Abstract
AIM: To further analyze the interaction of tupaia CD81 with hepatitis C virus (HCV) envelope protein E2.
METHODS: A tupaia CD81 large extracellular loop (CD81 LEL), which binds to HCV E2 protein, was cloned and expressed as a GST-fusion protein, and interaction of HCV E2 protein with a tupaia CD81 LEL was evaluated by enzyme-linked immunosorbent assay (EIA).
RESULTS: Although tupaia and human CD81 LEL differed in 6 amino acid changes, tupaia CD81 LEL was strongly recognized by anti-CD81 antibodies against human CD81 LEL conformation-dependent epitopes. Investigating LEL CD81-E2 interactions by EIA, we demonstrated that binding of tupaia CD81 LEL GST fusion protein to recombinant HCV E2 protein was markedly reduced compared to binding of human CD81 LEL GST fusion protein to recombinant HCV E2 protein.
CONCLUSION: These data suggest that the structural differences in-between the tupaia and human CD81 may alter the interaction of the large extracellular loop with HCV envelope glycoprotein E2. These findings may be important for the understanding of the mechanisms of binding and entry of HCV to PTHs.
Keywords: Hepatitis C virus E2 protein; Tupaia; CD81, Bind; Enzyme-linked immunosorbent assay
INTRODUCTION
Hepatitis C virus (HCV) is a major cause for posttransfusion and community-acquired hepatitis worldwide[1–4]. The majority of HCV-infected individuals develop chronic hepatitis that may progress to liver cirrhosis and hepatocellular carcinoma (HCC)[5]. Virion contains an approximately 9.5 kb long positive-strand RNA genome encoding for a single polyprotein containing 3010-3030 amino acids[6–8]. The polyprotein is cleaved co- and post-translationally by host cellular and viral proteases into at least ten different products. The predicted structural components of the virus comprise the core and two envelope glycoproteins: E1 and E2. The E2 protein is responsible for initiating viral attachment to receptor(s) on potential host cells due to its ability to bind to human cells[6,9]. CD81 has first been identified as a HCV E2 binding molecule by expression cloning by Pileri and colleagues[10]. The E2 binding site of CD81 is located within the LEL domain[10–13]. Further studies then demonstrated a key role of CD81 as a host entry factor for infection of human hepatoma cells with recombinant HCV pseudoparticle and tissue-culture-derived HCV (HCVcc)[14–18]. Furthermore, one very recent study has demonstrated evidence that CD81 is also involved in HCV infection of primary human hepatocytes using serum-derived HCV[19]. However, there appear to be subtle differences regarding the role of CD81 for productive infection of human hepatoma cells and human hepatocytes using serum-derived or recombinant virus[19–24].
Recently, a novel in vitro cell culture model system has been established for HCV with primary tupaia hepatocytes (PTHs)[25–27]. Using this cell culture system, we found that HCV E2 protein binding to PTHs and infection of PTHs with HCV could not be blocked with soluble CD81 and anti-CD81[25]. To further investigate the role of CD81 in the infection of PTHs with HCV, we cloned a tupaia CD81 large extracellular loop (LEL) and analyzed the interaction of tupaia CD81 with HCV E2 protein in vitro. The results indicate that changes occur in 6 amino acid residues of tupaia CD81 LEL and the ability of tupaia CD81 LEL to bind to HCV E2 is significantly decreased compared with human CD81 LEL.
MATERIALS AND METHODS
Reagents
Recombinant HCV E2 protein, and mouse anti-HCV E2 (3ES) were generously provided by M Houghton (Chiron Corp., Emeryville, CA). Human CD81 LEL (wild type and T163A mutant) and AGM CD81 LEL protein were kindly provided by Dr. S Levy (Department of Medicine, Stanford Medical School, Palo Alto, CA)[12]. Mouse monoclonal anti-CD81 antibodies 5A6 and 1D6 have been described elsewhere[11,12,28]. Glutathione S-transferase (GST) expression vector, PGEX-2T, was purchased from Amersham Pharmacia Bioteh Inc.
Cloning of tupaia CD81 LEL
Total cellular RNA was isolated from PTHs with RNeasy kits (Qiagen, GmbH, Hilden, Germany) according its manufacturer’s instructions. cDNA was transcribed from cellular RNA using rTth reverse transcriptase. Tupaia CD81 (TupCD81) LEL was amplified by PCR using Pfu DNA polymerase (Stratagene) and human CD81-specific primers covering the LEL domain (sense: 5’-TAACGGATCCAACAAGGACCAGATTGCCAAGGA-3’, anti-sense: TAACGAATTCACAGCTTCCCGGAGAAGAGCTC-3’). The PCR products were then cloned into pCR-Blunt II-TOPO (Invitrogen, Groningen, Netherlands). An EcoR I/BamH I fragment was subcloned into pGEX-2T (Amersham Pharmacia Biotech Inc.) with the same enzyme digestion. The resulting plasmid (named p2T-TupCD81 LEL) was sequenced using CD81-specific primers. Plasmids expressing African green monkey, and human CD81 LEL (both wild type and genetically modified T163A type) were kindly provided by Dr. S Levy and have been described previously.
Expression and purification of glutathione S-transferase (GST)-CD81 LEL fusion proteins
Expression and purification of GST-CD81 LEL fusion protein were performed as previously described[13]. Briefly, BL21 E. coli were transformed with plasmid DNA. Transfected E. coli were grown in 12 mL LB medium containing ampicillin (50 mg/L) at 30-32°C until the optical density (OD) value of the culture medium reached 0.6-0.8 at A600. Protein expression was then induced by adding isopropyl β-D-thiogalactoside (IPTG, 0.75 mmol/L). After cells grew for additional 3 h, they were lysed by sonication and the fusion proteins were purified using the MicroSpin GST purification module (Amersham Pharmacia Biotech). GST protein expressed in control plasmid pGEX-2T was purified in parallel. The purified proteins were analyzed using SDS-PAGE under non-reducing conditions, quantified, aliquoted, and stored at -80°C until use.
Test of HCV E2-CD81 LEL interaction
A 96-well microtiter plate was coated with GST-CD81 fusion protein (5 mg/L) at 4°C overnight. After washing 5 times with PBS and blocking with 4% dry milk (in PBS) at room temperature for 1 h, 50 μL of recombinant HCV E2 protein (with reciprocal dilutions, starting from 2 mg/L) was allowed to bind to CD81 LEL at 4°C overnight. The bound E2 was detected using anti-E2 (3E5; diluted at 1:2000 in PBS containing 1% Tween 20 and 1% BSA) and HRP-conjugated anti-mouse IgG (diluted at 1:5000 in PBS containing 1% Tween 20 and 1% BSA). GST protein and human and AGM CD81 LEL were tested as controls. For GST test, goat anti-GST (in dilution starting at 1:1000) (Amersham Pharmacia Biotech) was added, and incubated at room temperature for 1 h. The bound anti-GST was detected using HRP-conjugated anti-goat IgG. The bound HRP-conjugated second antibodies were quantified by colorimetric reaction with an Abbott OPD reagent kit (Abbott Laboratories, North Chicago, IL), and the OD value was measured at 490 nm on a Bio-Rad plate reader (Bio-Rad, Hercules, CA).
RESULTS
Tupaia CD81 LEL amino acid sequence
It has been demonstrated that the E2 binding site of CD81 is located within the LEL domain[10,12,13]. To study the E2-CD81 LEL interaction in vitro, tupaia CD81 LEL was cloned and sequenced. The deduced amino acid sequence of tupaia CD81 LEL cDNA showed mutations in 6 amino acid residues when compared with human CD81 LEL as previously showed[25]. These mutations were clustered around the E2 binding head subdomain (Figure 1), according to the 3D structure of human CD81 LEL[29].
Figure 1.

Putative structure of tupaia CD81. Tupaia CD81 LEL cDNA was cloned and sequenced as described in Materials and Methods. Deduced amino acid sequence was compared with that of human. The tupaia CD81 secondary structure was drawn according to the three-dimensional structure of human CD81 LEL, and changes in six amino acids of tupaia CD81 LEL were illustrated by black color.
Analysis of tupaia CD81 LEL-E2 interaction in vitro
To analyze the interaction between HCV glycoprotein E2 and tupaia CD81 LEL in vitro, tupaia CD81 LEL was expressed as a GST-fusion protein in E. coli. The correct expression and folding of in vitro-synthesized tupaia CD81 LEL were confirmed by analyzing the purified protein with non-reducing SDS-PAGE and immunoblot using anti-CD81 antibodies against conformation-dependent epitopes. As shown in Figure 2A and B, tupaia CD81 LEL was recognized by defined anti-human CD81 antibodies (5A6 and 1D6), similar to human or African green monkey CD81 LEL. The data suggest that folding of CD81 LEL-GST fusion protein is comparable to that of the native molecule, while slight differences might exist in the 3D structure of human and tupaia CD81 LEL since the staining intensity of TupCD81 was week compared with that of human CD81 (both wild and T163A mutation type) using antibody to human CD81 conformation-dependent epitopes. Interaction of tupaia CD81 LEL with recombinant E2 protein was analyzed with EIA, and compared with that of human- or African green monkey-derived CD81 LEL. Binding of tupaia CD81 LEL to HCV E2 protein was markedly reduced compared with the native or mutant human CD81 LEL (Figure 3).
Figure 2.

Expression and purification of soluble tupaia CD81 LEL using mouse monoclonal anti-human CD81 5A6 (A), and 1D6 (B) antibodies. Tupaia CD81 LEL was expressed and purified in E. coli as a GST fusion protein as described in Materials and Methods. Purified proteins were subjected to SDS-PAGE, and immunoblot using mouse monoclonal anti-human CD81 5A6 and 1D6 antibodies. Tup: tupaia; h: human; AGM: African green monkey; hCD81 LEL T163A: human CD81 containing a mutation of T to A at amino acid residue 163; 1: GST; 2: TupCD81 LEL-GST; 3: hCD81 LELT163A-GST; 4: AGMCD8 LEL-GST; 5: hCD81 LEL-GST.
Figure 3.
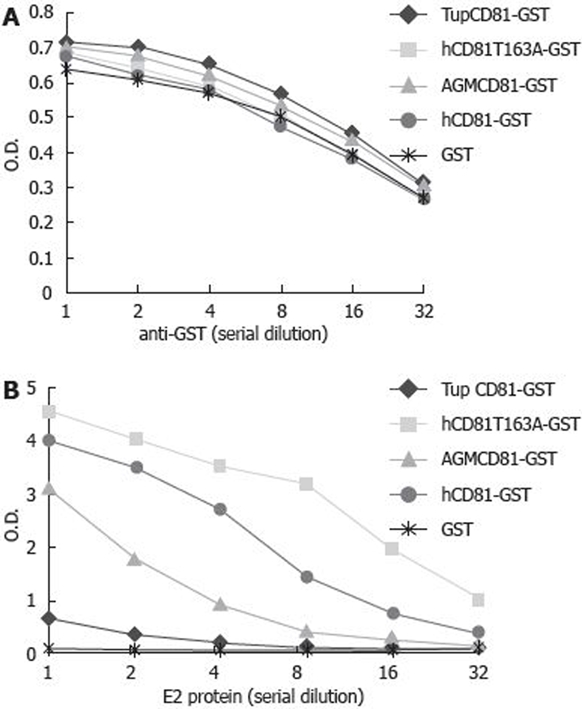
Interaction between tupaia CD81 LEL and HCV E2 protein. Plates were coated with 100 μL of recombinant GST or GST-CD81 LEL fusion proteins (5 mg/L), reciprocally diluted anti-GST antibody (A) (starting dilution at 1:1000) or HCV E2 protein (B) (starting concentration 2 mg/L) was added to the plates. Binding of anti-GST or HCV E2 protein was assessed as described in Materials and Methods. OD: optical density; Tup: tupaia; h: human; AGM: African green monkey; hCD81T163A: human CD81 containing a mutation of T to A at amino acid 165.
DISCUSSION
Recombinant HCV E2 could bind to tupaia, but not to rat hepatocytes in a dose-dependent manner and PTHs could be infected with HCV in vitro[25,26]. Furthermore, we demonstrated that both the binding of HCV E2 to PTHs, and infection of PTHs with HCV could not be blocked with soluble CD81 and anti-CD81 exhibiting the blocking ability of HCV E2 to bind to lymphoma cell lines, indicating that binding of E2 to PTHs and infection of tupaia hepatocytes with HCV might require additional or other molecules besides CD81. To further characterize the HCV E2-tupaia CD81 interaction, we cloned the HCV E2 binding domain of CD81, the CD81 LEL, and investigated the reaction of HCV E2 with tupaia CD81 in vitro.
CD81, a member of the super-family of tetra-spanins, comprises 4 transmembrane (TM1-4) and two extracellular loops. The HCV E2 binding domain locates in the large extracellular loop (LEL), which folds to form a mushroom-like 3D structure, and is stabilized by a number of specific interactions within the defined amino acid residues[29]. In the present study, the deduced amino acid sequence of tupaia CD81 LEL showed changes in only 6 amino acid residues compared to human CD81 LEL, while all the residues necessary for the 3D structure-stabilization were conserved, suggesting that soluble tupaia CD81 LEL folds in a manner comparable to human CD81 LEL. This hypothesis was confirmed by the cross interaction of tupaia CD81 LEL with anti-human CD81 antibodies against conformation-dependent epitopes (Figure 2).
It has been reported that HCV E2 binds to the head subdomain of CD81 LEL consisting of about 60 amino acid residues[29]. Distinct amino acid mutations can affect CD81-E2 interaction. African green monkey (AGM) CD81 LEL contains only 4 amino acid residues compared with human CD81 LEL, and shows reduced E2 binding[10–12]. Reduced E2-binding of AGM CD81 is due to a mutation of phenylalanine to leucine at the amino acid residue 186 (F186L)[12]. Interestingly, mutation of threonine to alanine at the amino acid residue 163 (T163A) can enhance E2-CD81 binding[12]. In this study, all the six mutant residues of tupaia CD81 LEL were clustered at its head subdomain which is the binding site for HCV E2, while the phenylalanine at residue 186, and the 4 cysteine residues, which are pivotal for human CD81-HCV E2 binding, were conserved in tupaia CD81. When purified tupaia CD81 LEL was tested for its binding to recombinant HCV E2 protein, only a mild E2-CD81 interaction was observed. By contrast, human CD81 LEL (both wild-type and genetically modified CD81 containing a T163A mutation) bound firmly to E2 protein. Interestingly, threonine at residue 163 of TupCD81 LEL changed into phenylalanine. The reduced E2 binding ability of tupaia CD81 was not due to the amount of protein coating the plates, since the GST activity of those fusion proteins was nearly similar. From the perspective of the 3D structure of CD81 LEL, mutations at amino acid residues 155 and 181 of tupaia CD81 LEL may be responsible for the reduced binding in Tupaia CD81 to HCV E2 protein[29]. The binding of HCV E2 to AGMCD81 LEL in this study different from a previous study[12]. This discrepancy might be due to the differences in the recombinant HCV E2 proteins. E2 used in this study is C-terminally truncated at amino acid 715, whereas other investigators used E2 C-terminally truncated at amino acid 661.
Although our studies were limited to evaluate CD81 LEL-E2 interaction with EIA, and need to be confirmed in model systems expressing full-length CD81 in transfected mammalian cell lines, the differences in HCV E2 binding between tupaia CD81 LEL and human CD81 LEL are consistent with our previous functional data assessing E2 binding to PTHs and HCV infection of PTHs in the presence of anti-CD81 antibodies. Taken together, these results indicate that although CD81 may play a functional role as a co-factor for entry of HCV into PTHs, it is likely that other or additional molecules besides CD81 play a key role in HCV entry into PTHs. These may include other identified HCV host factors including SR-BI[27] or Claudin-1[30]. Alternatively, other not yet identified host entry factors may mediate HCV-PTH interaction.
COMMENTS
Background
CD81, a member of the superfamily tetraspanins, comprises four transmembrane (TM1-4) and two extracellular loops. The HCV E2 binding domain locates in the large extracellular loop (LEL). The LEL folds to form a mushroom-like 3D structure, which is stabilized by a number of specific interactions within the defined amino acis residues.
Research frontiers
Hepatitis C virus (HCV) is a major cause for posttransfusion and community-acquired hepatitis worldwide. The majority of HCV-infected individuals develop chronic hepatitis that may progress to liver cirrhosis and hepatocellular carcinoma (HCC). The predicted structural components of HCV comprise the core and two envelope glycoproteins: E1 and E2. The E2 protein is responsible for initiating viral attachment to receptor(s) on potential host cells due to its ability to bind to human cells. HCV E2 could specifically bind to cell surface molecule CD81 expressed in lymphoma cells. Anti-HCV antibodies from chimpanzees that are protected against homologous HCV challenge by vaccination with envelope glycoproteins inhibit E2 binding to CD81, suggesting that CD81 represents a candidate receptor for HCV infection. Whether CD81-E2 interaction can mediate virion entry into host cells is unknown and has not been tested in a suitable model.
Innovations and breakthroughs
A novel in vitro cell culture model system has been established for HCV with primary tupaia hepatocytes (PTHs). Using this cell culture system, we found that HCV E2 protein binding to PTHs and infection of PTHs with HCV could not be blocked with soluble CD81 and anti-CD81. To further investigate the role of CD81 in the infection of PTHs with HCV, we cloned a tupaia CD81 large extracellular loop (LEL) and analyzed the interaction of tupaia CD81 with HCV E2 protein in vitro. The results indicate that changes occur in 6 amino acid residues of tupaia CD81 LEL and the ability of tupaia CD81 LE to bind to HCV E2 is significantly decreased compared with human CD81 LEL.
Applications
Tupaia CD81 has a reduced ability to bind to HCV E2 protein. HCV entry and infection of PTHs with HCV might occur through receptor(s) besides CD81.
Peer review
The manuscript is well written, but needs clarification of the importance of the found amino acid changes with regard to the interaction of tupaia CD81 and HCV E2 binding.
Footnotes
Supported by Grants from Health Department of Hubei Province (JX2B09), NSFC (30771911), National 973 Key Program (2009CB522502), and Tongji Hospital
Peer reviewer: Thomas Bock, PhD, Professor, Department of Molecular Pathology, Institute of Pathology, University Hospital of Tuebingen, D-72076 Tuebingen, Germany
S- Editor Li JL L- Editor Wang XL E- Editor Zheng XM
References
- 1.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 2.Alter HJ, Purcell RH, Shih JW, Melpolder JC, Houghton M, Choo QL, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- 3.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 4.Alter MJ, Kruszon-Moran D, Nainan OV, McQuillan GM, Gao F, Moyer LA, Kaslow RA, Margolis HS. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–562. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 5.Tong MJ, el-Farra NS, Reikes AR, Co RL. Clinical outcomes after transfusion-associated hepatitis C. N Engl J Med. 1995;332:1463–1466. doi: 10.1056/NEJM199506013322202. [DOI] [PubMed] [Google Scholar]
- 6.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 7.Rice CM. Flaviviridae: the viruses and their replication. In: BN Fields, DM Knipe, PM Howley., editors. Lippincott: Philadelphia; 1999. pp. 931–959. [Google Scholar]
- 8.Major ME, Feinstone SM. The molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 9.Flint M, McKeating JA. The role of the hepatitis C virus glycoproteins in infection. Rev Med Virol. 2000;10:101–117. doi: 10.1002/(sici)1099-1654(200003/04)10:2<101::aid-rmv268>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 10.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, et al. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 11.Flint M, Maidens C, Loomis-Price LD, Shotton C, Dubuisson J, Monk P, Higginbottom A, Levy S, McKeating JA. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J Virol. 1999;73:6235–6244. doi: 10.1128/jvi.73.8.6235-6244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higginbottom A, Quinn ER, Kuo CC, Flint M, Wilson LH, Bianchi E, Nicosia A, Monk PN, McKeating JA, Levy S. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J Virol. 2000;74:3642–3649. doi: 10.1128/jvi.74.8.3642-3649.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petracca R, Falugi F, Galli G, Norais N, Rosa D, Campagnoli S, Burgio V, Di Stasio E, Giardina B, Houghton M, et al. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J Virol. 2000;74:4824–4830. doi: 10.1128/jvi.74.10.4824-4830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 18.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, et al. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411–419. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Cormier EG, Tsamis F, Kajumo F, Durso RJ, Gardner JP, Dragic T. CD81 is an entry coreceptor for hepatitis C virus. Proc Natl Acad Sci USA. 2004;101:7270–7274. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Randall G, Higginbottom A, Monk P, Rice CM, McKeating JA. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J Virol. 2004;78:1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKeating JA, Zhang LQ, Logvinoff C, Flint M, Zhang J, Yu J, Butera D, Ho DD, Dustin LB, Rice CM, et al. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J Virol. 2004;78:8496–8505. doi: 10.1128/JVI.78.16.8496-8505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588–598. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akazawa D, Date T, Morikawa K, Murayama A, Miyamoto M, Kaga M, Barth H, Baumert TF, Dubuisson J, Wakita T. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J Virol. 2007;81:5036–5045. doi: 10.1128/JVI.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao X, Tang ZY, Klumpp B, Wolff-Vorbeck G, Barth H, Levy S, von Weizsacker F, Blum HE, Baumert TF. Primary hepatocytes of Tupaia belangeri as a potential model for hepatitis C virus infection. J Clin Invest. 2002;109:221–232. doi: 10.1172/JCI13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao XP, Tian ZF, Chen YC, Yang C, Tian DY, Yang DL, Hao LJ. [Infection of tupaia hepatocytes with hepatitis C virus in vitro] Zhonghua Ganzangbing Zazhi. 2005;13:805–807. [PubMed] [Google Scholar]
- 27.Barth H, Cerino R, Arcuri M, Hoffmann M, Schürmann P, Adah MI, Gissler B, Zhao X, Ghisetti V, Lavezzo B, et al. Scavenger receptor class B type I and hepatitis C virus infection of primary tupaia hepatocytes. J Virol. 2005;79:5774–5585. doi: 10.1128/JVI.79.9.5774-5785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy S, Todd SC, Maecker HT. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol. 1998;16:89–109. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 29.Kitadokoro K, Bordo D, Galli G, Petracca R, Falugi F, Abrignani S, Grandi G, Bolognesi M. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 2001;20:12–18. doi: 10.1093/emboj/20.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]


