Abstract
Background
A novel DNA phosphorothioate modification (DNA sulfur modification), in which one of the non-bridging oxygen atoms in the phosphodiester bond linking DNA nucleotides is exchanged by sulphur, was found to be genetically determined by dnd or dnd-counterpart loci in a wide spectrum of bacteria from diverse habitats. A detailed mutational analysis of the individual genes within the dnd locus in Streptomyces lividans responsible for DNA phosphorothioation was performed and is described here. It should be of great help for the mechanistic study of this intriguing system.
Results
A 6,665-bp DNA region carrying just five ORFs (dndA-E) was defined as the sole determinant for modification of the DNA backbone in S. lividans to form phosphorothioate. This provides a diagnostically reliable and easily assayable Dnd (DNA degradation) phenotype. While dndA is clearly transcribed independently, dndB-E constitute an operon, as revealed by RT-PCR analysis. An efficient mutation-integration-complementation system was developed to allow for detailed functional analysis of these dnd genes. The Dnd- phenotype caused by specific in-frame deletion of the dndA, C, D, and E genes or the enhanced Dnd phenotype resulting from in-frame deletion of dndB could be restored by expression vectors carrying the corresponding dnd genes. Interestingly, overdosage of DndC or DndD, but not other Dnd proteins, in vivo was found to be detrimental to cell viability.
Conclusion
DNA phosphorothioation is a multi-enzymatic and highly coordinated process controlled by five dnd genes. Overexpression of some proteins in vivo prevented growth of host strain, suggesting that expression of the gene cluster is strictly regulated in the native host.
Background
Most of the commonly found structural changes in DNA are due to methylation of selected bases. In some viral DNAs, certain bases may be hydroxymethylated or glucosylated [1-3]. Altered or unusual bases in DNA molecules often have significant physiological implications, such as DNA replication control, gene regulation, or protection of the respective organisms from invasion by foreign DNA [4].
In contrast to other types of DNA modification, S, lividans has a site-specific and stereo-selective sulfur modification on the DNA backbone termed phosphorothioation [5-7]. This sulfur modification occurs specifically between two guanine nucleotides in S.lividans [6,8]. The sulfur-modified DNA suffers double-stranded cleavage at the modification sites during normal and pulsed-field gel electrophoresis [6,9-13]. The Dnd phenotype was proven to be a peracid-mediated, oxidative and amine-catalysed reaction to form a phosphorothioated DNA backbone [6,14,15]. The Dnd phenotype can be overcome by replacing Tris with Hepes in the electrophoresis buffer or by adding a certain concentration of thiourea to Tris-containing buffers [14,15].
In S.lividans, this DNA sulfur modification was found to be determined by a dnd gene cluster carrying five open reading frames (ORFs, dndA-E) [5]. Homologous dnd gene clusters and/or Dnd phenotypes are found in many strains of Streptomyces, E. coli, Bacillus, Salmonella, Klebsiella, Enterobacter, Mycobacterium, Vibrio, Pseudomonas, Pseudoalteromonas, Hahella, Oceanobacter, Geobacter, Pelagibacter, Roseobacter, Mesorhizobium, Serratia, Acinetobacter, and Clostridium, as well as in certain Archaea and unidentified marine microbes, indicating that DNA sulfur modification is a widespread phenomenon in prokaryotes [16].
Here we attribute DNA phosphorothioate modification to a dnd gene cluster consisting of a 6,665-bp region of DNA carrying just five genes. We confirmed by transcriptional analysis that dndB-E constitute an operon, and made systematic in-frame deletion mutations within each gene or combinations of the five dnd genes before performing a series of complementation analyses to evaluate the roles of individual dnd genes in DNA sulfur modification.
Results
Identification of a minimal dnd region
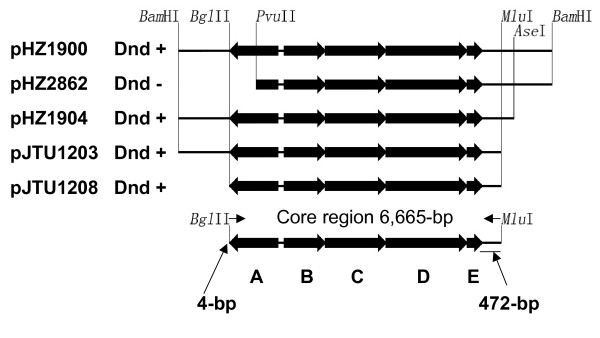
In an effort to precisely localize the region responsible for the Dnd phenotype and obtain unambiguous evidence on the genes involved in DNA phosphorothioation, we made a series of pHZ1900 derivatives by removing end segments from a ca. 10-kb fragment of DNA carrying some likely cis-acting elements using convenient restriction sites, thus identifying a core region carrying only five dnd genes. A combination of restriction fragments (Fig. 1) was incorporated into appropriate sites of integrative vector pSET152 [17] to produce four plasmids (pHZ1904 [5], pJTU1203, pHZ2862, and pJTU1208). Mediated by the attP site of Streptomyces phage ØC31 present on pSET152, these vectors can site-specifically integrate into the attB site in the chromosome of S. lividans ZX1 [9] after transfer by conjugation from E. coli ET12567/pUZ8002 into ZX1. The DNA of these ZX1-derivative strains was either degraded (Dnd+) or stable (Dnd-) during electrophoresis (Fig. 1). The minimal dnd region conferring the Dnd phenotype (Dnd+) was localised to a 6,665-bp fragment on pJTU1208. The left and right borders of the minimal dnd cluster are only 4-bp and 472-bp from the stop codons of dndA and dndE (Fig. 1), respectively, confirming that five genes are necessary and sufficient for DNA phosphorothioation.
Figure 1.
Localization of the boundaries for dnd gene cluster. pSET152-derivatives with the ability to confer Dnd (+ or -) phenotypes are indicated in line with their insert fragments. Five arrows from left to right represent five the ORFs of the dnd gene cluster (dndA-E). Directions of the arrows indicate the transcriptional directions of the genes.
Transcriptional analysis of the dnd genes
Bioinformatic analysis of the 6,665-bp region of pJTU1208 (GenBank accession number DQ075322) suggests that dndA and dndB-E are divergently transcribed. The facts that the 3' end of dndB and the 5' end of dndC overlap by 4 bp (ATGA, position 3,605 to 3,608), that the initiation codon (ATG) of dndD precedes the 3' end of dndC by 12 bp (5088-ATGCACCTGCATAA-5098), and that the initiation codon of dndE (ATG) is 9 bp upstream of the stop codon of dndD (ATGCCGTCTGA) strongly imply that the dndB-E might constitute an operon.
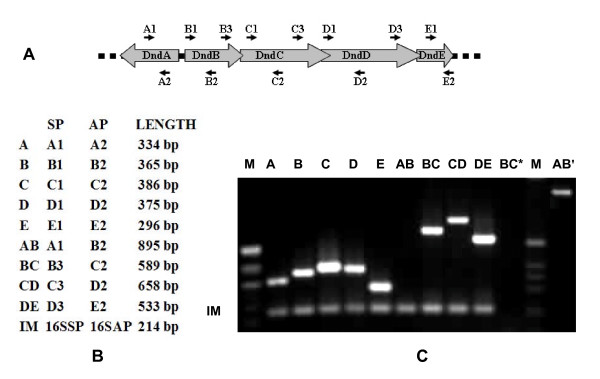
To prove divergent transcription of dndA and a hypothetical dndB-E operon, we performed a transcriptional analysis on the minimal dnd cluster by RT-PCR. RNA was extracted from S. lividans 1326 and amplified by RT-PCR using oligonucleotide primers depicted in Fig. 2A. The PCR products were fractionated by electrophoresis (Fig. 2C). As an internal control, 16S rRNA was amplified in all samples. The appearance of DNA bands (Fig. 2C), which were amplified using different sets of primers (Fig. 2A and 2B), unambiguously suggests that dndB-E are co-transcribed as a single operon in S. lividans 1326. The absence of DNA bands using primers A1 and B2 (Fig. 2C lane AB) suggests a lack of co-transcription in the region between A1 and B2, confirming independent transcription of dndA and dndB-E.
Figure 2.
RT-PCR analysis of the dnd genes transcripts. dnd gene transcripts were reverse transcribed and amplified. (A) Relative positions and directions of corresponding primers are marked with black arrows. (B) Amplification products with sense primer (SP), anti sense primer (AP) and their corresponding lengths. Intra-dnd gene amplification products are indicated as dnd gene names, while products of regions between dnd genes are named linking two corresponding genes such as AB. Amplification of 16S rRNA is used as an internal control marker (IM). (C) Electrophoresis of RT-PCR products. The amplification products are labeled as in Figure 2B. Reverse transcriptase inactivation (BC*) and without DNase treatment (AB') were carried out as negative and positive controls. DNA markers are labeled as "M".
A mutation-integration system for functional analysis of individual dnd genes
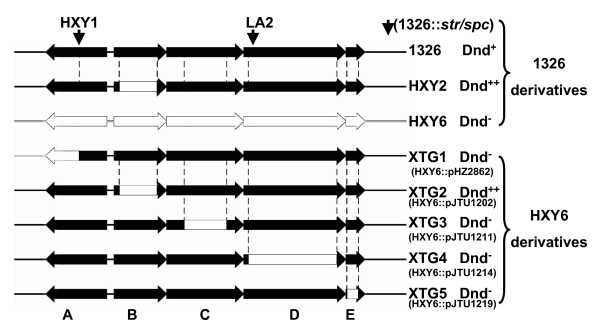
As demonstrated by the transcriptional analysis, dndB-E constitute an operon. We therefore inactivated each of the five dnd genes independently to examine their effect on the Dnd phenotype in terms of DNA phosphorothioation. Early experiments on disruption of dndA (mutant HXY1) and dndD (mutant LA2) by a str/spc cassette clearly abolished the Dnd phenotype [5] (Fig. 3) but could not provide unambiguous evidence for the function of dndD as insertion of antibiotic resistant genes could block expression of downstream gene(s) of an operon by a polar effect.
Figure 3.
dnd mutants. Black arrows represent dnd genes and their transcriptional directions. White blocks/arrows represent in-frame deletions in the corresponding genes.
We attempted to make in-frame deletions internal to individual dnd genes at their corresponding chromosomal loci to avoid polar effects. Apart from the dndB in-frame deletion mutant HXY2 [8] (Fig. 3), extensive efforts to obtain mutants specific to other dnd genes directly on the wild-type S. lividans 1326 chromosome failed for unknown reasons. We therefore attempted to develop a mutation-integration system by first generating a complete set of in-frame deletions of individual dnd gene in vitro in E. coli. These mutated dnd genes were then integrated back into the chromosome of S.lividans HXY6 (generated by targeted deletion of the complete dnd locus, [8]). A complete set of pSET152-derived integration plasmids with targeted in-frame deletions of the five dnd genes was generated by PCR and cloned into E. coli [detailed in Methods, pHZ2862 (651-bp deletion in dndA); pJTU1202 (729-bp deletion in dndB); pJTU1211 (819-bp deletion in dndC); pJTU1214 (1,704-bp deletion in dndD); and pJTU1219 (216-bp deletion in dndE), respectively]. These plasmids were introduced into HXY6 to obtain mutants XTG1-XTG5 with in-frame deletions in dndA-E in a uniform parental background.
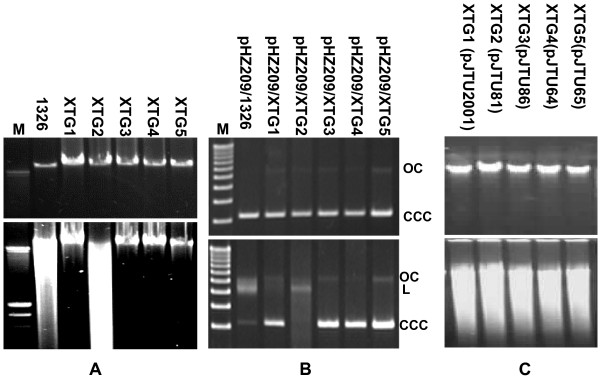
Isogenic mutant strains (XTG1-XTG5) were assayed for their Dnd phenotype. Interestingly, while the Dnd phenotype, as displayed by degradation of chromosomal (Fig. 4A) or plasmid pHZ209 (Fig. 4B) DNA isolated from strains XTG1, XTG3, XTG4, and XTG5 (corresponding to dndA, C, D, E) was clearly abolished, DNA isolated from XTG2 retained the Dnd phenotype, clearly showing that dndA, C, D, and E are all essential for DNA phosphorothioation. Single-stranded DNA modification, which should be indicated by shifting of the covalently closed circular (CCC) to the open circular (OC) form for plasmid pHZ209 DNA if cleaved by the electrophoresis buffer, was not observed with these mutants (Fig. 4B), as also found for HXY1 (data not shown).
Figure 4.
Dnd phenotype of 1326 and related dnd mutants. (A) Dnd phenotype of chromosomal DNA for 1326 and related dnd mutants. (B) Dnd phenotype of plasmids pHZ209 isolated from 1326 and related dnd mutants. (C) Dnd phenotype of chromosomal DNA from complemented dnd mutants. DNA was first treated with TAE (top panel) or peracetic acid TAE (bottom panel) before fractionation by electrophoresis in TAE with added thiourea. M: DNA markers; CCC: covalently closed circular plasmid; OC: open circular plasmid. L: linear plasmid.
A close comparison of the Dnd phenotypes displayed by the wild-type 1326 and the dndB mutant XTG2, however, revealed a clear difference. The degradation "smear" from the genomic DNA of XTG2 migrated much faster than that from wild-type strain 1326 (Fig. 4A). Smaller genomic DNA fragments, or more frequently degraded genomic DNAs, were observed in the mutant XTG2 than the wild-type strain 1326. In other words, the dndB mutation appeared to aggravate the Dnd phenotype, in agreement with the enhanced Dnd phenotype of another in-frame deletion mutant HXY2 [8]. On the other hand, degradation of the circular plasmid pHZ209, as shown by the relative intensities of the linearized pHZ209, appeared to be more intense from XTG2 than from 1326. Almost all the circular plasmid pHZ209 from XTG2 was degraded as linearized forms, but only about two-thirds of the circular plasmid pHZ209 from 1326 was linearized (Fig. 4B).
Rescue of the Dnd phenotype of dnd mutants by complementation
The first direct evidence that the Dnd phenotype, reflecting DNA phosphorothioation, involves the combined action of five independent proteins (DndA-E) comes from complementation experiments using plasmids expressing individual Dnd proteins. This was achieved by the construction of individual dnd gene expression plasmids using pHZ1272 [18], an E. coli-Streptomyces shuttle expression vector derived from pIJ6021 with a strong thiostrepton-inducible PtipA promoter [19]. Firstly, DNA fragments carrying individual dndA-E genes were cloned in-frame into pHZ1272 to generate expression plasmids (pJTU2001, carrying dndA; pJTU81, carrying dndB; pJTU86, carrying dndC; pJTU64, carrying dndD; and pJTU65, carrying dndE). Secondly, the expression plasmids were independently introduced by transformation into the corresponding mutant strains XTG1, 2, 3, 4, and 5 (with in-frame-deletions of dndA, B, C, D, and E, respectively). Even without induction of the PtipA promoter by addition of thiostrepton, strains XTG1, 3, 4, 5 carrying their counterpart expression plasmids recovered the Dnd phenotype of the wild-type strain 1326 (Dnd+), while XTG2 carrying pJTU81 (with a complete dndB gene) abolished enhanced Dnd phenotype (Dnd+) with recovery of the original Dnd phenotype (Dnd+) comparable with that of the wild-type strain 1326 (Fig. 4C).
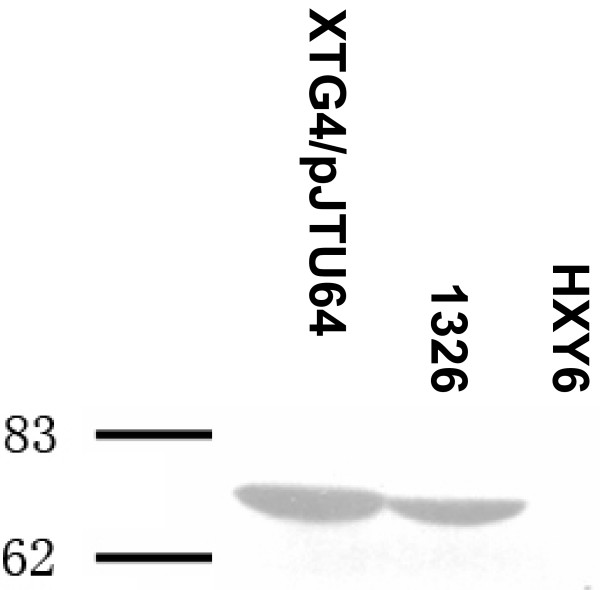
As additional evidence, we cloned dndD into pET15b to obtain an expression plasmid (pHZ2893) for the production of an N-terminal His-tag fusion protein. The purified DndD protein was then used for the production of rabbit anti-DndD polyclonal antibody. When we used this antibody to detect native DndD protein expression, we observed identical bands with a size of 74.6 KD in the expression strain XTG4/pJTU64, and wild-type S. lividans 1326 (Fig. 5). As a negative control, a 1326 derivative with complete deletion of the dnd gene cluster (HXY6) produced no signal in the corresponding position (Fig. 5). The protein size agrees well with our transcriptional analysis mentioned earlier and the DndD protein was correctly expressed in the complemented strain XTG4/pJTU64 (Fig. 5).
Figure 5.
Western blotting for detecting expression of Dnd proteins in S. lividans 1326 and derivative strains. Rabbit polyclonal antibody to DndD reacted with the protein extracted from wild-type S. lividans 1326 or strain XTG4/pJTU64 (a pHZ1272-derived dndD expression vector).
These results suggest that all of the mutations in XTG1–5 are dnd-specific and the Dnd proteins are correctly expressed in vivo. In the meantime, these expression plasmids carrying individual dnd genes could also rescue the Dnd phenotype of other corresponding mutants including HXY1 and HXY2 (data not shown). These complementation results from different mutants further support the conclusion that these mutations are not caused by secondary mutation(s) elsewhere on the chromosome.
Effect on cell viability by overdosage of Dnd proteins in vivo
The above complementation assays for all of the dnd mutants were tested without induction by the addition of thiostrepton. Because each individual dnd gene is under the control of the thiostrepton-inducible promoter PtipA in the expression plasmids, a feature which could provide us a tool for testing the effect of the over-expressed Dnd protein(s) in cells, we induced Dnd over-expression in the strains XTG1–5 carrying individual dnd gene expression plasmids by adding thiostrepton to a final concentration of 5 μg/ml in the normal culture medium. Surprisingly, XTG3/pJTU86 (carrying dndC) and XTG4/pJTU64 (carrying dndD) completely ceased growth, in sharp contrast to the normal growth of XTG1/pJTU2001 (carrying dndA), XTG2/pJTU81 (carrying dndB), and XTG5/pJTU65 (carrying dndE). This result suggests that over-expression of DndC or DndD proteins in vivo has a detrimental effect on cell viability.
Discussion
Early predictions of genes involved in DNA phosphorothioation and their organization as an operon within a region covering the cloned dnd gene cluster was mostly based on bioinformatic analysis, and no detailed experiments had been performed to provide direct evidence. We refined the conclusions by first minimizing the responsible region to a ca. 6.7-kb DNA fragment carrying only five genes, which still retained the ability to confer the Dnd phenotype on Dnd- hosts. We went on to confirm the expression of multiple and independent proteins encoded by an operon (dndB-E) using systematic mutagenesis either by targeted gene disruption and/or in-frame deletions internal to each protein. We then introduced individual engineered constructs, each containing only one specific gene under the control of a common promoter, into the above mutants. Reversion of the DNA shift from stable to degradation status or vice versa demonstrated unambiguously that DndA, C, D, and E are required in the biochemical pathway leading to the Dnd phenotype. The opposite effect of mutation in dndB to aggravate the Dnd phenotype was at least partly attributed to the changes of the sequence recognition specificity surrounding the modification sites [8].
The finding that excessive expression of DndC and DndD could affect cell growth and/or viability suggests that the in vivo level of the dnd system must be tightly regulated, consistent with our earlier observation that not all of the available sites could be modified [8]. This agrees well with the fact that the Dnd phenotype could only be detected when the entire dnd gene cluster was integrated into the chromosome or carried on a low copy-number plasmid in ZX1, but not on a plasmid with a copy-number of 10 or more [20]. All of these phenomena suggested that either an unknown mechanism is present in the cell to tightly control DNA phosphorothioation, or that over-expression of some of the proteins to override the regulation could be detrimental to the cells. We propose that the dosage of the Dnd proteins in the cells may not exceed the tolerable limit, and that the Dnd proteins must be balanced so as to be expressed in a highly coordinated manner in the cells. Therefore, simultaneous and/or unbalanced over-expression of one or even all four (dndB-E)of the dnd genes could seriously harm the cells, leading to inhibition of growth. The present study, showing that strongly induced expression of DndD and DndC, but not the other Dnd proteins, by the addition of thiostrepton, strongly suggests that these two proteins are the key determinants for the phenomenon. Being an IscS-like protein, DndA [21] was suggested to provide sulfur via its L-cysteine desulfurase activity and to catalyze iron-sulfur cluster assembly of DndC [22], probably by generating a persulfide (perhaps with the cysteine residue(s) in DndC or DndD) in the modification process. As such IscS-like proteins are also often required, as multi-functional proteins, for many other metabolic pathways [21], the detrimental effect by over-expression of DndC and DndD could be attributed to deprivation of DndA which is vital for primary metabolism. Thus, the fact that DndA function could not be substituted by other IscS homologs, at least in S. lividans analyzed here, might be due to a failure of proper persulfide formation, which could subsequently be delivered to target the DNA via DndC or DndD (not DndE because of its apparent lack of a .cysteine residue mediating persulfide formation). The exact mechanism of negative role of the over-expressed DndC and DndD proteins to cell viability remains, however, to be determined.
Conclusion
Genetic determination of the Dnd phenotype diagnostic for DNA sulfur modification in S. lividans was unambiguously attributed to a 6,665-bp DNA region carrying five dnd genes, with dndB-E constituting an operon and dndA transcribed divergently. Mutations in each of four dnd genes (dndA, C, D, and E) abolished the Dnd phenotype while mutation of dndB aggravated the Dnd phenotype. The Dnd phenotype of all mutants could be restored by complementation with the corresponding dnd gene, suggesting that they are essential for DNA sulfur modification. The fact that the cells ceased growth by overdosage of DndC or DndD in vivo suggests that the frequency of DNA phosphorothioate modification is under strict control in the native host.
Methods
Bacterial strains and plasmids
These are described in Additional file 1.
Methods and techniques
Standard methods for culturing cells, DNA cloning, PCR, Southern hybridization, and Western blotting were according to [23] in E. coli and [24] in Streptomyces. PCR primers are listed in Table 1 and PCR products successfully cloned into vectors were confirmed by sequencing. Mutants were confirmed by PCR and Southern hybridization. Tests of Dnd phenotype were described in [5,8] or [10,15].
Table 1.
primers used in PCR and RT-PCR
| Primer Name | Sequence (with the restriction enzyme sites underlined) | Enzyme site |
|---|---|---|
| A2 | ATCACCCCTTCCACCGAGAT | |
| A1 | ACTGGATGACCGCGGAGTTC | |
| B1 | GAGTACGTTTTTCCGGCCATCC | |
| B2 | TCCTTCAGCGCCTGCTCGAT | |
| B3 | CCAACACCGACTGGGAGGGG | |
| C1 | CAGAGATCGTCGAGGAGCTG | |
| C2 | GATCTTCAACCGCTCGGTGC | |
| C3 | CAGTATCGAACCATGACCCGG | |
| D1 | TGCGGCAAGACGACCCTGCT | |
| D2 | GTCGGCGAGCTGTTCCACCT | |
| D3 | CAGTGATCGACACCCCACTC | |
| E1 | ATGCCGTCTGAGATCACCAT | |
| E2 | ATAAGCAGCGTCTTGCCCAC | |
| 16S rRNA SP | AGTAACACGTGGGCAACTGC | |
| 16S rRNA AP | CTCAGACCAGTGTGGCCGGT | |
| xtg1 | CCGATCTTGTGCCCGCTGATG | |
| xtg2 | GCGCCTTAAGTCGTCCCTTGTTC | AflII |
| xtg3 | GAAGGTGTCTTAGATCTCCGG | BglII |
| xtg4 | CTGGCACGACAGGTTTCC | |
| xtg5 | AAGCACCGGTTCAAGACG | AgeI |
| xtg6 | GCCCAGGTCCGCAAGAA | |
| xtg7 | CTCGTGGTTGAGCGGGACTACGG | |
| xtg8 | CTGGCACCGGTCAAGCCTAGGTG | AgeI, AvrII |
| xtg9 | GGGACAGCCTAGGGGTGATC | AvrII |
| xtg10 | ACTGACCGCAGACCGCAAG | |
| wlr5 | CATATGGTGGGATCTTCTGCAGCT | NdeI |
| wlr6 | GGATCCTCAATGATGATGATGATGATGTGACTCTCCTCGCAGGTA | BamHI |
| wlr7 | CATATGAGCACCCCCAAGGCG | NdeI |
| wlr11 | GGATCCTTAGTGGTGGTGGTGGTGGTGTGCAGGTGCATCGGTGGTGA | BamHI |
| dnd-1 | AGAGATCACCACATATGCACCTGAGCACC | NdeI |
| dnd-2 | CAGCCGGATCCTGATCTCAG | BamHI |
| dndE-L | CACATATGCCGTCTGAGATCACC | NdeI |
| dndE-R | TAAGGCCTATTCGGCGGTGA |
Intensity of DNA bands was quantified from the fluorescence intensity using GeneTool software (Syngene).
Refinement of the limits of the dnd gene cluster
pHZ1900: a 10-kb BamHI fragment from pHZ825 was cloned into pSET152.
pJTU1203 or pJTU1204 (with opposite direction): a 7.9-kb MluI-EcoRI fragment from pHZ1904 was blunt-ended and cloned into the EcoRV site of pSET152.
pJTU1208: the 1.0-kb BglII fragment from pHZ1900 was inserted into the BamHI site of pBluescript II SK (+). Then a 0.3-kb SalI fragment of this plasmid was replaced with a 1.3-kb SalI fragment from pHZ1904 to generate pHZ2850, in which dndA accommodated in a 2.0-kb BamHI/BglII-SacI region. A 1.4-kb fragment from pHZ2850 generated by complete digestion with EcoRI and partial digestion with BglII was inserted into the EcoRI and BamHI sites of pSET152 to give pHZ2851. Finally, a 2.1-kb XbaI-SfiI fragment of pJTU1204 was replaced with a corresponding 0.8-kb fragment from pHZ2851, generating pJTU1208. Thus, in pJTU1208, the dnd gene cluster was shortened to the BglII site near the end of dndA, covering a 6,665-bp region.
pHZ2862 (also the vector for dndA deletion): a 2.0-kb PvuII fragment from pHZ1900 was cloned into the SmaI site of pBluescript II SK(+) to give pHZ2853, then a 6.5-kb SmaI-EcoRI fragment from pHZ1900 was used to replace the 0.7-kb corresponding fragment in pHZ2853 to give pHZ2861, in which dndB-E lay in a 7.8-kb SmaI/PvuII-EcoRI region. A 7.8-kb BamHI fragment from pHZ2861 was cloned into pSET152 to give pHZ2862.
Deletions within dnd genes
Vector construction for dndB deletion: using pHZ1904 as template, and xtg1 and xtg2 (with introduced AflII site) as primers, a 0.9-kb PCR product was amplified and cloned into pMD18-T (TaKaRa) to generate pJTU1201. Then, the 0.7-kb SfiI-AflII fragment from pJTU1201 was used to replace the 1.4-kb corresponding region in pHZ1904 to result in a dndB in-frame deletion vector, pJTU1202, in which a 729-bp DNA fragment was removed from dndB.
Vector construction for dndC deletion: after pHZ1904 was digested with SmaI and XbaI, a 5.0-kb fragment carrying dndC-E was introduced into the corresponding sites of pUC18 to generate pJTU1205. Using pJTU1205 as template, and xtg3 (with introduced BglII site) and xtg4 as primers, a 0.9-kb PCR product was amplified and cloned into pMD18-T to give pJTU1209. The 0.5-kb AflII-BglII fragment from pJTU1209 was used to replace the 1.3-kb corresponding region from pJTU1205 to generate pJTU1210 with an 819-bp in-frame deletion in dndC. The 4.8-kb AflII-XbaI fragment of pHZ1904 was replaced by the 4.0-kb AflII-XbaI fragment of pJTU1210 to generate pJTU1211, which carried dndC with an 819-bp in-frame deletion.
Vector construction for dndD deletion: using pJTU1205 as template, and xtg5 (with introduced AgeI site) and xtg6 as primers, a 0.5-kb PCR product was amplified and cloned into pMD18-T to give pJTU1212. The 0.4-kb BglII-AgeI fragment from pJTU1212 was used to replace the 2.1-kb corresponding region of pJTU1205 for generation of pJTU1213 with a 1704-bp in-frame deletion in dndD. The 4.8-kb AflII-XbaI fragment of pHZ1904 was replaced by the 3.1-kb AflII-XbaI fragment of pJTU1213 to generat pJTU1214, which carried dndD with a 1704-bp in-frame deletion.
Vector construction for dndE deletion: using pJTU1205 as template, and xtg7 and xtg8 (with introduced AgeI and AvrII sites) as primers, a 0.7-kb PCR product was amplified and cloned into pMD18-T to give pJTU1215. The 0.6-kb AgeI-MluI fragment from pJTU1215 was used to replace a 1.0-kb corresponding region of pJTU1205 to generate pJTU1217 with a 0.4-kb deletion traversing dndD and dndE. Using pJTU1205 as template, and xtg9 (with introduced AvrII site) and xtg10 as primers, a 1.0-kb PCR product was amplified and cloned into pMD18-T to give pJTU1216. The engineered 0.9-kb BstXI-AvrII fragment from pJTU1216 was used to replace a 0.7-kb corresponding region of pJTU1217 to generate pJTU1218 with a 216-bp in-frame deletion in dndE only. The 4.8-kb AflII-XbaI fragment of pHZ1904 was replaced by the 4.6-kb fragment corresponding fragment of pJTU1218 for to generate pJTU1219, which carried dndE with 216-bp in-frame deletion.
pHZ2862, pJTU1202, pJTU1211, pJTU1214, pJTU1219 were introduced into HXY6 by conjugation from E. coli ET12567 carrying pUZ8002 [25].
Construction of the expression vectors used in Streptomyces each carrying an independent dnd gene
dndA expression vector: a 1.2-kb engineered NdeI-BamHI fragment carrying dndA from pHZ882 was inserted into the corresponding sites of pHZ1272 to give pJTU2001.
dndB expression vector: using pHZ1904 as template, and wlr5 and wlr6 as primers, a 1.2-kb PCR product carrying dndB with introduced NdeI and BamHI sites (with C-terminal His-tag) was amplified and cloned into pMD18-T to give pJTU68. Then the corresponding NdeI-BamHI DNA fragment from pJTU68 was introduced into pHZ1272 between the restriction sites NdeI and BamHI to give pJTU81.
dndC expression vector: using pHZ1904 as template, and wlr7 and wlr11 as primers, a 1.5-kb PCR product carrying dndC with introduced NdeI and BamHI sites (with C-terminal His-tag) was amplified and cloned into pMD18-T to give pJTU72. Then dndC from pJTU72 was introduced into pHZ1272 between the restriction sites NdeI and BamHI to give pJTU86.
dndD expression vector: using pHZ1904 as template, and dnd-1 and dnd-2 as primers, a 2.0-kb PCR product carrying dndD with introduced NdeI and BamHI sites was amplified, digested with the corresponding enzymes and cloned into pET15b to generate pHZ2893. Then dndD from pHZ2893 was introduced into pHZ1272 between the restriction sites NdeI and BamHI to give pJTU64.
dndE expression vector: using pHZ1904 as template, and dndE-L and dndE-R as primers, a 0.4-kb PCR product carrying dndE with introduced NdeI site was amplified and cloned into pMD18-T to give pJTU180. Then dndE from pJTU180 was introduced into pHZ1272 after digestion with NdeI and BamHI to give pJTU65.
Over-expression and purification of DndD protein
After IPTG induction, E. coli BL21 (DE3) containing pHZ2893 over-expressed the DndD fusion protein with a His-tag at the N-terminal end. The fusion protein as inclusion bodies was further purified with an ÄKTA-fast protein liquid chromatography system (FPLC) (Amersham Pharmacia Biotech) and a 5-ml HiTrap chelating column (Amersham Pharmacia Biotech) under denaturing condition. The fusion protein was used for the production of rabbit anti-DndD polyclonal antibody.
RT-PCR analysis of dnd genes
RNA extraction was according to the standard protocol of RNeasy Protect Bacteria Midi Kit from Qiagen Co. Ltd. RT-PCR experiments were performed according to the standard protocol of OneStep RT-PCR Kit from the same company. Primers are listed in Table 1.
Authors' contributions
TX carried out most of the experiments. JL and ZW performed operon research and constructed dndA expression vector in S. lividans. Other expression vectors in S. lividans were constructed by SC and LW. They also overexpressed and purified DndD for DY to prepare anti-DndD polyclonal antibody. Work on HXY1, 2 was done by XH. pHZ1900 was constructed by AL. Plasmids from pHZ2850 to pHZ2983 were constructed by XZ. ZD oversaw the project. TX, ZW, SC and ZD wrote the paper. All authors discussed the results and assisted with editing of the manuscript.
Supplementary Material
Additional table 1. Table displaying bacterial strains and plasmids.
Contributor Information
Tiegang Xu, Email: tiegangxu@gmail.com.
Jingdan Liang, Email: jdliang@sjtu.edu.cn.
Shi Chen, Email: shi_chen@hms.harvard.edu.
Lianrong Wang, Email: lianrong@mit.edu.
Xinyi He, Email: xyhe@sjtu.edu.cn.
Delin You, Email: dlyou@sjtu.edu.cn.
Zhijun Wang, Email: wangzhijun@sjtu.edu.cn.
Aiying Li, Email: ayli@mail.ccnu.edu.cn.
Zhongli Xu, Email: xuzhongli2008@gmail.com.
Xiufen Zhou, Email: xfzhou@sjtu.edu.cn.
Zixin Deng, Email: zxdeng@sjtu.edu.cn.
Acknowledgements
We are very grateful to Prof. Sir David Hopwood, FRS for his continuous support and encouragement throughout this study for many years, and help for the editing of the manuscript. The authors wish to thank the National Science Foundation of China (NSFC), the Ministry of Science and Technology 973 and 863 programs, the Ministry of Education of China, the Shanghai Municipal Council of Science and Technology and Shanghai Leading Academic Discipline Project for research supports.
References
- Hattman S. Unusual modification of bacteriophage Mu DNA. J Virol. 1979;32(2):468–475. doi: 10.1128/jvi.32.2.468-475.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Specificity of the bacteriophage Mu mom+ -controlled DNA modification. J Virol. 1980;34(1):277–279. doi: 10.1128/jvi.34.1.277-279.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2'-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc Natl Acad Sci USA. 1983;80(24):7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70(3):830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XF, He XY, Liang JD, Li AY, Xu TG, Kieser T, Helmann JD, Deng ZX. A novel DNA modification by sulphur. Mol Microbiol. 2005;57(5):1428–1438. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, You D, Deng Z, Dedon PC. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3(11):709–710. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- Eckstein F. Phosphorothioation of DNA in bacteria. Nat Chem Biol. 2007;3(11):689–690. doi: 10.1038/nchembio1107-689. [DOI] [PubMed] [Google Scholar]
- Liang J, Wang Z, He X, Li J, Zhou X, Deng Z. DNA modification by sulfur: analysis of the sequence recognition specificity surrounding the modification sites. Nucleic Acids Res. 2007;35(9):2944–2954. doi: 10.1093/nar/gkm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Deng Z, Firmin JL, Hopwood DA, Kieser T. Site-specific degradation of Streptomyces lividans DNA during electrophoresis in buffers contaminated with ferrous iron. Nucleic Acids Res. 1988;16(10):4341–4352. doi: 10.1093/nar/16.10.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson P, Evans M. Novel post-replicative DNA modification in Streptomyces: analysis of the preferred modification site of plasmid pIJ101. Nucleic Acids Res. 1998;26(5):1248–1253. doi: 10.1093/nar/26.5.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boybek A, Ray TD, Evans MC, Dyson PJ. Novel site-specific DNA modification in Streptomyces: analysis of preferred intragenic modification sites present in a 5.7 kb amplified DNA sequence. Nucleic Acids Res. 1998;26(14):3364–3371. doi: 10.1093/nar/26.14.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser HM, Kieser T, Hopwood DA. A combined genetic and physical map of the Streptomyces coelicolor A3(2) chromosome. J Bacteriol. 1992;174(17):5496–5507. doi: 10.1128/jb.174.17.5496-5507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Deng Z, Hopwood DA, Kieser T. Streptomyces lividans 66 contains a gene for phage resistance which is similar to the phage lambda ea59 endonuclease gene. Mol Microbiol. 1994;12(5):789–797. doi: 10.1111/j.1365-2958.1994.tb01065.x. [DOI] [PubMed] [Google Scholar]
- Ray T, Mills A, Dyson P. Tris-dependent oxidative DNA strand scission during electrophoresis. Electrophoresis. 1995;16(6):888–894. doi: 10.1002/elps.11501601149. [DOI] [PubMed] [Google Scholar]
- Ray T, Weaden J, Dyson P. Tris-dependent site-specific cleavage of DNA. FEMS Microbiol Lett. 1992;75(2-3):247–252. doi: 10.1111/j.1574-6968.1992.tb05425.x. [DOI] [PubMed] [Google Scholar]
- He X, Ou HY, Yu Q, Zhou X, Wu J, Liang J, Zhang W, Rajakumar K, Deng Z. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol. 2007;65(4):1034–1048. doi: 10.1111/j.1365-2958.2007.05846.x. [DOI] [PubMed] [Google Scholar]
- Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116(1):43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Yang R, Hu Z, Deng Z, Li J. Construction of Escherichia coli-Streptomyces shuttle expression vectors for gene expression in Streptomyces. Chin J Biotechnol. 1998;14(1):1–8. [PubMed] [Google Scholar]
- Takano E, White J, Thompson CJ, Bibb MJ. Construction of thiostrepton-inducible, high-copy-number expression vectors for usein Streptomyces spp. Gene. 1995;166(1):133–137. doi: 10.1016/0378-1119(95)00545-2. [DOI] [PubMed] [Google Scholar]
- Li A. Ph.D thesis. Huazhong Agricultural University; 2000. Molecular Genetic Analysis of an Unusual DNA Modification in Streptomyces lividans. [Google Scholar]
- Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat Chem Biol. 2006;2(4):185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- You D, Wang L, Yao F, Zhou X, Deng Z. A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry. 2007;46(20):6126–6133. doi: 10.1021/bi602615k. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. 3. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Kieser T, Bibb JM, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich: John Innes Foundation; 2000. [Google Scholar]
- MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene. 1992;111(1):61–68. doi: 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional table 1. Table displaying bacterial strains and plasmids.