Abstract
Heat shock protein 90 is emerging as an important target in cancer chemotherapy. In a program directed towards identifying novel chemical probes for Hsp90, we found 4-hydroxy-3-(2-hydroxynapthalene-1-yl)phenyl)benzene sulfonamide as an Hsp90 inhibitor with very weak activity. In this report we present a new and general method for the synthesis of variety of analogs around this scaffold and discuss their structure activity relationships.
Molecular chaperones are a class of proteins that are responsible for the correct folding and maturation of several cellular client proteins.1 One among those chaperones is the heat shock protein 90 (Hsp90), which recently emerged as a very important and validated target in several diseases including cancer, neurodegeneration, viral, fungal and microbial infection.2,3 Hsp90 chaperone activity is found to be essential for the stability and function of a number of conditionally expressed signaling proteins, as well as mutated, chimeric, and/or overexpressed signaling proteins that promote cell growth and survival.3 The client proteins found to interact with Hsp90 include Raf-1, mutant BRaf, IGF-IR, Akt, HER2, C-Met, C-Kit, mutant EGFR and others.4,5 These client members are found to play crucial role in various types of cancer2 and to undergo proteosome-mediated degradation when Hsp90 activity is inhibited. 6, 7
The natural product geldanamycin (GM) and its synthetic analog (17-AAG) were found to bind to the ATP binding site on the N-terminal domain of the Hsp90 and inhibit ATP-dependent chaperone activities.8 Only the synthetic derivative 17-AAG is currently being evaluated for clinical use. Though Hsp90 is expressed abundantly in most tissues, studies employing GM, 17-AAG and other compounds have concluded that cancer cells are more sensitive to Hsp90 inhibition compared to normal cells.9 This conclusion is further supported by the studies of Kamal et al,10 who demonstrated that tumor cell Hsp90 is found to be in an activated complex with co-chaperones, whereas Hsp90 in normal tissue resides in a free and uncomplexed state. These observations gave the impetus to develop novel and effective small molecule Hsp90 inhibitors, and further affirm the notion that Hsp90 inhibition leads to selective killing of cancer cells over normal cells.
Structure based design and high-throughput (HTS) screening efforts are underway to identify novel chemical scaffolds that can potentially generate lead structures for further development.11 So far, this work has produced at least two clinical candidates (PU-H7112, CNF202413) and several other preclinical candidates from the pyrazole/ isoxazole class of compounds demonstrating successful Hsp90 activity in both in vitro and in vivo tumor models.14
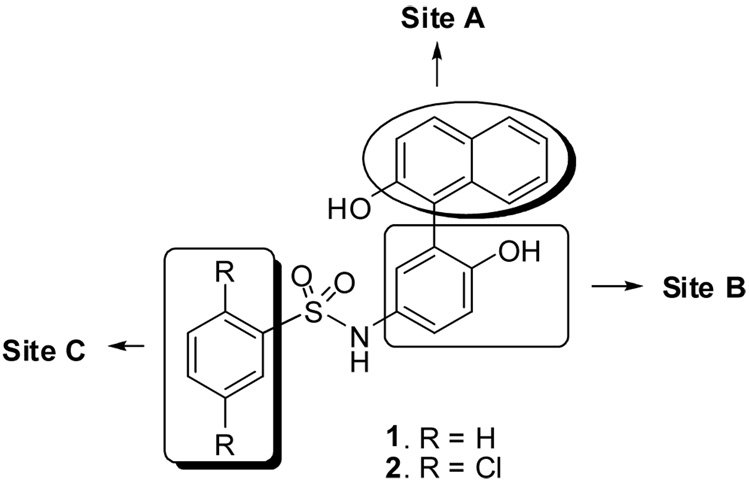
Under the MLSCN program,15 we recently identified aminoquinolines by HTS16 as a novel class of Hsp90 inhibitors. The work was complemented by ligand-based design and virtual screening exercises. Within this context, structures 1 and 2 (Figure 1) were previously identified as Hsp90 blockers by means of virtual screening performed by Barril et al.17 The compounds were found to show low micromolar activity in the malachite green assay18 and subsequently in an FP assay.19 However these analogs showed very poor activity in a cell growth inhibition assay (2: IC50 = 29 µM). Moreover, a very limited SAR study on the 4-hydroxy-3-(2-hydroxynapthalene-1-yl)phenyl)-benzene sulfonamide scaffold was reported.17 This prompted us to perform a more expanded SAR evaluation on this scaffold and to devise a synthetic method amenable to generating a variety of analogs. For instance, only derivatives on the terminal phenyl ring extending from the sulfone unit were reported; the naphthalene and middle rings remaining untouched.
Figure 1.
Potential sites for modification and SAR study
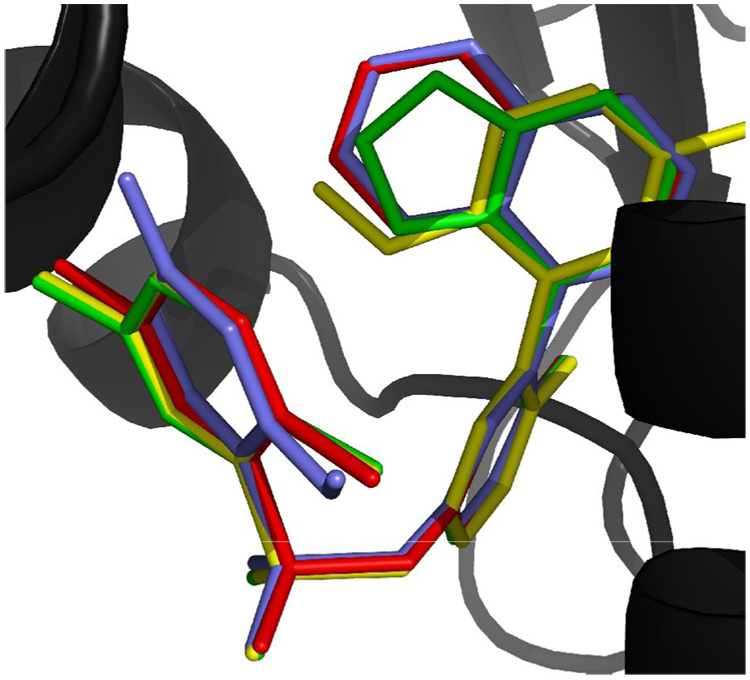
Previous crystallographic investigation of 2 by Barril et al. reveals that its naphthalene ring is buried deep within the ATP binding pocket in the N-terminal region of Hsp90.17 Compound 2 follows the general pharmacophore established by other known Hsp90 inhibitors (Figure 2): contact with the hydrophobic cluster by the naphthalene moiety and participation in a hydrogen bonding network with residues Thr184 and Asp93 (the numbering refers to Hsp90α). Analysis of the available steric space and augmentation of the hydrophobic contacts and hydrogen bonds were considered during the optimization of the arylsulfonamide series.
Figure 2.
Docking of 6c (Blue), 13 (Yellow), 17 (Green) at N-terminal ATP binding site of Hsp90 (Black, PDB: 2bz5). Docking poses are overlapped with co-crystallized pose of compound 2 (Red). Figure shows that the docking poses were able to reproduce most heavy-atom positions of the crystal pose. Hydrogen, water, and residue atoms were not displayed. Coordinates for poses can be provided upon request.
The possible sites for modification are outlined in Figure 1. Site A: The 2-napthol ring could be replaced with 2-phenol, 3,4-benzodioxane-2-phenol and 2-hydroxy-carbazole. The latter was not only considered able to provide bulkiness to fill the cavity observed in the X-ray model, but also scaffold hydrophilicity which can promote increased compound solubility. Likewise, crystal structure analysis suggested modifications could be performed on site B by replacing the OH group with NH2, F etc. In addition, more than one functional group can be added to the middle ring, while the sulfonamide tail can be moved to other positions on the same ring. At site C with a limited SAR, there is room for modification such as replacing the two chlorines with hydroxyl groups combined with modification at sites A and B to explore potential activity enhancement.
At this juncture, we recognized the lack of a general synthetic strategy for preparing the contemplated compounds. Only one synthetic method making use of preformed N-arenesulfonoyl-p-benzoquinonimines as starting materials had been reported in a Russian journal.20 Unfortunately, the procedure is unsuitable to a wide range of quinones. Thus, we envisioned an alternative and versatile synthetic method for 4-hydroxy-3-(2-hydroxynapthalene-1-yl)phenyl)benzene-sulfonamide. Below, we show that this method is applicable to a variety of analogs by means of a split pool synthetic approach as shown in Schemes 2–3. A preliminary SAR for these analogs is also described.
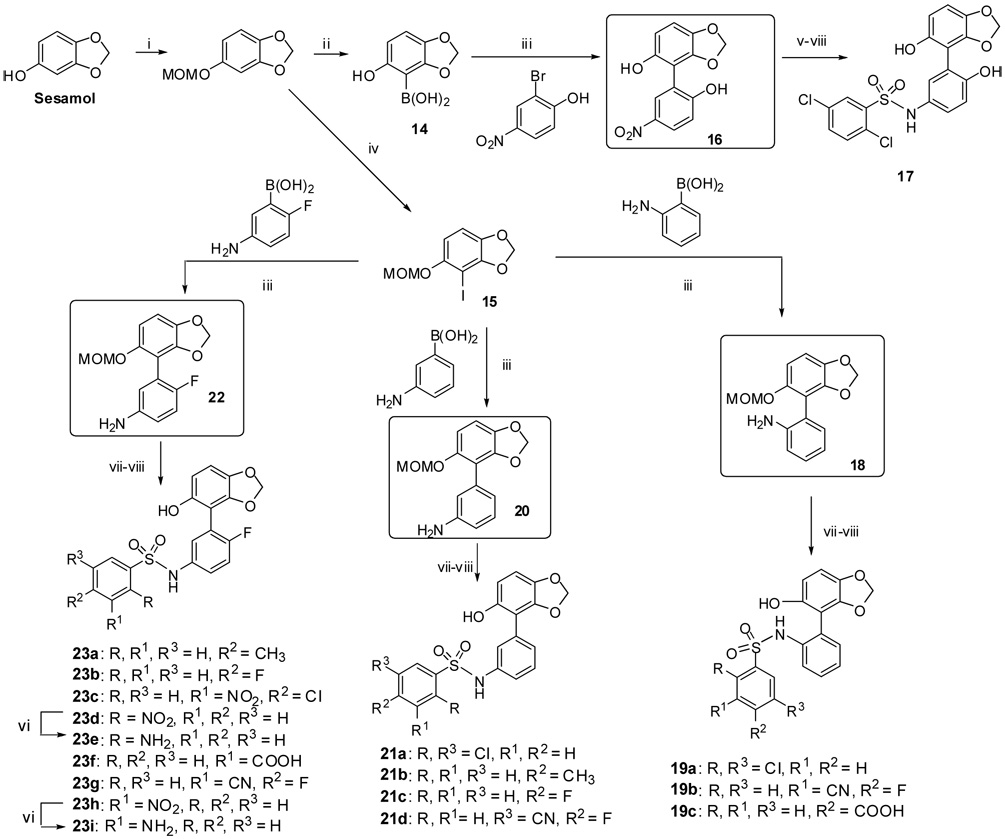
Scheme 2.
Reagents and conditions: i. NaH, MOMCl, DMF, 0 °C-RT, 98%, ii. n-BuLi, THF, B(OMe)3, −78 °C, then 10% HCl, RT tech-grade (50%). iii. 15 (1 eq, 2 mmol), boronic acids (1.8–2 eq), Pd (PPh3)4 (5–10 mol%) , K2CO3,(3–3.3 eq), DME:EtOH:H2O (3:2:1 mL), Microwave initiator, 15 min., 160 °C, 90–95% (except for the 16 which only obtained in 20% yield based on recovered SM), iv. n-BuLi, THF, I2, 0 °C-RT v. Boc2O, DMAP, THF, Reflux, 3–4 h, 95%, vi. H2, 10%Pd-C, MeOH:EtOAc (6:2 mL), 12 Psi, 1 h, 80–85%, vii. ArSO2Cl, Py, 100% viii. 2M HCl,THF:MeOH, 90%.
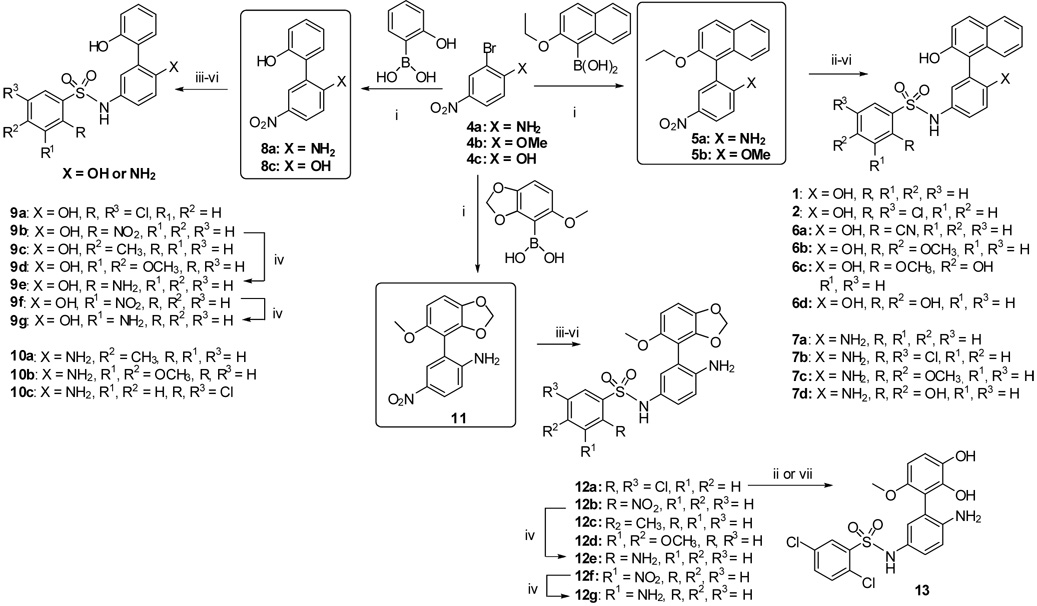
The synthesis of 1–2 and their analogs is depicted in Scheme 1. Thus, 2-bromo-4-nitroanisole or 2-bromo-4-nitroanilines (4a or 4b) were subjected to the Suzuki-Miyaura cross coupling reaction21 with 2-ethoxy-1-napthalene boronic acid in a microwave initiator for 15 minutes to produce product 5 in 95–98% yield. To our surprise such a cross coupling strategy does not seem to have been previously reported for unsymmetrical 2,2'-binapthols and or 1,1'-biphenols, though palladium-mediated procedures have been employed to generate self-coupled symmetrical 1,1'-binapthols or 2,2'-biphenols.22 The methyl-and ethyl-ethers were deprotected by treating 5a and 5b with borontribromide in dichloromethane solution, then the resulting hydroxyl and amino groups (from 5a) or two hydroxyls (from 5b) were protected with Boc-groups before reducing the nitro group by hydrogenation with 10%-palladium on carbon. The resulting amine was subsequently treated with substituted arylsulfonylchlorides in pyridine to provide Boc-protected sulfonamides. The latter were subjected to 2%-trifluoracetic acid in dichloromethane to provide final products 1–2, 6–7.23
Scheme 1.
Reagents and conditions: i. 4a or 4b (1 eq), boronic acids(1.8–2 eq), Pd (PPh3)4 (5–10 mol%), K2CO3,(3–3.3 eq), DME:EtOH:H2O (3:2:1 mL), Microwave initiator, 15 min., 160 °C, 95–98% ii. BBr3, CH2Cl2, −78 °C-RT, 1 h, 85–90% iii. Boc2O, DMAP, THF, Reflux, 3–4 h, 95%, iv. H2, 10%Pd-C, MeOH:EtOAc (6:2 mL), 12 Psi, 1 h, 80–85%, v. ArSO2Cl, Py, 100% vi. 2%TFA in CH2Cl2, RT, 90%. vii. aq. HBr, 80 °C, 65%.
The palladium cross coupling strategy appeared promising for generating a variety of building blocks such as biphenol (8) and aminophenol ether (11). Indeed both these intermediates were synthesized analogous to 5 and carried forward by similar synthetic steps for 1 to provide 9a–g, 10a–c and 12a–g. Selective demethylation of 12a with either borontribromide or aqueous hydrogen bromide yielded only methylendioxy ring opened product 13. As part of our study, we decided to prepare 17, a benzodioxane moiety as a replacement for the naphthalene core of 2. To this end, 5-hydroxybenzo-1,3-dioxo-4-yl-boronic acid (14) was synthesized albeit in very low yield and in a poor quality as shown in Scheme 2. Coupling 14 with 2-bromo-4-nitrophenol by the same Suzuki procedure provided intermediate 16 in very low yield. Nevertheless, this was smoothly carried through to the final product 17 as shown in Scheme 2.
Docking calculations were performed to determine if proposed synthesized analogues (6c, 13 and 17) could attain a binding pose similar to that of the known arylsulfonamide 2 (Figure 2). Schrodinger’s GLIDE24 docking protocol was employed, while structure 2BZ5 was obtained from the Protein Data Bank (PDB).25 Schrodinger’s Protein Preparation protocol was employed, and the receptor geometry was optimized around compound 2 using the OPLS2001 force field. Crystal water 150 was retained since it appeared to be a conserved interaction in all Hsp90 co-crystals examined in the PDB.8,11a,12,17 Crystal water 286 was retained since no residue had any direct interaction with the side C and sulfonamide region. All other crystal waters were removed. The optimization was constrained to 0.3 Å for all atoms. Standard docking precision was performed, in which non-planar amide bond conformations were penalized, and van der Waals radii were scaled to a factor of 0.8. Each compound structure was allowed to generate up to 5 poses. Strain correction terms were applied. No constraints were applied during the docking. The Glide docking poses for 2 and its analogues were energy-rescored using MMGBSA.26 The top scoring MMGBSA pose of each analogue was overlaid with the crystallographic pose in Figure 2, while the resulting scores are listed in Table 1. Although the Glide score and the MMGBSA rescore favor different compounds, the overall overlap shows that the analogues take advantage of the same contacts as 2.
Table 1.
Docking scores for compound 2 and higher affinity compounds
| Compound | MMGBSA(kcal/mol) | Gscore(kcal/mol) |
|---|---|---|
| 2 | −44.6 | −8.1 |
| 6c | −42.1 | −8.6 |
| 13 | −41.3 | −7.1 |
| 17 | −40.4 | −7.9 |
Based on the published X-ray crystal data of Barril et al and our computational docking results, we decided to retain the 3,4-dioxane moiety and prepare derivatives by altering other parts of the molecule. Difficulties in preparation of good quality boronic acid 14 and the low yields in coupling reactions with it prompted us to revise the synthetic strategy shown in Scheme 2. A known iodide building block 1527 was prepared in large quantity starting from sesamol, and this was coupled to various commercially available boronic acids by Suzuki-coupling to generate intermediates 18, 20, 22 in excellent yields. All of the compounds were subjected to synthetic steps similar to those used in Scheme 1 to generate final products 19, 21 and 23 as shown in Scheme 2.
All synthesized compounds were tested in a fluorescent polarization (FP) assay28 which measures the interaction of small molecule ligands with fluorescently labeled GM and displaces the latter from the ATP binding site of Hsp90. To our surprise, we have not identified compounds more active than 2. Apart from the analogs tabulated in Table 2, other analogs have shown no inhibitory activity at the maximum concentration tested (50µM). Only structural modifications at the terminal phenyl ring (6a–6d) appear to be tolerated and thereby retain activity. Replacing the OH on the middle ring with amine decreased the activity (7b–d). Substituting the 2-napthol unit with the less bulky 2-phenol moiety completely eliminated activity (9b–g and 10a–b), except for 9a which showed about 20-fold less activity than 2. However, replacing it with the 3,4-methylenedioxy-2-phenol moiety was well tolerated and decreased the activity by approximately 1.5-2-fold (17). This observation reinforces the suggestion made by the docking model that substituents equal in size or bulkier than the 2-napthol ring might be better in this region to fill the hydrophobic cavity. Surprisingly, however, 13 exhibits a sub-micromolar IC50 approximately equal to the IC50 of 2.
Table 2.
| Compd. | IC50 (µM) | Compd. | IC50(µM) | ||
|---|---|---|---|---|---|
| FP | SKBr3 | FP | SKBr3 | ||
| 1 | 4.5 | na | 9a | 19.4 | na |
| 2 | 0.70 | 6.5 | 13 | 0.55 | 6.0 |
| 6a | 4.5 | na | 17 | 1.7 | 4.4 |
| 6b | 2.5 | na | 21a | 14.5 | na |
| 6c | 1.3 | 9.7 | 21b | 31.3 | na |
| 6d | 1.8 | >50 | 21c | 19.6 | na |
| 7b | 9.4 | na | 23a | 28.0 | na |
| 7c | 25.0 | na | 23c | 26.0 | na |
| 7d | 9.5 | na | 23g | 27.0 | na |
Mean of 2 independent determinations.
The other compounds which did not show any inhibitory effect up to 50 µM concentration are not shown in this table
na = not tested in SKBr3 cells
The compounds presented in Table 2 were also tested in the Western blot assay by examining client protein (HER2, Raf-1, PRAS40) degradation. Disappointingly, none of the analogs showed any promising effect, except 2 and 13, both of which possess IC50 values of approximately 20 µM in HER2 degradation. However neither had any effect on Raf-1 and PRAS40. Though, it is difficult to hypothesize that these analogs bind to Hsp90 protein with high affinity, but do not enforce disruption of client member association, but it could be speculated to their poor permeability properties which might cause them to be less potent in cell-based WB-assay. To support this hypothesis, we predicted ADME properties for the best active compounds by QikProp software.29 As shown in Table 3, the compounds 2, 6c, 13 and 17 are out side of the projected range.
Table 3.
| 2 | 6c | 13 | 17 | Ranges (nm/sec) |
|
|---|---|---|---|---|---|
| <25 poor, | |||||
| Caco2 | 242 | 84.9 | 81.8 | 249.7 | >500 great |
| <25 poor, | |||||
| MDCK | 439.3 | 35.1 | 128.8 | 491.9 | >500 great |
Schrodinger’s Qikprop employed for these calculations
Highlighted numbers in red denote compound’s property value borders range value of 95% of known drugs.
The best active sulfonamides (2,6c, 6d, 13 and 17) in FP assay were also tested in cell growth inhibition assay against HER2 over-expressing SKBr3 breast cancer cells.30 As shown in Table 2, these derivatives exhibited only low micromolar IC50 values in this cell line, however, these values were only about 1.4-3.1-fold (except for 6d) less to IC50 values noted by a known pyrazole inhibitor.31 Nonetheless, the potencies of 2, 6c, 13 and 17 are at least 2.6-11-fold less in antiproliferative assay compared to the in vitro binding (FP) assay. Further studies are necessary to explain the activity differences in two assays, but partly could be attributed to their poor cell permeability properties as predicted by QikProp.
In conclusion, a ligand-based design strategy centered on the sulfonamide scaffold was conceived to develop effective Hsp90 inhibitors. Although the anticipated nanomolar activity was not observed by examining synthetic representatives of this scaffold, a new and versatile synthesis was developed. The procedure should not only enable us to prepare a number of analogs for SAR, but also may be generally applicable for the synthesis of unsymmetrical biphenols and binapthols, namely 5, 8, 11, 16, 18, 20 and 22. The SAR study suggests that still more work needs to be done to manipulate the naphthalene moiety and also the middle phenyl ring of the scaffold, an effort that will be exerted in due course.
Acknowledgements
This work was supported by the US National Institutes of Health 1 U54 HG003918-02 and 1R03MH076499-01 and encouraged by Professor Dennis Liotta (Emory University)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Whitesell L, Lindquist SL. Nat. Rev. Can. 2005;5:761–767. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 2.Solit DB, Chiosis G. Drug Discovery Today. 2008;13:38–43. doi: 10.1016/j.drudis.2007.10.007. and reference cited their in. [DOI] [PubMed] [Google Scholar]
- 3.Neckers L. J. Biosci. 2007;32:517–530. doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
- 4.Picard D. Cell Mol. Life. Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shames DS, Minna JD. Proc. Natl. Acad. Sci. USA. 2008;105:1389–1390. doi: 10.1073/pnas.0711993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mimnaugh EG, Chavany C, Neckers L. J. Biol. Chem. 1996;271:22796–22801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 7.Sepp-Lorenzino L, Ma Z, Lebwohl DE, Vinitsky A, Rosen N. J. Biol. Chem. 1995;270:16580–16587. doi: 10.1074/jbc.270.28.16580. [DOI] [PubMed] [Google Scholar]
- 8.Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 9.Chiosis G, Huezo H, Rosen N, Mimnaugh E, Whitesell L, Neckers L. Mol. Cancer Ther. 2003;2:123–129. [PubMed] [Google Scholar]
- 10.Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ. Nature. 2003;425:407. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 11.(a) Cheung K-MJ, Matthews TP, James K, Rowlands MG, Boxall KJ, Sharp SY, Maloney A, Roe SM, Prodromou C, Pearl LH, Aherne GW, McDonald E, Workman P. Bioorg. Med. Chem. Lett. 2005;15:3338–3343. doi: 10.1016/j.bmcl.2005.05.046. [DOI] [PubMed] [Google Scholar]; (b) Gopalsamy A, Shi M, Golas J, Vogan E, Jacob J, Johnson M, Lee F, Nilakantan R, Petersen R, Svenson K, Chopra R, Tam MS, Wen Y, Ellingboe J, Arndt K, Boschelli F. J. Med. Chem. 2008;51:373–375. doi: 10.1021/jm701385c. [DOI] [PubMed] [Google Scholar]; (c) Avila C, Hadden MK, Ma Z, Kornilayev BA, Ye Q-Z, Blagg BS. Bioorg. Med. Chem. Lett. 2006;16:3005–3008. doi: 10.1016/j.bmcl.2006.02.063. [DOI] [PubMed] [Google Scholar]; (d) Barluenga S, Wang C, Fontaine J-G, Aouadi K, Beebe K, Tsutsumi S, Neckers L, Winssinger N. Angew. Chem. Int. Ed. Engl. 2008;47:4432–4435. doi: 10.1002/anie.200800233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Immormino RM, Kang Y, Chiosis G, Gewirth DT. J. Med. Chem. 2006;49:4953–4960. doi: 10.1021/jm060297x. [DOI] [PubMed] [Google Scholar]
- 13.Kasibhatla SR, et al. J. Med. Chem. 2007;50:2767–2778. doi: 10.1021/jm050752+. [DOI] [PubMed] [Google Scholar]
- 14.Sharp SW, et al. Molecular Cancer Therapeutics. 2007;6:1198–1211. doi: 10.1158/1535-7163.MCT-07-0149. [DOI] [PubMed] [Google Scholar]
- 15.(a) http://nihroadmap.nih.gov/molecularlibraries/; (b) http://www.emory.edu/chemical-biology/index.html.
- 16.Ganesh T, Min J, Thepchatri P, Du Y, Li L, Lewis L, Wilson L, Fu H, Chiosis G, Dingledine R, Liotta D, Snyder JP, Sun A. Bioorg. Med. Chem. 2008;16:6903–6910. doi: 10.1016/j.bmc.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barril X, Brough P, Drysdale M, Hubbard RE, Massey A, Surgenor A, Wright L. Bioorg. Med. Chem. Lett. 2005;15:5187. doi: 10.1016/j.bmcl.2005.08.092. [DOI] [PubMed] [Google Scholar]
- 18.Rowlands MG, Newbatt YM, Prodromou C, Pearl LH, Workman P, Aherne W. Anal. Biochem. 2004;327:176–183. doi: 10.1016/j.ab.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 19.Howes R, et al. Anal. Biochem. 2006;350:202–213. doi: 10.1016/j.ab.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Avdeenko AP. Zhurnal Organicheskoi Khimii. 1989;25:2375–2381. [Google Scholar]
- 21.For a recent review on Suzuki-Miyaura cross coupling reaction, please see Kotha S, Lahiri K. Eur. J. Org. Chem. 2007:1221–1236.
- 22.Torii S, Tanaka H, Morisaki K. Tetrahedron. Lett. 1985;26:1655–1658. [Google Scholar]
- 23.Synthesis of 2 (Typical procedures): 2-Bromo-4-nitroanisole (4b)(425 mg, 1.8 mmol), 2-ethoxy-napthalene-1-ylboronic acid (800 mg, 2eq), Pd(PPh3)4 (100 mg, 5 mol%), K2CO3 (735 mg, 3eq) were charged into a microwave reaction vessel. To this vessel, a three solvent mixture (5 ml) (DME: EtOH: H2O, 3:2:1) was added and then the reaction mixture was irradiated in Biotage-microwave initiator for 15 min. Reaction mixture was portioned between ethyl acetate and water. The organics were separated and washed with water, brine, dried over Na2SO4 and concentrated (usual work up). The crude mass obtained was purified by chromatography over silica gel, eluting with 5–8% ethyl acetate in hexanes to provide 2-ethoxy-1-(2-methoxy-5-nitrophenyl)naphthalene (5b) (580 mg, 98%). 1H NMR (400 MHz, CDCl3):δ 8.35 (dd, 1H), 8.21 (d, 1H), 7.91 (d, 1H), 7.84 (m, 1H), 7.36 (m, 4H), 7.08 (d, 1H), 4.12 (q, 2H), 3.77 (s, 3H), 1.24 (t, 3H). 13C NMR (100 MHz):δ 163.2, 153.9, 141.5, 133.3, 130.2, 129.2, 128.7, 128.4, 126.9, 126.7, 125.5, 124.6, 123.9, 119.9, 115.1, 110.7, 65.3, 56.4, 15.2. HRMS Calc. for C19H18N4O, 324.12263; observed 324.12303 (M+H). To a solution of 5b (525 mg, 1.62 mmol) in dichloromethane (15 ml), BBr3 (3eq) was added at −78 °C and the reaction mixture was brought to room temperature over night. Reaction was quenched by addition to ice-cold water and the product was extracted with ethyl acetate. Organics were on usual work gave black residue, which was purified by silica gel chromatography by eluting with 20–30% ethyl acetate in hexanes to provide diol, 1-(2-hydroxy-5-nitrophenyl)naph-thalen-2-ol (400 mg, 87%). 1H NMR (400 MHz, CDCl3):δ 8.23 (dd, 1H), 8.18 (d, 1H), 7.83 (d, 1H), 7.80 (dd, 1H), 7.37 (m, 2H), 7.26 (m, 1H), 7.21 (d, 1H), 7.14 (d, 1H). 13C NMR (100 MHz):δ 160.3, 151.9, 141.8, 133.1, 131.9, 129.4, 128.9, 128.7, 128.0, 126.5, 124.4, 123.8, 121.7, 118.1, 117.0, 112.5. HRMS Calc. for C16H12N4O, 282.07583; observed 282.07608 (M+H). To the above diol-compound (400 mg, 1.43 mmol) in tetrahydrofuran (10 ml) was added (Boc)2O (4.3 mmol, 3eq), DMAP (cat), Et3N (3.6 mmol, 2.5eq) and the reaction mixture was refluxed for 4 hrs. Reaction was cooled and filtered through a short silica gel column eluting with 10% ethyl acetate in haxanes. The filtrate was concentrated to give the Boc-protected product (700 mg, 99%). 1H NMR (400 MHz, CDCl3):δ 8.35 (dd, 1H), 8.28 (d, 1H), 7.94 (d, 1H), 7.88 (d, 1H), 7.52-7.38 (m, 5H), 1.36 (s, 9H), 1.14 (s, 9H). The above compound (650 mg, 1.35 mmol) was dissolved in EtOAc: MeOH (6:2, 6 ml) and was added Pd-C (10% on charcoal) and hydrogenated at 12 Psi for 45 min. Filtered off the catalyst through short silica gel column by eluting with dichloromethane. The filtrate was concentrated give amino-product (490 mg, 80%) which was used for next reaction. 1H NMR (400 MHz, CDCl3):δ 7.84 (d, 1H), 7.80 (dd, 1H), 7.61 (d, 1H), 7.39 (m, 3H), 7.06 (d, 1H), 6.72 (dd, 1H), 6.61 (d, 1H), 1.39 (s, 9H), 1.07 (s, 9H). HRMS Calc. for C26H30NO6, 452.20576; observed 452.20676 (M+H). To the solution of above amine (40 mg, 0.088 mmol) in pyridine (1 ml) was added 2,5-dichlorobenzenesulfonyl chloride (36 mg, 1.4eq) and stirred for 8 hrs. Removed the pyridine and the residue was dissolved in dichloromethane (2 ml) and was added TFA (0.3 ml) and stirred the reaction mixture at room temperature for 8 hrs. The reaction mixture was diluted with ethyl acetate and worked up as usual. The crude mass obtained was purified by silica gel preparative-TLC eluting with 0.8% methanol in dichloromethane to provide 2 (35 mg, 90%). 1H NMR (400 MHz, CDCl3):δ 7.91 (d, 1H), 7.82 (d, 1H), 7.78 (d, 1H), 7.42 (m, 2H), 7.34 (m, 2H), 7.21 (m, 2H), 6.98 (d, 2H), 6.95 (d, 1H). 13C NMR (100 MHz):δ 153.4, 151.7, 137.5, 134.3, 133.8, 133.0, 132.8, 131.9, 131.4, 129.7, 129.3, 128.6, 128.4, 127.7, 127.6, 127.0, 124.2, 123.7, 120.7, 118.0, 117.7, 113.1. HRMS Calc. for C22H15Cl2NO4S, 459.00934; observed 459.009343. Similar procedures were employed to make other compounds. 1H NMR data (recorded on Unity-400 MHz in CDCl3) for selected compounds is given below. 5a. 8.15 (dd, 1H), 8.0 (d, 1H), 7.92 (d, 1H), 7.83 (dd, 1H), 7.36 (m, 4H), 6.76 (d, 1H), 4.10 (q, 2H), 1.24 (t, 3H). 6b. 7.69 (dd, 1H), 7.64 (d, 1H), 7.30-7.12 (m, 5H), 7.0 (s, 1H), 6.94 (d, 1H), 6.88 (d, 1H), 6.85 (d, 1H), 6.42 (dd, 2H), 3.84 (s, 3H), 3.78 (s, 3H). 6c. 7.80 (m, 2H), 7.40 (d, 1H), 7.33 (m, 2H), 7.24-7.15 (m, 2H), 7.10 (m, 1H), 7.0 (d, 1H), 6.80 (d, 1H), 6.54 (bs, 1H), 6.43 (dd, 1H), 6.34 (d, 1H), 3.71 (s, 3H). 6d. 7.69 (m, 2H), 7.38 (d, 1H), 7.21 (m, 2H), 7.13 (m, 2H), 6.88 (m, 1H), 6.83 (dd, 2H), 6.30 (dd, 1H), 6.26 (d, 1H). 7b. 7.91 (d, 1H), 7.77 (d, 2H), 7.42 (dd, 1H), 7.36-7.27 (m, 3H), 7.20 (d, 1H), 7.12 (dd, 1H), 6.97 (dd, 2H), 6.81 (d, 1H), 6.75 (d, 1H). 8a. 7.94 (m, 2H), 7.15 (t × d, 1H), 7.08 (d, 1H), 6.87 (t, 2H), 6.64 (d, 1H). 8c. 8.18 (d, 1H), 8.06 (dd, 1H), 7.24 (m, 2H), 6.94 (m, 3H). 9a. 7.89 (s, 1H), 7.40 (s, 2H), 7.22 (t, 2H), 6.95 (m, 6H), 6.80 (dd, 1H). 10a. 7.58 (d, 2H), 7.24 (m, 2H), 7.20 (d, 1H), 7.03 (dd, 1H), 6.96 (m, 3H), 6.77 (d, 1H), 6.68 (d, 1H). 10c. 7.92 (m, 1H), 7.43 (m, 2H), 7.28 (m, 1H), 7.0-6.96 (m, 6H), 6.90 (d, 1H), 6.72 (d, 1H). 11. 8.0 (m, 2H), 6.80 (d, 1H), 6.72 (d, 1H), 6.42 (d, 1H), 5.93 (d, 2H), 3. 72 (s, 3H). 12a. 7.97 (dd, 1H), 7.53 (t, 2H), 7.21 (m, 3H), 6.84 (d, 1H), 6.52 (d, 1H), 5.90 (dd, 2H), 3.68 (s, 3H). 13. 7.92 (s, 1H), 7.41 (s, 2H), 6.98 (m, 3H), 6.86 (d, 1H), 6.72 (d, 1H), 6.44 (d, 1H), 3.59 (s, 3H). 16. 8.32 (d, 1H), 8.08 (d, 1H), 7.00 (d, 1H), 6.68 (d, 1H), 6.40 (d, 1H), 5.89 (s, 2H). 17. 7.89 (dd, 1H), 7.36 (m, 2H), 7.16 (d, 1H), 7.0 (dd, 1H), 6.82 (d, 1H), 6.63 (d, 1H), 6.38 (d, 1H), 5.82 (s, 2H). 19a. 7.49 (dd, 2H), 7.28 (m, 1H), 7.19 (m, 3H), 6.98 (d, 1H), 6.48 (d, 1H), 6.22 (d, 1H), 5.62 (dd, 2H). 21a. 7.86 (t, 1H), 7.25 (m, 3H), 7.18 (d × t, 1H), 7.12 (t, 1H), 6.93 (d × q, 1H), 6.45 (d, 1H), 6.19 (d, 1H). 5.70 (s, 2H). 22. 6.93 (t, 1H), 6.73 (d, 1H), 6.66 (m, 2H), 5.90 (d, 1H), 4.98 (s, 2H), 3.30 (s, 3H). 23a. 7.62 (d, 2H), 7.16 (d, 2H), 7.12 (m, 2H), 7.0 (m, 2H), 6.66 (d, 1H), 6.36 (d, 1H), 5.85 (s, 2H). 23c. 8.21 (d, 1H), 7.80 (dd, 1H), 7.61 (d, 1H), 7.10 (m, 3H), 6.67 (d, 1H), 6.34 (d, 1H), 5.88 (s, 2H). All the synthetic products showed satisfactory analytical (13C NMR, MS) data, consistent with the assigned structure.
- 24.(a) Schrodinger LLC. Schrodinger Ref: Glide, version 4.5. New York, NY: 2007. [Google Scholar]; (b) Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shae DE, Francis P, Shenkin PS. J. Am. Chem. Soc. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 25.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.(a) Schrödinger LLC. Prime, version 1.6. New York, NY: 2007. [Google Scholar]; (b) Schrödinger LLC. Maestro, version 8.0. New York, NY: 2007. [Google Scholar]; (c) Huang N, Kalyanaraman C, Bernacki K, Jacobson MP. Phy. Chem. Chem. Phys. 2006;8:5166–5177. doi: 10.1039/b608269f. [DOI] [PubMed] [Google Scholar]
- 27.Weeratunga G, Jaworska-Sobiesiak A, Horne S, Rodrigo R. Can. J. Chem. 1987;65:2019–2023. [Google Scholar]
- 28.Du Y, Rodina A, Aguirre J, Felts S, Dingledine R, Fu H, Chiosis G. J. Biomol. Screen. 2007;12:915–924. doi: 10.1177/1087057107306067. [DOI] [PubMed] [Google Scholar]
- 29.Schrodinger LLC. QikProp, version 3.0. New York, NY: 2005. [Google Scholar]
- 30.The SKBr3 breast cancer cells were grown in RPMI-1640 medium supplemented with 10% FBS. 1250 cells in 50 µl of culture medium were plated in 384-well microtiter plates (Costar) and allowed to attach for overnight. After treated with either compounds or vehicle (DMSO) for 72 hr, the viability of the cells was measured by CellTiter-Blue (Promega). Briefly, 10µl of CelTiter-Blue were added and incubated at 37°C for 4 hr. The fluorescence intensity (FI) was measured using the Analyst HT plate reader (Molecular Devices) with an excitation at 545 nm and emission at 595 nm. The IC50 was calculated as the compound concentration that inhibits cell viability by 50% compared with vehicle control wells. The data reported are average values from four replicates. IC50 values were determined using a nonlinear regression analysis as implemented in Prism 4.0 (Graphpad Software) O'Brien J, et al. Eur. J. Biochem. 2000;267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x.
- 31.A known pyrazole derivative was resynthesized as reported in reference 11a (compound 16a from reference 11a) and used as a positive control in the biological testing. This analog has shown IC50 value 1.6, 3.1 µM in FP and cell growth inhibition (against SKBr3 cells) assays respectively