Abstract
The UV light-induced synthesis of UV-protective flavonoids diverts substantial amounts of substrates from primary metabolism into secondary product formation and thus causes major perturbations of the cellular homeostasis. Results from this study show that the mRNAs encoding representative enzymes from various supply pathways are coinduced in UV-irradiated parsley cells (Petroselinum crispum) with two mRNAs of flavonoid glycoside biosynthesis, encoding phenylalanine ammonia-lyase and chalcone synthase. Strong induction was observed for mRNAs encoding glucose 6-phosphate dehydrogenase (carbohydrate metabolism, providing substrates for the shikimate pathway), 3-deoxyarabinoheptulosonate 7-phosphate synthase (shikimate pathway, yielding phenylalanine), and acyl-CoA oxidase (fatty acid degradation, yielding acetyl-CoA), and moderate induction for an mRNA encoding S-adenosyl-homocysteine hydrolase (activated methyl cycle, yielding S-adenosyl-methionine for B-ring methylation). Ten arbitrarily selected mRNAs representing various unrelated metabolic activities remained unaffected. Comparative analysis of acyl-CoA oxidase and chalcone synthase with respect to mRNA expression modes and gene promoter structure and function revealed close similarities. These results indicate a fine-tuned regulatory network integrating those functionally related pathways of primary and secondary metabolism that are specifically required for protective adaptation to UV irradiation. Although the response of parsley cells to UV light is considerably broader than previously assumed, it contrasts greatly with the extensive metabolic reprogramming observed previously in elicitor-treated or fungus-infected cells.
Previous studies using parsley cells (Petroselinum crispum) treated with a pathogen-derived elicitor revealed an extensive reprogramming of both primary and secondary metabolism at the gene expression level (1–3). This finding raised the question as to whether or not other external signals caused similarly drastic responses. UV light was expected to be a comparatively mild stimulus, because the induction of the general phenylpropanoid and flavonoid glycoside pathways and the consequential accumulation of UV-protective flavonoids so far have been regarded as the only major metabolic response of cultured parsley cells to UV irradiation (4, 5). The two sequentially acting pathways converting phenylalanine to 4-coumaroyl-CoA and 4-coumaroyl-CoA to flavonoid end products (see Fig. 1) are induced by transcriptional activation both in cultured cells (6) and the UV-protective epidermis of parsley leaves (7, 8). The only other UV-light response of parsley cells observed so far at the transcriptional level is the simultaneous repression of various cell cycle-related activities (9).
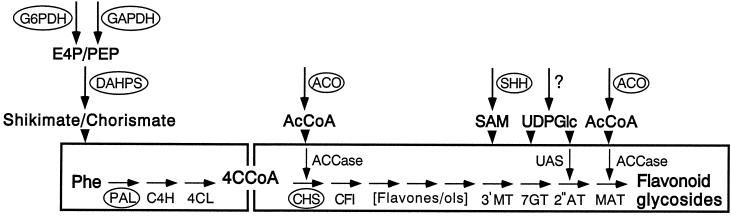
Figure 1.
Scheme indicating the enzymatic steps and the primary metabolites serving as substrates for the biosynthesis of variously substituted flavone and flavonol glycosides in UV-irradiated parsley cells. Abbreviated substrates and intermediates are: erythrose 4-phosphate (E4P), phosphoenolpyruvate (PEP), acetyl-CoA (AcCoA), S-adenosyl-l-methionine (SAM), UDP-glucose (UDPGlc), phenylalanine (Phe), and 4-coumaroyl-CoA (4CCoA). Highlighted enzymes are: G6PDH, GAPDH, DAHPS, ACO, SHH, PAL, cinnamate 4-hydroxylase (C4H), 4-coumarate:CoA ligase (4CL), ACCase, CHS, S-adenosyl-l-methionine:flavone/flavonol 3′-O-methyltransferase (3′MT), UDP-glucose:flavone/flavonol 7-O-glucosyltransferase (7GT), UAS, UDP-apiose:flavone/flavonol 7-O-glucoside 2′′-O-apiosyltransferase (2′′AT), and malonyl-CoA:flavonoid glycoside 6′′-O-malonyltransferase (MAT). Early, partially divergent oxidative steps in flavone and flavonol formation are summarized as [flavones/ols].
Earlier attempts to test whether UV light also induced the supply pathways providing the various substrates for the rapid and massive production of flavonoid glycosides were made before the availability of the sensitive tools of molecular biology and thus were confined to enzyme activity measurements (4). With one exception, these studies gave no indication of light-induced changes in enzyme activities from primary metabolism, probably because of the occurrence of multiple isoforms with different regulatory properties as well as the pre-existing high levels above which comparatively small changes were difficult to detect.
The exception was acetyl-CoA carboxylase (ACCase), which was strongly induced in tight coordination with the enzymes of the flavonoid glycoside pathway. ACCase previously had been considered to be an enzyme solely of primary metabolism (10) and had not been expected to be so tightly regulated with phenylpropanoid secondary metabolism. It therefore was speculated that the enzyme might occur in distinct isoforms, one of which would be a true member of the group of enzymes constituting the coordinately regulated flavonoid glycoside pathway (4), although initial attempts failed to demonstrate the existence of isoenzymes (11). However, if the occurrence of stimulus-specific isoforms were a general phenomenon, why were similar, large effects on other supply reactions of flavonoid biosynthesis not detected, and would such a mechanism not require numerous independently regulated isoforms for many enzymes and for a large variety of conditions? To clarify these points, we have reinvestigated the interrelationship of flavonoid biosynthesis and its supply pathways in UV-irradiated parsley cells at the mRNA level, using previously generated cDNAs (2) as well as an acyl-CoA oxidase (ACO) cDNA as hybridization probes.
The first indication that a supply pathway might indeed be coregulated with flavonoid biosynthesis came from the independent observation that the mRNA encoding 3-deoxy-d-arabinoheptulosonate 7-phosphate synthase (DAHPS), an enzyme of the phenylalanine-generating shikimate pathway, was induced by light in cultured parsley cells (12). We now confirm and extend this result by comparing the mRNA induction patterns for DAHPS, ACO, and several other enzymes of primary and secondary metabolism with those obtained for phenylalanine ammonia-lyase (PAL) and chalcone synthase (CHS), the first committed steps of the general phenylpropanoid and flavonoid glycoside pathways, respectively (see Fig. 1). The promoter of the ACO gene whose UV-light responsiveness previously has been demonstrated under the provisional term LF53 (13) was selected for functional and structural comparison with the well-established CHS gene promoter. The data indicate both a tightly coordinated and a highly selective induction process that distinguishes the UV-light response in both respects from the much more drastic pathogen defense response.
Materials and Methods
All materials and methods have been described: maintenance, treatment, and harvest of parsley (Petroselinum crispum) cell suspension cultures (9, 14, 15); cDNA and genomic cloning, sequencing, and further analysis (16); sources and characteristics of all cDNAs used (2) except the newly prepared ACO cDNA; RNA-blot analysis (9); and protoplasting, transformation, and transient expression assays (13, 17).
Results
Cloning and Analysis of an ACO cDNA.
A small cDNA fragment that initially had been mistaken, because of immunological crossreactivity, for a partial PAL cDNA (18) was used to screen two independent λGT11 cDNA libraries. These libraries were generated by using RNA from cultured parsley cells that had been treated either for 6 h with UV-containing white light or for 5 h with elicitor. A total of 30 “UV clones” and 10 “elicitor clones” were plaque-purified, and the three longest cDNA inserts were selected for sequencing. All of them were identical in their overlapping portions and apparently represented different regions from the same corresponding mRNA. A full-size cDNA was not obtained and therefore was reconstructed from three overlapping fragments.
The deduced amino acid sequence was compared with the sequences available in the database and showed 83% identity (88% similarity) with an ACO from Arabidopsis thaliana (19) and about 80% identity with several established or putative ACO proteins from other sources. All of them, including the parsley protein, contain a conserved N-terminal nonapeptide sequence motif (PTS2) required for targeting to the peroxisomal matrix (20). Based on this high degree of similarity, we consider the protein encoded by this cDNA to be an ACO. The deduced protein has a calculated relative molecular mass of 77,824.
UV-Light Induction of mRNAs from Supply Pathways of Flavonoid Biosynthesis.
The ACO cDNA and all other cDNAs encircled in Fig. 1 were used to determine the extent and timing of UV light-induced mRNA accumulation in previously dark-grown, cultured parsley cells. For comparison, several cDNAs encoding enzymes with no apparent function in flavonoid glycoside biosynthesis, including bergaptol O-methyl-transferase, cinnamyl alcohol dehydrogenase, and caffeoyl-CoA 3-O-methyltransferase from unrelated phenylpropanoid pathways, also were analyzed.
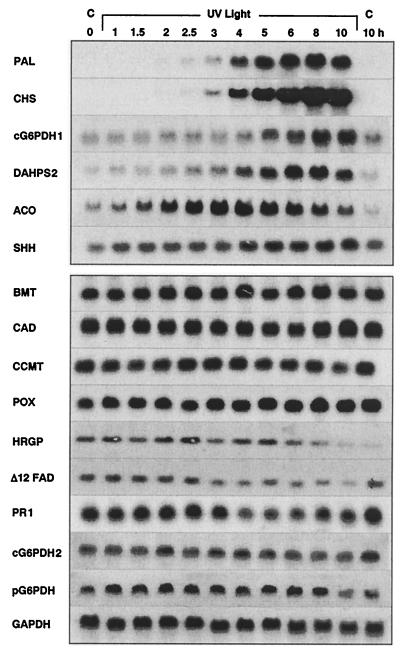
The results (Fig. 2) demonstrate that PAL and CHS mRNAs were strongly induced from undetectable levels with an apparent lag of about 1.5–2 h, as expected (6). One of the three glucose 6-phosphate dehydrogenase (G6PDH) mRNA isoforms analyzed, as well as the DAHPS2 and ACO mRNAs, also were strongly induced, though from appreciable background levels and with somewhat different time courses. One selected S-adenosyl-homocysteine hydrolase (SHH) mRNA was induced to a lesser extent, again from a high background level, whereas all other tested mRNAs remained unaffected.
Figure 2.
Representative results from RNA blot analyses. cDNA probes were used as indicated for hybridization with RNA samples taken at various time points after onset of UV irradiation. C, Dark control. Abbreviations not given in Fig. 1 are as follows: bergaptol O-methyl-transferase (BMT), caffeoyl-CoA 3-O-methyltransferase (CCMT), cinnamyl alcohol dehydrogenase (CAD), Δ12 fatty acid desaturase (Δ12FAD), hydroxyproline-rich glycoprotein (HRGP), anionic peroxidase (POX), and pathogenesis-related protein 1 (PR1).
Repression by Fungal Elicitor.
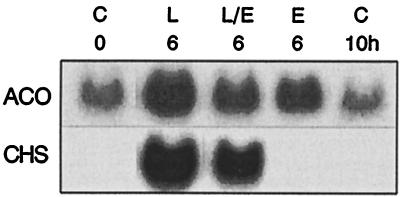
We previously have demonstrated the rapid reversal of UV light-mediated CHS mRNA induction by subsequent addition of elicitor (15). One of the coincuded mRNAs from supply pathways, ACO, whose gene promoter was available for analysis (see below), was selected to test whether it responded in a similar manner. Fig. 3 shows that this was indeed the case, although in contrast to CHS, the ACO mRNA level was again moderately high already in untreated cells, and that ACO mRNA also was induced by elicitor, though to a lesser extent than by light. Allowing for these differences, the responses to UV light or a combination of UV light and elicitor were essentially similar for the two mRNAs.
Figure 3.
Comparison of ACO and CHS mRNA levels in parsley cells treated for 6 h either with UV light alone (L), fungal elicitor alone (E), or UV light followed after 4 h by elicitor (L/E). C, Untreated control.
Functional and Structural Comparison of the ACO and CHS Gene Promoters.
This result prompted us to extend a previous study using the TATA-proximal portion (245 bp upstream from the transcription start site) of the ACO gene promoter (previously termed LF53; ref. 13) in combination with the β-glucuronidase (GUS) reporter gene to demonstrate its UV-light responsiveness in transformed parsley protoplasts. While the ACO/GUS construct was induced 11-fold by UV light, and a similar CHS/GUS construct 50-fold (13), the induction was now shown to be reduced to 3- and 4-fold, respectively, when elicitor was added simultaneously with UV light under otherwise identical conditions.
In view of this functional similarity in transient expression assays, we determined the nucleotide sequence of the ACO gene promoter and compared it with the corresponding regions of the CHS and several other previously investigated gene promoters. Fig. 4 shows that the TATA-proximal promoter region contains various sequences with high similarity to previously determined cis-acting elements in the UV light-responsive CHS and PAL gene promoters (18, 21, 22) as well as in the elicitor-responsive WRKY1 (23) and several other gene promoters.
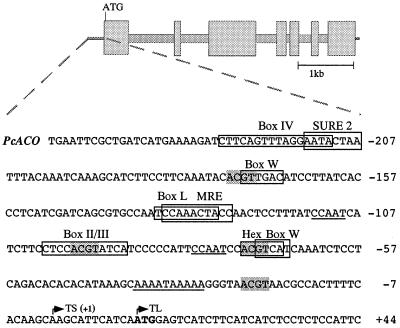
Figure 4.
Nucleotide sequence of the TATA-proximal ACO promoter region. Boxes and shading indicate putative or established cis-acting elements, including the ACGT core of Box II, Hex (hexanucleotide), MRE (Myb-recognition) and sugar-responsive elements (SURE). Boxes II/III and IV as well as MRE, the consensus version of box I, are functionally important elements in the parsley CHS gene promoter, boxes L and W in the PAL1 and WRKY1 gene promoters, respectively. See text for further explanations. Possible CCAAT and TATA boxes are underlined and the transcription (TS) and translation (TL) start sites marked with arrows.
Discussion
These results indicate a limited, yet considerably broader, response of parsley cells to UV irradiation than previously deduced from studies at the enzyme activity level. Analysis at the mRNA level now revealed that one representative each from all but one [glyceraldehyde 3-phosphate dehydrogenase (GAPDH), see below] of the various tested supply pathways responded in a manner similar to PAL and CHS (representing the two phenylpropanoid pathways), although from much higher background levels. The supply pathways included those generating phenylalanine and acetyl-CoA, the two precursors of the basic carbon skeleton from which all flavone and flavonol glycosides are derived, as well as S-adenosyl-l-methionine, the donor of methyl groups for final structural variations through 3′-O-methylation (5). Not tested was the pathway leading to UDP-glucose, whose precise nature is unknown for heterotrophically growing, sucrose-fed parsley cells.
The seeming exception of GAPDH from this induction behavior may be explained by the occurrence of differentially regulated isoforms (24), only one of which was analyzed here. In fact, most of the mRNAs/enzymes from the supply pathways outlined in Fig. 1 occur in multiple isoforms with different regulatory properties (2). It is therefore highly probable that the arbitrarily chosen isoform of GAPDH happened to be a light-insensitive one, whereas UV light-responsive isoforms were selected by chance for DAHPS and SHH, and that both types were among the three G6PDH isoforms analyzed, one being strongly responsive and two not responsive at all. A particularly well-studied example for such a kind of differential behavior is PAL, which occurs in four isoforms in parsley (25), only three of which are induced by UV light (26).
In view of this complexity at the isoform level, it would have been particularly interesting to extend these studies to the corresponding enzyme activities and intermediary metabolites. However, only the general phenylpropanoid and flavonoid glycoside pathways (bold frames in Fig. 1) are easily accessible to such analyses. Strong induction of their constituting enzymes, as well as the flavonoid end products, was readily demonstrated in UV-irradiated cultured parsley cells, largely because in all cases the preinduction levels were very low or undetectable (5). In the case of CHS, it was even possible to mathematically relate the shapes of the individual induction curves for all steps in the sequence of events to one another, from gene transcription via mRNA amounts and enzyme activities to flavonoid glycoside accumulation, indicating that each step is the immediate consequence of the preceding one (6).
Such a relationship is difficult to unequivocally prove for enzymes and intermediates of primary metabolism, including the supply pathways of flavonoid biosynthesis, largely because of their subcellular functional and spatial diversity. If the example of G6PDH in elicitor-treated parsley cells (2) is generally applicable, even strong induction of one or a few isoforms in connection with a given metabolic switch might be obscured at the overall enzyme activity level by the simultaneous repression of other isoforms with different physiological roles. By comparison, gene-specific hybridization probes offer very sensitive tools for analyzing induced metabolic changes at the level of individual mRNA isoforms, although the actual fluxes and conversion rates of metabolites in vivo cannot be deduced from such indirect measurements in vitro, neither at the mRNA nor at the enzyme activity level.
These considerations concerning the occurrence of multiple isoenzymes with distinct physiological roles may have a bearing on the previous assignments of ACCase (4) and UDP-apiose synthase (UAS; ref 27) as bona fide members of the flavonoid biosynthetic pathway purely on the basis of their induction behavior and metabolic relatedness. Using the same criteria, ACO, G6PDH1, DAHPS2, and SHH likewise would qualify as members of this pathway. However, all of them have been shown to be strongly induced by elicitor as well (this study and ref. 2), which represses flavonoid biosynthesis (15). Hence, they are very unlikely to be true members of the flavonoid glycoside pathway. Rather, we conclude from the present results that UV light selectively induces all those primary as well as secondary metabolic activities that are directly and indirectly required for flavonoid glycoside formation, and that those of the induced enzymes that do not belong to the flavonoid biosynthetic pathway proper are recruited from other, more complex regulatory circuits.
Thus, the involvement of ACCase and UAS in flavonoid biosynthesis as well as unrelated metabolic pathways, e.g., fatty acid (28) and cell-wall biosynthesis (29), respectively, is comparable to the multiple metabolic roles of all enzymes depicted outside the bold frames in Fig. 1. An important difference may lie in the low background activities of ACCase and UAS as compared with the other enzymes and mRNAs from primary metabolism under the experimental conditions used. Because cDNA probes for ACCase and UAS from parsley are not available, clarification of their relatedness to flavonoid and other biosynthetic pathways must await further studies in this direction. In any event, the apparent combinatorial flexibility of the various metabolic activities analyzed here and previously (2) may argue against too categorical delimitations of pathways.
The notion of flexible combinations of primary and secondary metabolic pathways is further supported by the data on the ACO gene promoter, whose TATA-proximal region was shown to contain sequences previously identified as functional cis-acting elements in UV light-responsive promoters, including the parsley PAL and CHS gene promoters [box L and boxes I (i.e., MRE, Myb-recognition element), II, III, and IV, respectively; refs. 18 and 21], and in the elicitor-responsive WRKY1 gene promoter (box W; ref. 23). In accord with these structural similarities, the relative expression modes of the ACO and CHS gene promoters in transformed parsley protoplasts were essentially similar to one another as well as to those observed for the ACO and CHS mRNA accumulation patterns in intact parsley cells. A sugar-responsive element (30) may indicate additional modes of regulation of the ACO gene besides the responses to UV light and elicitor/infection. For example, β-oxidation in plants, including the reaction catalyzed by ACO, has important roles not only in the degradation of stored lipids, but also in developmental and stress-induced processes, including jasmonate synthesis (31), senescence, and fatty acid homeostasis (32). Thus, although a detailed functional analysis of the ACO gene promoter is still lacking, the observed structural features support the notion that the encoded enzyme serves multiple roles in metabolism.
In conclusion, the metabolic changes induced in UV-irradiated parsley cells appear to extend to a considerable degree from secondary into primary metabolism, but are not nearly as drastic as recently reported for elicitor-treated parsley cells. Upon elicitor treatment, all of more than 40 tested mRNAs increased or decreased strongly (2), including all those arbitrarily selected mRNAs that were now shown to remain unaffected by UV light. Thus, the UV-light response is either much more selective, e.g., confined to the generation of a certain number of primary metabolites and their conversion to UV-protective flavonoids, or much more subtle, such that only drastic changes are clearly detectable. In either case, we have demonstrated a broad spectrum of flexible metabolic adaptiveness to environmental hazards, from the extensive reprogramming during the pathogen defense response (2) to the predominant or even exclusive induction of those pathways that are directly and indirectly involved in the accumulation of a single major class of protective agents in response to UV irradiation. These results confirm, for the interplay of primary and secondary metabolism, the general notion of a wide-ranging metabolic plasticity that has been observed in a large variety of conditions (e.g., ref. 33).
Acknowledgments
We thank Drs. Robert Cormack and Gunnar Plesch for valuable experimental advice and Drs. Nikolaus Amrhein and Paul Rushton for helpful comments on the manuscript. Financial support was received from Fonds der Chemischen Industrie.
Abbreviations
- ACCase
acetyl-CoA carboxylase
- ACO
acyl-CoA oxidase
- DAHPS
3-deoxy-d-arabinoheptulosonate 7-phosphate synthase
- PAL
phenylalanine ammonia-lyase
- CHS
chalcone synthase
- SHH
S-adenosyl-homocysteine hydrolase
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- UAS
UDP-apiose synthase
- G6PDH
glucose 6-phosphate dehydrogenase
Footnotes
Data deposition: The ACO gene sequence has been deposited in the GenBank database (accession no. AF 202987).
References
- 1.Somssich I E, Bollmann J, Hahlbrock K, Kombrink E, Schulz W. Plant Mol Biol. 1989;12:227–234. doi: 10.1007/BF00020507. [DOI] [PubMed] [Google Scholar]
- 2.Batz O, Logemann E, Reinold S, Hahlbrock K. Biol Chem. 1998;379:1127–1135. doi: 10.1515/bchm.1998.379.8-9.1127. [DOI] [PubMed] [Google Scholar]
- 3.Somssich I E, Hahlbrock K. Trends Plant Sci. 1998;3:86–90. [Google Scholar]
- 4.Ebel J, Hahlbrock K. Eur J Biochem. 1977;75:201–209. doi: 10.1111/j.1432-1033.1977.tb11518.x. [DOI] [PubMed] [Google Scholar]
- 5.Hahlbrock K, Scheel D. Annu Rev Plant Physiol Mol Biol. 1989;40:347–369. [Google Scholar]
- 6.Chappell J, Hahlbrock K. Nature (London) 1984;311:76–78. [Google Scholar]
- 7.Schmelzer E, Jahnen W, Hahlbrock K. Proc Natl Acad Sci USA. 1988;85:2989–2993. doi: 10.1073/pnas.85.9.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu S C, Hahlbrock K. Z Naturforsch. 1992;47:591–600. [Google Scholar]
- 9.Logemann E, Wu S C, Schröder J, Schmelzer E, Somssich I E, Hahlbrock K. Plant J. 1995;8:865–876. doi: 10.1046/j.1365-313x.1995.8060865.x. [DOI] [PubMed] [Google Scholar]
- 10.Volpe J J, Vagelos P R. Annu Rev Biochem. 1973;42:21–60. doi: 10.1146/annurev.bi.42.070173.000321. [DOI] [PubMed] [Google Scholar]
- 11.Egin-Bühler B, Loyal R, Ebel J. Arch Biochem Biophys. 1980;203:90–100. doi: 10.1016/0003-9861(80)90156-3. [DOI] [PubMed] [Google Scholar]
- 12.Henstrand J M, McCue K F, Brink K, Handa A K, Herrmann K M, Conn E E. Plant Physiol. 1992;98:761–763. doi: 10.1104/pp.98.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tovar Torres J, Block A, Hahlbrock K, Somssich I E. Plant J. 1993;4:587–592. [Google Scholar]
- 14.Kombrink E, Hahlbrock K. Plant Physiol. 1986;81:216–221. doi: 10.1104/pp.81.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lozoya E, Block A, Lois R, Hahlbrock K, Scheel D. Plant J. 1991;1:227–234. [Google Scholar]
- 16.Douglas C, Hoffmann H, Schulz W, Hahlbrock K. EMBO J. 1987;6:1189–1195. doi: 10.1002/j.1460-2075.1987.tb02353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Block A, Dangl J L, Hahlbrock K, Schulze-Lefert P. Proc Natl Acad Sci USA. 1990;87:5387–5391. doi: 10.1073/pnas.87.14.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lois R, Dietrich A, Hahlbrock K, Schulz W. EMBO J. 1989;8:1641–1648. doi: 10.1002/j.1460-2075.1989.tb03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooks M K, Kellas F, Graham I A. Plant J. 1999;20:1–13. doi: 10.1046/j.1365-313x.1999.00559.x. [DOI] [PubMed] [Google Scholar]
- 20.Subramani S. J Biol Chem. 1996;271:32483–32486. doi: 10.1074/jbc.271.51.32483. [DOI] [PubMed] [Google Scholar]
- 21.Schulze-Lefert P, Dangl J L, Becker-Andre M, Hahlbrock K, Schulz W. EMBO J. 1989;8:651–656. doi: 10.1002/j.1460-2075.1989.tb03422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiβhaar B, Armstrong G A, Block A, da Costa e Silva O, Hahlbrock K. EMBO J. 1991;10:1777–1786. doi: 10.1002/j.1460-2075.1991.tb07702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eulgem T, Rushton P J, Schmelzer E, Hahlbrock K, Somssich I E. EMBO J. 1999;17:4689–4699. doi: 10.1093/emboj/18.17.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin W, Gierl A, Saedler H. Nature (London) 1989;339:46–48. [Google Scholar]
- 25.Appert C, Logemann E, Hahlbrock K, Schmid J, Amrhein N. Eur J Biochem. 1994;225:491–499. doi: 10.1111/j.1432-1033.1994.00491.x. [DOI] [PubMed] [Google Scholar]
- 26.Logemann E, Parniske M, Hahlbrock K. Proc Natl Acad Sci USA. 1995;92:5905–5909. doi: 10.1073/pnas.92.13.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardiner S E, Schröder J, Matern U, Hammer D, Hahlbrock K. J Biol Chem. 1980;255:10752–10757. [PubMed] [Google Scholar]
- 28.Ohlrogge J B, Jaworski J G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- 29.O'Neill M A, Warrenfeltz D, Kates K, Pellerin P, Doco T, Darvill A G, Albersheim P. J Biol Chem. 1996;271:22923–22930. doi: 10.1074/jbc.271.37.22923. [DOI] [PubMed] [Google Scholar]
- 30.Grierson C, Du J S, Zabala De Torres M, Kyle B, Smith C, Holdsworth M, Bevan M. Plant J. 1994;5:815–926. doi: 10.1046/j.1365-313x.1994.5060815.x. [DOI] [PubMed] [Google Scholar]
- 31.Wasternack C, Parthier B. Trends Plant Sci. 1997;8:302–307. [Google Scholar]
- 32.Eccleston V S, Ohlrogge J B. Plant Cell. 1998;10:613–621. doi: 10.1105/tpc.10.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guyer D, Patton D, Ward E. Proc Natl Acad Sci USA. 1995;92:4997–4500. doi: 10.1073/pnas.92.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]