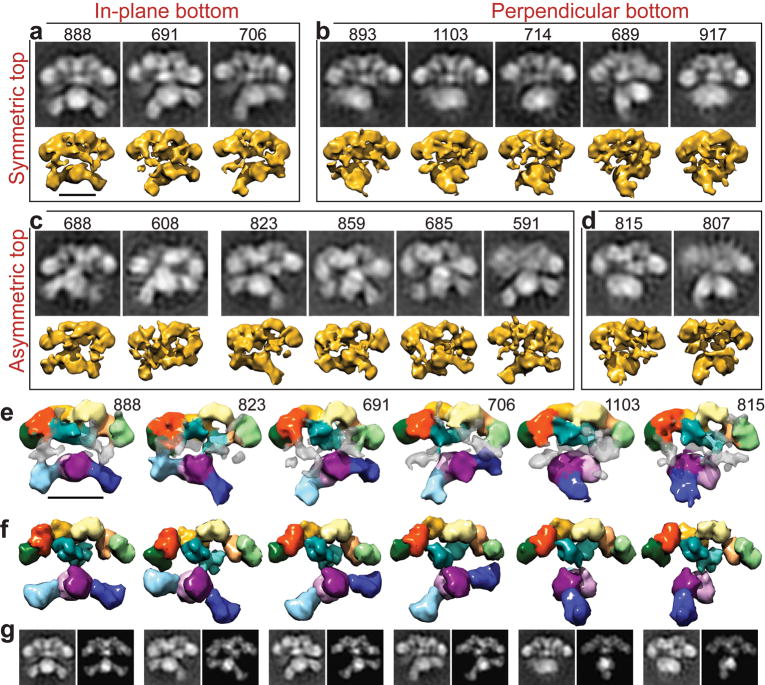
The conformational variability of FAS was characterized using a FAS mutant (Δ22-FAS) bearing a 22-residue deletion in the linker between the ACP and TE domains. The shortened tether was expected to restrict mobility of the TE domain and simplify alignment and classification of single-particle images without affecting the activity of other FAS components (the only effect of the mutation is to slightly slow down the chain-termination step catalyzed by the TE)11, while increasing the probability of imaging the previously undetected TE domain (Supplementary Fig. 1 online). To avoid oversampling conformations related to the slowed catalysis of acyl chain release, the Δ22-mutant FAS was examined in the absence of substrates. Images of the Δ22-mutant FAS were collected as tilted pairs so that 3D structures could be calculated by the random conical tilt reconstruction method12, maximizing the information garnered about each observed conformation. Reference-free image alignment and classification were used to separate particles into the minimum number of groups necessary to describe structural variability in reasonable detail. A total of 16 distinct FAS conformations were identified in 2D projection and 3D structures were calculated for each (Fig. 3a-d). FAS appears to adopt a continuum of conformations and further image subdivision was possible, but at the cost of diminishing the resolution of the resulting 3D structures (Supplementary Fig. 2 online).
Figure 3.
Conformational variability of the Δ22-FAS in the absence of substrates. (a-d) Single particle images were classified (black and white panels) and corresponding 3D structures calculated (yellow). The number of particles in each class is indicated above its 2D class average. The domain arrangements in the upper portion of the structure range from predominantly symmetric (a,b) to strongly asymmetric (c,d). The lower domains are arranged with respect to the upper domains either in parallel, swinging from right to left (a,c), or swiveling about the narrow “waist” into a perpendicular arrangement (b,d). (e) 3D structures of the Δ22-FAS mutant were colored as in Figure 2 to indicate the regions that could be fitted with structures of the KS, MAT, DH, ER, KR, and SD. Regions of density that were not fitted (transparent grey) may accommodate the TE and/or ACP domains. (f) Atomic structures of individual domains were fitted into several RCT structures and filtered to match the resolution of the EM structures. (g) 2D projections of these fitted atomic structures (right image in each pair) closely resemble the 2D class averages (left image in each pair, also in a-d) that correspond to each of the 3D RCT reconstructions (directly above each pair in e). Scale bars represent 100 Å.
The remarkable diversity among the observed FAS conformations could be described according to three main criteria: [i] in-plane rotation between the top and bottom portions of the structure (Fig. 3a,c) [ii] re-organization resulting in progressive asymmetry in the upper half of the structure (Fig. 3c and Supplementary Video 1 online), and [iii] off-plane rotation between the top and bottom portions (Fig. 3b,d). A nearly symmetric conformation with approximately equal sized reaction chambers (Fig. 3a, class #888) closely resembles the conformation in the 4.5 Å X-ray structure7. FAS is well preserved in the EM reconstructions (Supplementary Fig. 3 online) and its domain organization can be interpreted by comparison with the model based on the X-ray structure of FAS. Fitting atomic structures of the DH, ER, and KR domains requires only an adjustment from their positions in the crystal structure (Fig. 3e-g). A large density is apparent adjacent to the KR monomers at each end of the upper portion of the EM reconstructions. In homologous modular polyketide synthase (PKS) systems the KR domain is stabilized by partnering with a structural subdomain13. The analogous structural domain (SD) of the metazoan FAS was predicted to have a fold resembling that of SAM-dependent methyltransferases14 and we fitted the human histamine methyltransferase structure15 into this part of the EM reconstructions (also SD in Fig. 2). Comparison of the different EM 3D structures indicates that the position of the SD is variable, likely explaining the partial absence of density attributable to the SD in the 4.5 Å X-ray structure7. Densities corresponding in size and location to those expected for the ACP (Fig. 2b) and TE domains are also apparent in the EM reconstructions, with the TE domain often positioned within an open reaction chamber or in front of a closed one (Supplementary Fig. 1 online).
Consistent with the highly structured nature of the linker that connects the MAT and KS domains in FAS [Protein Data Bank accession code (PDB): 2jfd] and a modular PKS16, fitting of atomic structures indicates that the MAT and KS domains maintain their relative positions, with the entire lower portion of the FAS structure rotating in-plane as a unit (Fig. 3a,c). In molecules where the upper and lower sections were orientated perpendicular to each other (Fig. 3b,d) the MAT densities that extend above and below the plane of the molecule appear to have been compressed, a familiar complication when imaging negatively stained molecules17. However, the bi-lobed density representing each KS-MAT didomain could still be identified and used to determine the relative angle between the top and bottom portions of the FAS structure.
Effect of substrates and point mutations on FAS conformation
To expand on a previous analysis of the effect of substrates and catalytic state on the conformation of FAS6, three mutants, Δ22-FAS (slower product release)11, H878A-FAS (DH activity compromised)18, and C161Q-FAS (KS activity compromised)19, were imaged in the presence of substrates. Because different mutations should variously affect specific catalytic steps, changes in the distribution of FAS conformations could be expected following the addition of substrates. Independent analysis of each mutant image data set produced a range of class averages revealing that all mutants sample the same conformations observed in the absence of substrates (Supplementary Fig. 4 online). This implies that the conformations we have documented most likely represent a full range of domain motions sufficient to enable all reactions catalyzed by FAS.
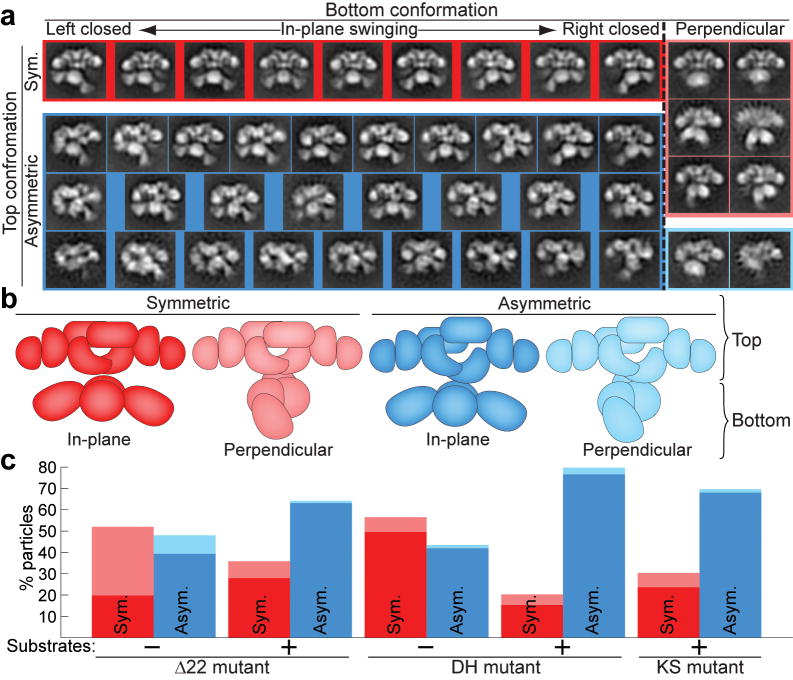
To avoid the possible introduction of subjective bias in comparison of conformation distributions, images from all FAS data sets were combined, simultaneously aligned as one group, and then separated into 50 different classes. Finer partition into more classes was facilitated by the larger size of the combined data set and the smaller number of images required for 2D analysis (Fig. 4a). The conformations observed in each class were then categorized (e.g., asymmetric or symmetric, perpendicular or in-plane)(Fig. 4b), and only on completion of this analysis were the origin of the particles in each conformation decoded.
Figure 4.
Distribution of FAS conformations is altered in the presence of substrates. (a) The Δ22-FAS and H878A (DH) mutants were imaged without substrates and these mutants and the C161Q (KS) mutant were imaged in the presence of substrates. Particles from all five data sets were classified together into 50 groups. After discarding 6 classes of grossly misaligned or distorted particles (3.4% of particles, not shown), the remaining 44 classes were categorized into those with symmetric (red) and asymmetric (blue) conformations in the upper β-carbon processing section (Top conformation) and those with perpendicular (faded colors) or in-plane conformations in the lower MAT-KS2-MAT section (Bottom conformation). The in-plane conformations exhibited by the lower section are arranged according to the degree of rotation of the lower section: from left-swinging (Left closed) to right-swinging (Right closed). For simplicity, class averages that show an opening between the DH, ER, and KR domains in the left half of the structure were mirrored so that the opening always appears in the right half of the structure. (b) Cartoon representation of each conformation colored according to a. (c) After categorization of classes, the numbers of particles from each FAS preparation in each category were determined. Bars are colored according to conformations as in a and b.
The most appreciable change resulting from addition of substrates was a marked increase in the fraction of molecules displaying an asymmetric arrangement of the β-carbon processing domains (Fig. 4c). Of the three FAS mutants prepared with substrates, the DH mutant has the strongest preference for asymmetry in the upper portion (80% asymmetric), suggesting that this arrangement may facilitate interactions required for β-carbon processing. The Δ22 and KS mutants also show a preponderance of asymmetric upper domains (70% and 64% asymmetric, respectively) when imaged in the presence of substrates. In fact, the Δ22 and KS mutants exhibit nearly identical conformation distributions, possibly reflecting the fact that both are compromised in the chain elongation step (by KS inactivity and by slow elongation from 16 to 18 carbon atoms resulting from reduced TE activity11, respectively). To further confirm that the arrangement of the β-carbon processing domains was directly influenced by catalytic activity (addition of substrates) the DH mutant that showed the largest preference for asymmetric arrangement was imaged in the absence of substrates. This resulted in a 2-fold decrease in the number of DH mutant molecules with asymmetrically arranged β-carbon processing domains (Fig. 4c). Finally, whereas the proportion of molecules showing a perpendicular disposition of the top and bottom portions of the FAS structure varies for each mutant, this conformation is always more prevalent when the upper portion is symmetric, suggesting a possible correlation between catalytic activity and in-plane arrangement of the top and bottom portions of the FAS structure (see below).
Although we consider it very unlikely, we cannot completely rule out the possibility that the changes we observe upon addition of substrates might have resulted from selective adsorption of certain molecular conformations rather than from conformational changes in FAS as it engages in catalysis.
Domain movements and implications
The insight gained from EM reconstructions of different FAS conformations was leveraged by fitting high-resolution structures of individual domains into the EM maps (Fig. 3e-g). The most prominent domain rearrangements result from movement about the narrow connection that separates the upper and lower portions of the FAS structure, which are covalently held together by the linker connecting each of the MAT-DH domain pairs. By analogy with the structure of a PKS KS-MAT module20, the FAS MAT catalytic domain (PDB: 2jfd) is followed by a linker region (likely composed of residues from Val823 up to the absolutely conserved Trp842) that threads between the structured pre-MAT linker and the KS domain and immediately precedes a region rich in proline, glycine, and serine residues. This sequence likely represents an unstructured, solvent-exposed region, well suited to function as a flexible tether. Major flexibility at this “hinge” permits two distinct motions: a pendulum-like swinging of the MAT-KS2-MAT module from side to side and a swiveling motion perpendicular to the plane of the upper portion of the structure.
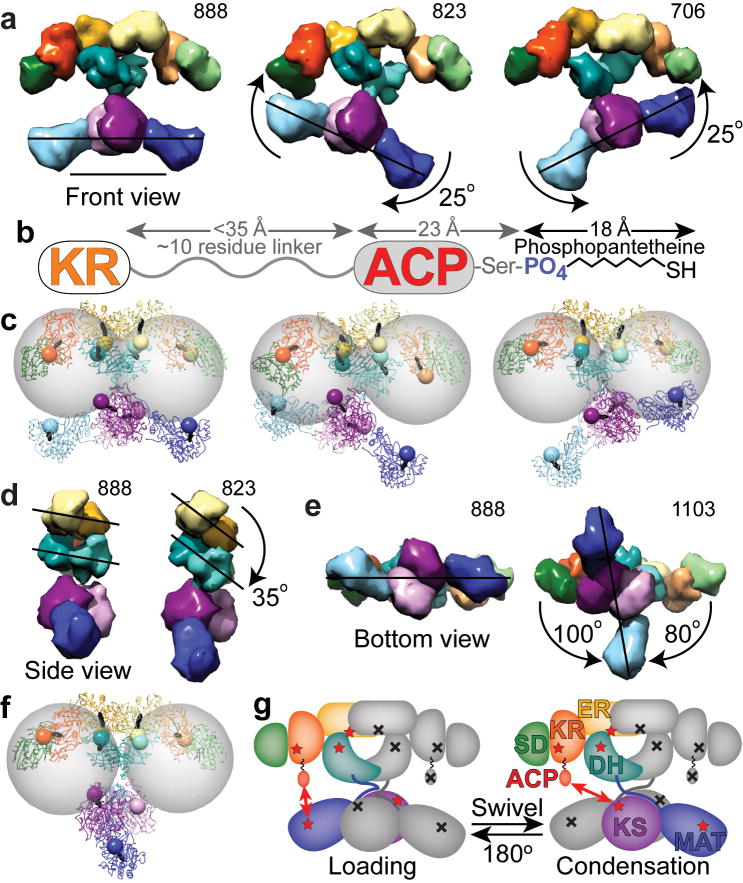
With the upper and lower portions of the FAS structure in the same plane, a swinging motion of the bottom as a rigid unit results in changes of up to ∼25° in the angle between the upper and lower portions of FAS (Figs. 3e-f and 5a). This swinging motion is considerably larger than the ∼7° upward or downward movement suggested by the asymmetric reaction chambers of the FAS crystal structure7 and results in synchronized cycling between open and closed reaction chamber conformations. Thus, the two reaction chambers might simultaneously engage in different activities. Substrate loading and chain extension reactions require close contacts between domains in the upper and lower sections and it is unlikely that the ACP and associated phosphopantetheine moiety could access the KS and MAT domains in a fully open reaction chamber (Figs. 5b,c). This suggests that chamber closing is required for the substrate loading and chain extension reactions, while the open chamber might be capable of only β-carbon processing and chain termination. Since the TE domain appears to be excluded from a closed chamber, the chain-termination reaction likely occurs preferentially in the open-chamber conformation, when free access to the ACP is facilitated.
Figure 5.
Changes in domain position bring catalytic domains into proximity of the ACP to facilitate catalytic interactions. (a) Lower portion swings relative to upper portion. (b) Based on crystal and NMR structures of the human and rat ACP domains36,40, and from analysis of sequence conservation among metazoan FAS, the KR-ACP linker likely consists of approximately 10 residues between Lys2109 through Arg2120 (rat FAS numbering). When fully extended this linker could be up to 35 Å in length. The ACP is approximately 23 Å long from its N-terminus to the phosphopantetheinylated Ser2151. The phosphopanthetheine must extend approximately 18 Å from the ACP to reach the active site within each catalytic domain. (c) After roughly modeling a rigid, extended phosphopantetheine into each active site pocket, the phosphate was rendered as an 8 Å radius sphere. Gray spheres of 55 Å radius indicate the distance that the ACP domains could reach from a fixed tether point at the C-terminus of the KR. (d) Side view of FAS with KR and SD removed from one subunit revealing rotation of the DH and ER domains. (e,f) Lower portion swivels relative to upper portion. (gi) Full 180° swiveling of the lower portion of the structure occurs during each catalytic cycle to explain the FAS activity of a heterodimer composed of a wild-type subunit (colored domains with red stars) partnered with a mutant subunit lacking all 7 functionalities (indicated by gray domains with black crosses). Domains of FAS are colored as in Figure 2. Scale bar in a represents 100 Å.
Our interpretation of rearrangements of the β-carbon processing domains in the top portion of the FAS structure was based on fitting of DH2, ER2, KR, and SD domains into the 3D EM reconstructions (Fig. 5d and Supplementary Fig. 3b online). Comparing the position of the fitted domains to their original positions in the X-ray model of FAS revealed correlated motion and suggested that the changes observed in the top portion of the FAS EM reconstructions could be partially explained by rotation of the domains as a rigid unit. However, such rigid motion was insufficient to explain the degree of asymmetry apparent in the majority of FAS molecules imaged in the presence of substrates. Although certain projections of the top portion of the FAS X-ray model would resemble the asymmetric projections observed in this study (Figs. 3c,d, 4a, and Supplementary Video 1 online), the largely symmetric arrangement of domains in the X-ray model is in sharp contrast with the clearly asymmetric nature of the corresponding portion of several of the 3D FAS reconstructions presented here (Supplementary Fig. 3c online). The asymmetrically arranged β-carbon processing domains in the presence of substrates is further supported by consideration of a previously published 3D cryo-EM reconstruction of FAS (Fig. 2b) that was also determined from particles imaged in the presence of substrates6.
The asymmetric arrangement of the β-carbon processing domains favored in the presence of substrates opens up a space about 15 Å in diameter between the DH, ER, and KR domains of one reaction chamber while closing contacts between the same domains in the opposite chamber. The access points to the DH and ER active sites face this asymmetric opening21 and dilation could facilitate alternating ACP access to the DH and ER domains in either chamber (Fig. 5c). Interestingly, the asymmetric conformation of the top was most prevalent when fatty acid synthesis was arrested during dehydration (Fig. 4c, DH mutant with substrates) suggesting that this structural rearrangement is particularly relevant to β-carbon processing and that varying accessibility of the ACP may provide a mechanism for coordinating β-carbon processing and substrate loading/condensing between the two reaction chambers.
Configurations in which the upper and lower portions of the FAS structure swivel by ± 80-100° with respect to each other (Fig. 5e) must be facilitated by flexibility at the narrow connection between them formed by extended linkers between the MAT and DH domains. In the resulting perpendicular configuration the MAT active sites are positioned out of reach of the ACP and would be unable to support substrate loading (Fig. 5f). Moreover, the fraction of molecules with a perpendicular conformation is significantly reduced upon addition of substrates. This is most apparent for the Δ22 mutant, in which a shortened ACP-TE linker must interfere with free rotation of the lower domains or allow the TE domain to participate in protein-protein interactions that transiently stabilize the perpendicular conformation. Taken together, these observations suggest that the perpendicular configuration might not be directly relevant for catalysis. However, its detection as a possible intermediate between two states in which the upper and top portions of the FAS structure have flipped appears to illuminate a critical aspect of FAS function.
Discussion
The single conformations captured in our previous cryo-EM study6 and in the 4.5 Å X-ray structure of the metazoan FAS7 could not account for a substantial body of biochemical evidence demonstrating that both ACP domains in a FAS homodimer can make functional contacts with the MAT and KS domains of either subunit. We previously suggested that FAS subunits might associate in either of two conformations, one with the polypeptides crossed over in the center of the molecule, the other with the subunits arranged back-to-back6. It now seems clear that FAS must switch between these two alternative modes of subunit association. The present EM results provide direct evidence that FAS can adopt a configuration in which the MAT-KS2-MAT module comprising the bottom of the FAS structure is rotated by 80-100° relative to the upper section of the molecule. Swiveling of the lower section in either direction by ∼90° restores particles to the familiar conformation in which the upper and lower sections are in the same plane and would bring the ACP domains within reach of either of the KS and MAT domains in the lower section depending on the direction of rotation. The ability of FAS to swivel between back-to-back and crossed-over configurations would explain the long-standing observation that the bifunctional dibromopropanone reagent cross-links the phosphopantetheine of one subunit with the KS active-site cysteine of the same or opposite subunits9.
Swiveling would explain how a FAS heterodimer in which one wild-type subunit is paired with a subunit compromised by mutations in all seven functional domains (WT/7KO-FAS) can synthesize fatty acids22. In the WT/7KO-FAS heterodimer the single functional ACP is forced to interact with KS and MAT domains in the same subunit that can only be accessed from opposite reaction chambers7. Thus, the ability of the WT/7KO-FAS heterodimer to synthesize palmitate at a significant rate implies that the mutant must be capable of extremely rapid 180° swivel motions between every substrate loading and condensation event (Fig. 5g). Improved crystal structures of porcine FAS in the NADPH-bound and free states were recently published23 and provide atomic coordinates for most of FAS (the ACP and TE domains were not resolved). In agreement with our conclusion, the authors suggest that the loading/condensation portion of FAS may swivel by 180° relative to the β-carbon processing portion. However, they speculate that the swiveling may involve a further coiling of the linkers rather than uncoiling that we believe could be more favorable.
Interestingly, the wild-type FAS should in principle be able to function without swiveling if each ACP would always cooperate with the same MAT and KS domains. That the flexible structural organization of FAS allows for alternative routes for substrate delivery and chain extension suggests that this apparent redundancy offers some inherent benefit. Indeed, kinetic analysis of a panel of FAS heterodimeric mutants in which only one of the two ACP domains was functional revealed that heterodimers retaining both options for substrate delivery and chain extension enjoy a significant catalytic advantage over those that rely on only a single option. For the wild-type FAS, the availability of alternative routes for substrate delivery and chain extension was estimated to contribute ∼20% to the overall rate of fatty acid biosynthesis8.
The FAS structures characterized in this study illustrate the conformational flexibility of the protein and reveal specific long-range domain rearrangements ostensibly essential for catalytic activity. The conformational changes we observed (Fig. 3), as well as the catalytic interactions they facilitate (Fig. 5), are illustrated in an animation assembled from the gallery of 3D structures (Supplementary Video 2 online). The animation shows how a swinging motion of the substrate loading and condensation domains results in cycling between open- and closed-chamber conformations, while swiveling allows an ACP to alternately engage the β-carbon processing domains of the same subunit and the loading and condensation portions of both subunits. Swinging and swiveling are apparently coordinated with rearrangement of β-carbon processing domains, possibly resulting in asynchronous chain extension and β-carbon processing reactions between the two reaction chambers
The metazoan and fungal FAS systems represent two contrasting alternative solutions to the problem of shuttling reaction intermediates between multiple catalytic sites in a single protein. The fungal FAS has invested almost 50% of its total mass in supportive infrastructure. Structural inserts interspersed both within and between catalytic domains serve to position the active sites of the constituent enzymes so that they can be readily accessed by the mobile ACP domain4,5. In contrast, the metazoan FAS is remarkably parsimonious. Its catalytic domains are almost completely uninterrupted by non-catalytic domains and the connecting regions account for a very small portion of the total mass. Success of the metazoan FAS design depends on maintaining an extraordinary degree of flexibility in the structure to ensure productive interactions between the ACP and its catalytic partners.
Methods
Protein expression and purification
Mutagenesis, expression, and purification of rat FAS mutants, H878A (DH), C161Q (KS) and Δ22, was described previously11,18,19.
Specimen preparation and electron microscopy
FAS aliquots (1-5 mg ml-1) were stored at -80°C in 0.25 mM potassium phosphate (pH 7.0), 1 mM DTT, 1 mM EDTA, and 10% (v/v) glycerol. For EM sample preparation, FAS aliquots were diluted to a final concentration of 10-15 ng μl-1 in reaction buffer (55 mM potassium phosphate (pH 7.0), 1.1 mM DTT, 20 μM acetyl-CoA, 100 μM malonyl-CoA, 180 μM NADPH, 1 mM Tris (pH 8.0)) or mock reaction buffer lacking malonyl-CoA, acetyl-CoA, (replaced with 1:5 dilution of 3 mM HCl) and NADPH24. Samples were prepared by placing 3-5 μl of FAS reaction mixture on a continuous carbon-coated EM specimen grid (300 mesh, Cu/Rh, Ted Pella) that had been freshly glow-discharged, incubating for ∼1 min, and staining with 1% (w/v) uranyl acetate using a double carbon layer technique25. Specimens prepared with substrates were automatically imaged at ∼15 e- Å-2 using the Leginon26 package in MSI-Raster mode to run a Tecnai F20 microscope (FEI) operating at 120 kV. Images were recorded at 50,000× magnification on Gatan or Teitz 4096 × 4096 CCD cameras resulting in a pixel size of 2.26 Å or 1.63 Å, respectively, on the object scale. Untilted and 55° tilt-pair images of the Δ22 mutant without substrates were collected manually using low dose conditions (each exposure ∼18 e- Å-2) and recorded on SO163 film (Kodak). Micrograph negatives were digitized using an SCAI scanner (Zeiss) and binned to a final pixel size of 4.2 Å on the object scale.
Processing of single particle images
All image processing was carried out with routines implemented using the SPIDER and Web software packages (Version 13)27. The defocus of each micrograph and CCD frame was computed, power spectra were visually evaluated, and incorrectly estimated defocus values were recalculated manually using Web. Particles from untilted micrographs and CCD frames with estimated defocus between approximately 200 and 600 nm were selected for further processing. Individual FAS molecules imaged in the presence of substrates were selected from CCD frames by template matching28 using projections of a previously calculated FAS cryo-EM density map6. Incorrectly selected particles were removed from the data set by visual inspection. Tilted micrographs were assessed on an optical diffractometer to ensure that the image was entirely underfocused. Particles in tilt-pair micrographs were selected manually and interactively using TiltPicker, a program currently under development the Automated Molecular Imaging group at The Scripps Research Institute (software available at www.appion.org). All individual particle images were windowed, ramped, normalized. In addition, particles from CCD frames were interpolated to a final pixel size of 4.2 Å for direct comparison with images of the Δ22-FAS mutant without substrates. Images were then band-pass filtered, retaining information between 21 Å and 330 Å, and a soft-edged circular mask was applied to remove information at the corners of the images.
Particles were reference-free aligned29, classified using correspondence analysis and hierarchical ascendant clustering30, and then subjected to an additional reference-free alignment within each more homogenous group. Due to conformational heterogeneity in the data set, this preliminary alignment of FAS particles yielded variable results and was repeated 10 times to widely survey the possible structural arrangements. Similar groups from each classification round were then combined by an additional round of classification, and particles were further reference-free aligned within these super-classes. The resulting class averages were used as references to initiate multi-reference classification followed by reference-free alignment within each class. This multi-reference/reference-free procedure was repeated 20 times with the resulting class averages used as references for the next iteration. Finally, particles within each class were reference-aligned to their final reference-free class average using an alignment radius optimized for each group of particles. This procedure is summarized in Supplementary Figure 5 (online).
An initial round of this alignment/classification routine was used to remove particles that were consistently assigned to poorly aligned classes of particles. After removal of these particles, the Δ22, KS, and DH mutants with substrates and the Δ22 and DH mutants without substrates were composed of 35902, 17319, 15369, 13847, and 11586 single particle images, respectively. These remaining particles were subjected to a second round of alignment/classification producing 2D class averages that enabled a rudimentary comparison of the particle distributions between data sets. To more objectively compare conformation distributions between data sets, all particle images were merged into a single data set and subjected to the alignment/classification routine. After manually categorizing each class of FAS conformation, particles of each FAS variant belonging to each category were identified. Particle distributions were displayed with Matlab 7.5.
For the Δ22 mutant without substrates, the final in-plane rotational alignment parameters were used to produce random conical tilt (RCT) reconstructions31. These reconstructions were improved through 6 iterations of shift refinement to center the tilted particle images followed by a single 5° angular search.
Fitting domain structures into density maps
Structures of DEBS KS3 (PDB: 2qo3)20, human MAT (PDB: 2jfd), human mitochondrial ER (PDB: 1zsy), tylosin KR1 (PDB: 2z5l)32, and a homology model of DH33 after removal of the N-terminal residues that likely are part of the MAT-DH linker, were initially positioned according to the FAS crystal structure7. The sequence between the DH and ER domains (Glu1166 to Gln1520) was submitted to HHpred34 to identify the human histamine methyltransferase (PDB: 2aot)15. This structure was positioned adjacent to the tylosin KR domain by using sequence-based structural alignment implemented in UCSF Chimera35 to superimpose the Rossman folds of the KR structural subdomain and the methyltransferase. After subtracting this multi-model structure from the cryo-EM map6, the rat ACP (PDB: 2png)36 was fitted into small protruding densities located just below the KR domain using COAN37.
This FAS model was aligned to each of the 3D EM structures (calculated using the RCT method) using the “fit in map” tool in Chimera and then the domain positions were manually adjusted to improve the fit into each conformation while maintaining appropriate domain orientations and contacts. For this procedure, the MAT-KS2-MAT, ER2, and DH2 could be easily docked as rigid units. The KR-SD didomain was initially treated as a single structural unit, and the positions of individual domains were then further adjusted.
To indicate the locations of the substrate binding pockets of each enzyme and approximately define (within ∼8 Å radius) the region where the ACP would dock, fully extended phosphopanthetheine ligands were inserted into each active site through the deep binding cleft leading to the active site within each domain. In some cases homologous structures with bound CoA were available (PDB: 1pn438 and 2g2z39). Sequence-based structural alignment was used to position the phosphopanthetheine ligands of these structures. Models and animated structures were rendered using Chimera.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Acknowledgments
We thank Dr. Andrzej Witkowski for helpful discussions. We also acknowledge the National Resource for Automated Molecular Microscopy for assistance with data collection. The work was supported by a research fellowship F32 DK080622 (to EJB) and grant RO1 DK16073 (to SS) from the National Institutes of Health.
Footnotes
Author Contributions: E.J.B performed all experiments and data analysis. S.S. provided purified FAS. All authors contributed to designing experiments, interpreting results and writing the manuscript.
References
- 1.Sul HS, Smith S. Fatty acid synthesis in eukaryotes. In: Vance DE, V JE, editors. Biochemistry of lipids, lipoproteins and membranes. Elsevier; 2008. pp. 155–190. [Google Scholar]
- 2.Kuhajda FP, et al. Fatty acid synthesis: a potential selective target for antineoplastic therapy. Proc Natl Acad Sci USA. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loftus TM, et al. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science. 2000;288:2379–81. doi: 10.1126/science.288.5475.2379. [DOI] [PubMed] [Google Scholar]
- 4.Jenni S, et al. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science. 2007;316:254–61. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- 5.Lomakin IB, Xiong Y, Steitz TA. The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell. 2007;129:319–32. doi: 10.1016/j.cell.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Asturias FJ, et al. Structure and molecular organization of mammalian fatty acid synthase. Nat Struct Mol Biol. 2005;12:225–32. doi: 10.1038/nsmb899. [DOI] [PubMed] [Google Scholar]
- 7.Maier T, Jenni S, Ban N. Architecture of mammalian fatty acid synthase at 4.5 A resolution. Science. 2006;311:1258–62. doi: 10.1126/science.1123248. [DOI] [PubMed] [Google Scholar]
- 8.Rangan VS, Joshi AK, Smith S. Mapping the functional topology of the animal fatty acid synthase by mutant complementation in vitro. Biochemistry. 2001;40:10792–9. doi: 10.1021/bi015535z. [DOI] [PubMed] [Google Scholar]
- 9.Witkowski A, et al. Dibromopropanone cross-linking of the phosphopantetheine and active-site cysteine thiols of the animal fatty acid synthase can occur both inter- and intrasubunit. Reevaluation of the side-by-side, antiparallel subunit model. J Biol Chem. 1999;274:11557–63. doi: 10.1074/jbc.274.17.11557. [DOI] [PubMed] [Google Scholar]
- 10.Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J Struct Biol. 2004;147:247–58. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Joshi AK, Witkowski A, Berman HA, Zhang L, Smith S. Effect of modification of the length and flexibility of the acyl carrier protein-thioesterase interdomain linker on functionality of the animal fatty acid synthase. Biochemistry. 2005;44:4100–7. doi: 10.1021/bi047856r. [DOI] [PubMed] [Google Scholar]
- 12.Radermacher M. The three-dimensional reconstruction of single particles from random and non-random tilt series. J Electron Microsc Tech. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- 13.Keatinge-Clay AT, Stroud RM. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure. 2006;14:737–48. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat Prod Rep. 2007;24:1041–72. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton JR, Sawada K, Nishibori M, Cheng X. Structural basis for inhibition of histamine N-methyltransferase by diverse drugs. J Mol Biol. 2005;353:334–44. doi: 10.1016/j.jmb.2005.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci U S A. 2006;103:11124–9. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Y, et al. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J Mol Biol. 2006;355:1048–65. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 18.Joshi AK, Smith S. Construction, expression, and characterization of a mutated animal fatty acid synthase deficient in the dehydrase function. J Biol Chem. 1993;268:22508–13. [PubMed] [Google Scholar]
- 19.Witkowski A, Joshi AK, Lindqvist Y, Smith S. Conversion of a beta-ketoacyl synthase to a malonyl decarboxylase by replacement of the active-site cysteine with glutamine. Biochemistry. 1999;38:11643–50. doi: 10.1021/bi990993h. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem Biol. 2007;14:931–43. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen ZJ, et al. Structural enzymological studies of 2-enoyl thioester reductase of the human mitochondrial FAS II pathway: new insights into its substrate recognition properties. J Mol Biol. 2008;379:830–44. doi: 10.1016/j.jmb.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 22.Joshi AK, Rangan VS, Witkowski A, Smith S. Engineering of an active animal fatty acid synthase dimer with only one competent subunit. Chem Biol. 2003;10:169–73. doi: 10.1016/s1074-5521(03)00023-1. [DOI] [PubMed] [Google Scholar]
- 23.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–22. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 24.Smith S, Abraham S. Fatty acid synthase from lactating rat mammary gland. Methods Enzymol. 1975;35:65–74. doi: 10.1016/0076-6879(75)35139-2. [DOI] [PubMed] [Google Scholar]
- 25.Tischendorf GW, Zeichhardt H, Stoffler G. Determination of the location of proteins L14, L17, L18, L19, L22, L23 on the surface of the 5oS ribosomal subunit of Escherichia coli by immune electron microscopy. Mol Gen Genet. 1974;134:187–208. doi: 10.1007/BF00267715. [DOI] [PubMed] [Google Scholar]
- 26.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 28.Rath BK, Frank J. Fast automatic particle picking from cryo-electron micrographs using a locally normalized cross-correlation function: a case study. J Struct Biol. 2004;145:84–90. doi: 10.1016/j.jsb.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 30.Bretaudiere JP, Frank J. Reconstitution of molecule images analysed by correspondence analysis: a tool for structural interpretation. J Microsc. 1986;144:1–14. doi: 10.1111/j.1365-2818.1986.tb04669.x. [DOI] [PubMed] [Google Scholar]
- 31.Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–36. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- 32.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 33.Pasta S, Witkowski A, Joshi AK, Smith S. Catalytic residues are shared between two pseudosubunits of the dehydratase domain of the animal fatty acid synthase. Chem Biol. 2007;14:1377–85. doi: 10.1016/j.chembiol.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 34.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Ploskon E, et al. A mammalian type I fatty acid synthase acyl carrier protein domain does not sequester acyl chains. J Biol Chem. 2008;283:518–28. doi: 10.1074/jbc.M703454200. [DOI] [PubMed] [Google Scholar]
- 37.Volkmann N, Hanein D. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J Struct Biol. 1999;125:176–84. doi: 10.1006/jsbi.1998.4074. [DOI] [PubMed] [Google Scholar]
- 38.Koski MK, Haapalainen AM, Hiltunen JK, Glumoff T. A two-domain structure of one subunit explains unique features of eukaryotic hydratase 2. J Biol Chem. 2004;279:24666–72. doi: 10.1074/jbc.M400293200. [DOI] [PubMed] [Google Scholar]
- 39.Oefner C, Schulz H, D'Arcy A, Dale GE. Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr D Biol Crystallogr. 2006;62:613–8. doi: 10.1107/S0907444906009474. [DOI] [PubMed] [Google Scholar]
- 40.Bunkoczi G, et al. Mechanism and substrate recognition of human holo ACP synthase. Chem Biol. 2007;14:1243–53. doi: 10.1016/j.chembiol.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.